Abstract
Polarized Wnt signaling along the primary body axis is a conserved property of axial patterning in bilaterians and prebilaterians, and depends on localized sources of Wnt ligands. However, the mechanisms governing the localized Wnt expression that emerged early in evolution are poorly understood. Here we find in the cnidarian Hydra that two functionally distinct cis-regulatory elements control the head organizer-associated Hydra Wnt3 (HyWnt3). An autoregulatory element, which mediates direct inputs of Wnt/β-catenin signaling, highly activates HyWnt3 transcription in the head region. In contrast, a repressor element is necessary and sufficient to restrict the activity of the autoregulatory element, thereby allowing the organizer-specific expression. Our results reveal that a combination of autoregulation and repression is crucial for establishing a Wnt-expressing organizing center in a basal metazoan. We suggest that this transcriptional control is an evolutionarily old strategy in the formation of Wnt signaling centers and metazoan axial patterning.
Keywords: regulatory DNAs, cis-regulation, enhancer, molecular evolution, axis formation
The establishment and patterning of the primary body axis is fundamental to metazoan body plan development, a conserved feature of which is the spatially restricted expression of Wnt genes at the posterior end (1, 2). In cnidarians and bilaterians, Wnt genes are expressed in the blastopore and equivalent regions, and this localized Wnt expression is critical for organizing the primary body axis (1–3). Polarized Wnt expression in the sponge Amphimedon embryo (4) provides evidence that the origin of the Wnt signaling center is at the base of metazoan evolution. However, the regulation of localized Wnt expression is largely unknown.
In basal metazoans, the axial role of Wnt/β-catenin signaling has been extensively studied in the cnidarian freshwater polyp Hydra (5–9), which has a single body axis, the oral–aboral axis, with a head at the oral end and a foot at the aboral end. The head of the adult Hydra is classically defined to consist of the upper part carrying the hypostome, a domelike structure with the mouth opening in its center, and the lower part with the tentacles. Axial patterning in Hydra is controlled by the head organizer, located in the apical tip of the hypostome (10, 11). It has been hypothesized that the organizer patterns the body along the oral–aboral axis through diffusible short-range autocatalytic activators and long-range inhibitors (12, 13). Although the molecular identity of these theoretical factors is still uncertain, Wnt/β-catenin signaling has been postulated to encompass the activator (3).
Hydra Wnt (HyWnt) genes are expressed at the apical tip of the hypostome, whereas the transcriptional components of Wnt/β-catenin signaling Hydra Tcf (HyTcf) and nuclear Hydra β-catenin (Hyβ-catenin) are more broadly distributed along the oral–aboral axis, with higher levels in the hypostome than in the body column (5–7, 14). These genes are also induced when a new head organizer is formed during asexual reproduction by budding and during head regeneration (5). The sufficiency of Wnt/β-catenin signaling for providing the head organizer activity has been recently demonstrated by overexpression of a constitutively active form of Hyβ-catenin (9).
A putative master Wnt ligand in Hydra axial patterning is HyWnt3, being expressed at the earliest phase of head regeneration and stimulating head organizer formation (7). The coup-regulation of HyTcf and Hyβ-catenin during head organizer formation suggests that they are involved in HyWnt3 transcriptional regulation. Indeed, elevated activation of Wnt/β-catenin signal induces ectopic HyWnt3 expression (6), and autoregulation through the Wnt/β-catenin circuit has been proposed to activate and maintain HyWnt3 expression (6), although a direct molecular evidence is missing. A simple autoregulation although cannot explain the restriction of HyWnt3 expression to the head organizer, as HyTcf and Hyβ-catenin are broadly expressed and seem to be required for the narrower HyWnt3 expression (3, 5, 15). Thus, the mechanism for HyWnt3 regulation remains to be discovered.
By extensively using transgenic Hydra, we analyzed cis-regulation of HyWnt3. We identified two functionally distinct cis-regulatory elements in the HyWnt3 promoter that are responsible for the head organizer-specific HyWnt3 expression. An autoregulatory element interacts with the HyTcf/Hyβ-catenin transcriptional complex and induces gene expression in a broad domain in the head. By contrast, a repressor element is necessary and sufficient for restriction of the expression to the head organizer. These results demonstrate that a combination of autoregulation and repression has a crucial function for the establishment and maintenance of the localized HyWnt3 expression in the head organizer.
Results
HyWnt3 Upstream Promoter Sequence Reproduces the Endogenous Expression in the Head Organizer Region.
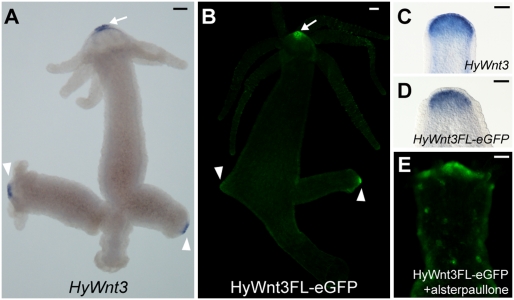
To dissect the molecular mechanisms regulating HyWnt3 expression in the head organizer, we sought to identify HyWnt3 cis-regulatory elements. A 2,149-bp fragment (HyWnt3FL) upstream of the HyWnt3 translation initiation site was isolated and analyzed for its ability to drive expression in transgenic Hydra polyps. Injection of the HyWnt3FL with an EGFP reporter gene (HyWnt3FL-EGFP) into Hydra embryos produced transgenic polyps, in which EGFP expression was exclusively observed at the apexes of the adult hypostome and developing buds, reflecting the endogenous expression of HyWnt3 (Fig. 1 A and B). As demonstrated by in situ hybridization the reporter expression was also activated in the apical tip of head-regenerating animals (Fig. 1 C and D), in which the head organizer is restored. EGFP fluorescence signals were not detectable in regenerating tips as a result of the delay in protein maturation. Previous studies showed that HyWnt3 is activated by high Wnt/β-catenin signaling caused by alsterpaullone treatment, which inhibits the activity of glycogen synthase kinase-3β (GSK-3β) involved in β-catenin degradation in Hydra (6). In agreement with the endogenous responsiveness, HyWnt3FL-EGFP was ectopically activated in multiple spots along the body column following alsterpaullone treatment (Fig. 1E). Thus, in all experimental settings, HyWnt3FL-EGFP activity mimicked endogenous HyWnt3 expression, demonstrating that HyWnt3FL contains the regulatory elements required for HyWnt3 expression in the head organizer.
Fig. 1.
HyWnt3 promoter reproduces the endogenous expression. (A and B) Expression of HyWnt3 mRNA (A) and HyWnt3FL-EGFP transgene (B) in the adult Hydra. HyWnt3FL-EGFP is activated exclusively in the HyWnt3-expressing cells in the apical tips of the adult hypostome (arrow) and developing buds (arrowheads). (C and D) Induction of HyWnt3 and HyWnt3FL-EGFP in head-regenerating animals at 3 h after head removal, visualized by in situ hybridization for HyWnt3 (C) and EGFP (D). (E) Ectopic activation of HyWnt3FL-EGFP reporter in the body column at 48 h after alsterpaullone treatment for 24 h. (Scale bars: 100 μm.)
HyWnt3 Regulatory Region Consists of Activator and Repressor Modules.
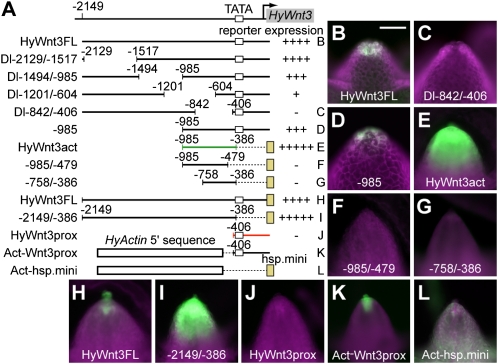
To identify the sequences within HyWnt3FL responsible for the head organizer-specific expression, a series of internal deletions of HyWnt3FL was generated and analyzed for their activity in transgenic polyps (Fig. 2 and Figs. S1 and S2). The deletion of −2,129 to −985 bp did not significantly change reporter expression (Dl−2129/−1517 and Dl−1494/−985; Fig. 2A and Fig. S2). In contrast, removal of −1,201 to −604 bp (Dl−1201/−604) strongly reduced reporter expression, and deletion of −842 to −406 bp (Dl−842/−406) eliminated expression (Fig. 2 A and C). Generation of multiple transgenic lines for each of the deletion construct and an independent transformation marker Hydra Actin (HyActin)–red fluorescent protein (RFP) reporter gene in the same construct ensured that the hypostome cells carried the HyWnt3 EGFP reporter gene under examination (Fig. 2 and Fig. S1). Thus, the −1,201 to −406 bp region contains the sequences essential for HyWnt3 expression in the head organizer. Further, the −985 bp upstream sequence (−985) drove reporter expression at the apex of the hypostome (Fig. 2D), indicating that the crucial sequences for HyWnt3 transcription are located in the −985 to −406 bp region.
Fig. 2.
Identification of HyWnt3act and HyWnt3rep cis elements for HyWnt3 expression. (A) Schematic diagram of deletion constructs used for the identification of the HyWnt3act (green) and HyWnt3rep (red) elements. The numbers indicate positions from the translation start site. Reporter expression activities of the constructs are indicated (Right). (+++++, Augmented levels of expression; ++++, WT levels of expression; +++, moderately reduced expression; ++, severely reduced expression; +, expression detectable; −, expression undetectable.) The white and yellow boxes indicate putative TATA boxes of the HyWnt3 promoter (TATA; −378, −368, and −360 bp) and the Hyhsp70 minimal promoter (hsp.mini), respectively. (B–L) EGFP expression in the hypostome of transgenic animals with the reporter construct HyWnt3FL (B and H), Dl−842/−406 (C), −985 (D), HyWnt3act (E), −985/−479 (F), −758/−386 (G), −2149/−386 (I), HyWnt3prox (J), Act-Wnt3prox (K), and Act-hsp.mini (L). Ectodermal or endodermal lines are shown in B–D or E–L, respectively. Reporter constructs lacking the HyWnt3 proximal promoter sequence exhibit dramatic expansion of expression (E and I). In contrast, reporter constructs involving it displayed restricted expression to the head organizer (B, D, H, and K). (Scale bar: 100 μm.)
We next tested whether a fragment encompassing the −985 to −406 bp region was sufficient to drive expression in the hypostome. The −985 to −386 bp fragment, which we named HyWnt3act enhancer element, was linked to the core promoter from the Hydra heat shock protein 70 (Hyhsp70) gene (hsp.mini) and EGFP reporter gene, and analyzed for its activity. The hsp.mini was demonstrated to be a functional core promoter in our heterologous reporter analysis (Fig. S1). The HyWnt3act element induced reporter expression in the hypostome (Fig. 2E). Surprisingly, HyWnt3act also directed ectopic reporter expression in the lower part of the head, whereas HyWnt3FL activity was limited to the tip of the hypostome when examined using the hsp.mini reporter construct (Fig. 2H). This observation indicates that the downstream region of −386 bp is necessary to limit HyWnt3 expression to the head organizer region (as detailed later). We also noticed that the HyWnt3act element is active only in endodermal epithelial cells, indicating that the HyWnt3 expression in ectodermal cells also requires the downstream region of −386 bp because −985 drove reporter expression both in the ectoderm and endoderm (Fig. 2 and Fig. S2). Loss of reporter activity of −985/−479 and −758/−386 demonstrated that HyWnt3act was a minimal enhancer in our studies (Fig. 2 F and G).
The cis-regulatory analysis of HyWnt3act suggested a possible negative regulation of HyWnt3 by the proximal sequence of HyWnt3FL. Indeed, the −2,149 to −386 bp fragment (−2149/−386), which included the entire 5′ sequence of HyWnt3FL but lacked the proximal sequence (Fig. 2A), drove similarly increased reporter expression (Fig. 2I). Thus, the proximal sequence of HyWnt3FL seems to be essential for HyWnt3 repression outside of the head organizer region. A −406 bp HyWnt3 proximal promoter region (HyWnt3prox) on its own did not drive reporter expression (Fig. 2J). However, we speculated that, if HyWnt3prox is responsible for HyWnt3 repression in cells outside of the head organizer region, it might be capable of repressing the activity of a given promoter that is normally active outside of the head organizer. To address this, we made use of the HyActin promoter, which is active in the entire animal (16). When the HyActin 5′ sequence (−1,300 to −221 bp HyActin promoter lacking its putative core promoter) was fused to the HyWnt3prox sequence (Act-Wnt3prox; Fig. 2A), this chimeric fragment activated reporter expression only in the apical end of the hypostome (Fig. 2K), which is the same pattern as that resulting from HyWnt3FL (Fig. 2H). In combination with other heterologous promoters (e.g., the Hyhsp70 core promoter), the HyActin 5′ sequence (Act-hsp.mini) directed uniform expression as the full-length HyActin promoter does (Fig. 2L). This result demonstrated that HyWnt3prox was compatible with the HyActin 5′ regulatory activity, but was able to induce reporter expression only in the head organizer. We therefore conclude that HyWnt3prox has a repressive function on gene expression in cells distant from the head organizer, and HyWnt3 expression can be locally restricted exclusively by this repressor element (HyWnt3rep).
HyWnt3act Requires Direct Inputs by T Cell-Specific Factor and Other Transacting Factors.
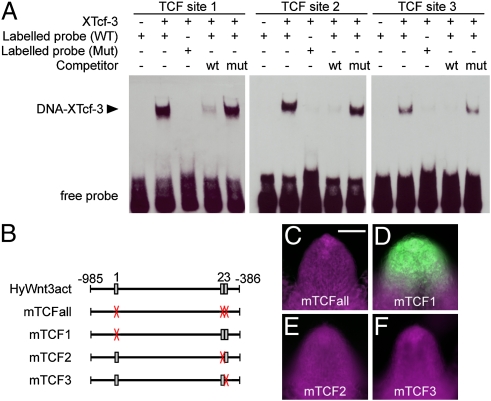
An autoregulatory positive feedback loop of Wnt/β-catenin signaling has been previously suggested to control HyWnt3 expression (5, 6, 14). To investigate whether HyWnt3act is under direct Wnt/β-catenin signaling control, we surveyed HyWnt3act for T cell-specific factor (TCF) binding sites and found three candidate sites (Fig. 3B). To examine whether Tcf protein binds to these sites, we performed EMSA (Fig. 3A). Each of the three sites showed binding affinity for recombinant Xenopus Tcf-3 (XTcf-3). This XTcf-3 binding was inhibited by competition with a TCF-binding site from the Xenopus siamois promoter, which was confirmed to interact with XTcf-3 (17). Consistently, mutation of the TCF sequences in the probe abolished binding, demonstrating a sequence-specific interaction of recombinant XTcf-3 with the TCF sites from HyWnt3act enhancer.
Fig. 3.
Direct regulation of HyWnt3act by Tcf. (A) EMSA shows binding of recombinant His-tagged XTcf-3 protein to the Tcf sites 1, 2, and 3. (B) Schematic diagram of the HyWnt3act element with the location of TCF-binding sites indicated. Boxes and X (red) indicate WT and mutant sites, respectively. (C–F) Requirement of the Tcf sites in Hydra in vivo. Mutation of all Tcf sites (C) abolished reporter activity. Tcf sites 2 (E) and 3 (F), but not 1 (D), were essential for the activity. (Scale bar: 100 μm.)
Having defined the TCF sites as potential binding sequences for transcriptional activation of HyWnt3, we tested their function in vivo. We introduced mutations into the TCF-binding sequences defined by EMSA and analyzed the effects on the HyWnt3act enhancer activity in transgenic Hydra (Fig. 3 B–F). Simultaneous mutation of all three TCF-binding sites (mTCFall) resulted in complete loss of reporter activity (Fig. 3C). Subsequently, individual mutations of the TCF sites revealed that the sites 2 and 3 are essential for the HyWnt3act function (Fig. 3 D–F). Thus, we concluded that HyWnt3 expression requires TCF binding sites 2 and 3 located in the HyWnt3act enhancer for Tcf-dependent induction.
Importantly, the −758/−386 fragment, which lacks a 5′ 227-bp region of HyWnt3act but retains the TCF sites 2 and 3, failed to drive reporter expression (Fig. 2G), indicating that other transactivators may interact with the 5′ end of HyWnt3act and act in concert with Tcf to control the expression of HyWnt3. A bioinformatic analysis of the 227-bp region reveals the existence of further putative transcriptional activators, e.g., CREB/CRE, Forkhead, and LIM (Table S1). CREB/CRE binding proteins have been reported to be activated early on during head regeneration (18–20), and they are therefore potential coactivators of HyWnt3.
β-Catenin Is a Direct Regulator of HyWnt3.
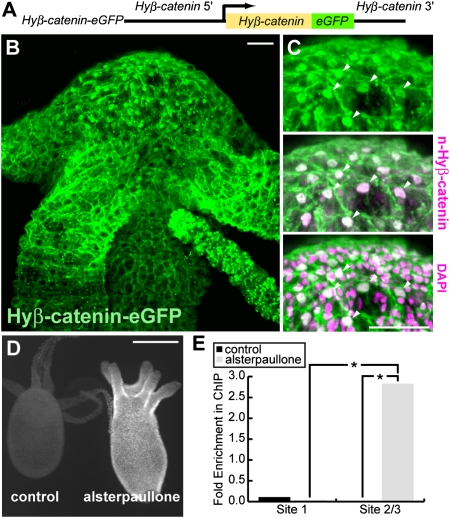
Physical interaction of Hyβ-catenin and HyTcf in vitro has been demonstrated previously (5, 8), suggesting that Hyβ-catenin is recruited to TCF-binding sequences to activate transcription in cooperation with HyTcf. To test whether this mechanism also applies to the control of HyWnt3 expression, we generated a Hyβ-catenin-EGFP transgenic Hydra, in which expression of EGFP-tagged Hyβ-catenin is driven by the Hyβ-catenin promoter (Hyβ-catenin-EGFP; Fig. 4A). In these animals, Hyβ-catenin-EGFP colocalized with the native Hyβ-catenin, and consistent with previous experiments using anti–Hyβ-catenin antibody (14), nuclear accumulation of Hyβ-catenin-EGFP was higher in the hypostome than in the body column (Fig. 4 B and C). Alsterpaullone treatment led to stabilization and increased nuclear accumulation of the Hyβ-catenin-EGFP throughout the animal (Fig. 4D), similar to the behavior of endogenous Hyβ-catenin.
Fig. 4.
Hyβ-catenin signaling directly regulates HyWnt3. (A) Schematic diagram of the Hyβ-catenin-EGFP construct. (B and C) Distribution of Hyβ-catenin-EGFP protein. (B) Hyβ-catenin-EGFP is associated with the cell membranes and localized in the nuclei of cells at higher levels in the hypostome than in the body column. (C) Hyβ-catenin-EGFP is colocalized with the endogenous Hyβ-catenin detected with antinuclear Hyβ-catenin antibody, in the nuclei of cells at the hypostome (arrowheads). (D) Hyβ-catenin-EGFP was stabilized and accumulated throughout the animal upon alsterpaullone treatment. Hyβ-catenin-EGFP transgenic animals were treated without (control) and with alsterpaullone for 24 h. (E) ChIP analysis detecting interaction of Hyβ-catenin and the HyWnt3 promoter. The fragment containing sites 2/3 was enriched in ChIP of alsterpaullone-treated Hyβ-catenin-EGFP transgenic animals compared with nontreated animals. In contrast, site 1 was never enriched upon treatment (*P < 0.05; n = 3). (Scale bars: 50 μm.)
To test whether Hyβ-catenin is present at the identified TCF-binding sites in vivo, we performed ChIP assay using the Hyβ-catenin-EGFP polyps and an anti-GFP antibody (Fig. 4E). In this ChIP experiment, the TCF-binding sites 2 and 3 were analyzed as a single sequence (site 2/3) because they are closely positioned in the promoter. In intact polyps, we failed to detect enrichment of the immunoprecipitation of the fragments containing the TCF site 1 or site 2/3 (Fig. 4E). This probably happens because the HyWnt3-expressing cells in the hypostome represent only a small fraction of the epithelial cells in the animal, and this could obstruct the attempt to identify binding of Hyβ-catenin complex in the HyWnt3 promoter. In contrast, alsterpaullone treatment led us to detect the enrichment of immunoprecipitated sites 2/3 but not site 1 (Fig. 4E). These results confirm the contribution of sites 2 and 3 for HyWnt3 regulation and demonstrate the interaction of Hyβ-catenin and the TCF sites in vivo. Overall, HyWnt3act is directly regulated by the Hyβ-catenin/HyTcf transcriptional complex.
Wnt/β-Catenin Activation Suppresses HyWnt3rep Function.
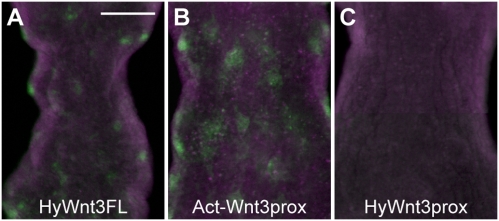
We next asked how HyWnt3rep is controlled within the head organizer region. Considering the localized activity of HyWnt3FL or Act-Wnt3prox to the apical end of the hypostome, the function of HyWnt3rep might be suppressed in the head organizer and such a suppression might involve factors acting in the head organizer region. A potential candidate is Wnt/β-catenin signaling itself. To examine whether HyWnt3rep is regulated by Wnt/β-catenin signaling, we treated the Act-Wnt3prox transgenic animals with alsterpaullone. In this way, ectopic HyWnt3 expression foci are generated, as a result of ubiquitous Wnt signaling activation. Interestingly, this treatment of Act-Wnt3prox animals led to EGFP expression in a spotted pattern in the body column (Fig. 5B), which was very similar to the responsiveness of HyWnt3FL (Fig. 5A). In contrast, the HyWnt3prox did not show such an ectopic expression (Fig. 5C). These results suggest that the HyWnt3prox function is suppressed at high levels of Wnt signaling; thereby, the HyActin 5′ sequence activity is “de-repressed” and becomes activated in the body column. This also means that the de-repression is independent of the TCF-binding sequences within the HyWnt3act element, and thus controlled by other transcription factors. From this experiment, we conclude that HyWnt3rep is sensitive to high levels of Wnt/β-catenin signaling activation so that it becomes suppressed, thereby allowing HyWnt3 gene expression in the head organizer region.
Fig. 5.
Derepression of HyWnt3rep by Wnt activation. (A–C) EGFP reporter activities of transgenic Hydra carrying HyWnt3FL (A), Act-Wnt3prox (B), and HyWnt3prox (C) after 48 h for 24-h alsterpaullone treatment. Act-Wnt3prox showed ectopic EGFP expression in the body column in a spotted pattern similar to that resulting from HyWnt3FL but not HyWnt3prox. (Scale bar: 100 μm.) This figure is a composite of multiple panels.
Discussion
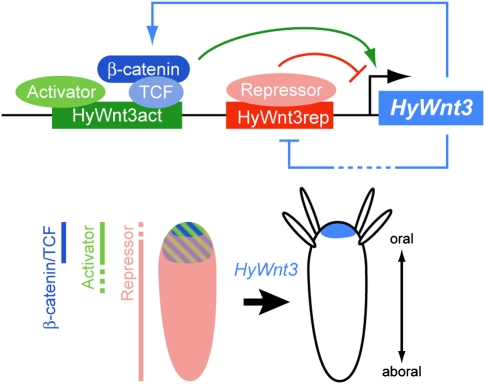
How Wnt signaling centers are formed and maintained is an important but unsettled question. By using transgenic Hydra, we analyzed cis-regulatory mechanisms of HyWnt3 transcription and found that it represents a key level in the formation of such a center. We found that in the Hydra head organizer HyWnt3 is regulated by two functionally distinct cis-regulatory elements located in the HyWnt3 promoter (Fig. 6). One of them, HyWnt3act, acts as an autoregulatory element mediating direct Wnt/β-catenin signaling. This demonstrates that Wnt/β-catenin signaling directly regulates HyWnt3. When it has been expressed, HyWnt3 can maintain its own expression by creating a positive feedback loop (Fig. 6). This regulatory system fits well with the pattern formation model proposed by Gierer and Meinhardt (3, 12).
Fig. 6.
Model for the transcriptional regulation of the head organizer-specific HyWnt3 expression. Schematic diagram of the mechanistic model governing HyWnt3 in the head organizer. HyWnt3 is controlled by two distinct cis-regulatory elements, the HyWnt3act (green) and HyWnt3rep (red) elements, which are positively (light blue arrow) and negatively (light blue bar) regulated by Wnt/β-catenin signaling. The β-catenin/Tcf complex (blue) and putative activators (light green) bind to HyWnt3act, and their combinatorial inputs act in HyWnt3 transcription (green arrow). Potential repressors (pink) bind to HyWnt3rep and inhibit HyWnt3 expression (red bar). Presumed distribution or activity of the β-catenin/Tcf and activators is in and below the head, and that of repressors is absent from the organizer region along the body axis. Their positive and negative regulation restricts HyWnt3 expression (light blue) to the head organizer region. Note that, in addition to the transcriptional level, there are also inhibitory inputs by HyDkk1/2/4 inhibition in the gastric region (36).
In addition to the other Wnt genes (7, 21), Brachyury and Goosecoid define the posterior, blastopore signaling center (22, 23) in Hydra and other cnidarians. There is evidence that Wnt signaling centers in bilaterians are also regulated by Wnt ligands themselves. Reminiscent of the Hydra hypostome, Wnt genes are expressed in the posterior growth zone in arthropods and vertebrates (24). It is not known yet whether the same genetic cascades act in maintaining Wnt expression. In zebrafish, a Brachyury/Wnt loop acts in Wnt3a and Wnt8 expression in the posterior growth zone (25), which is not present in insects (26, 27). A positive feedback regulation by Wnt/β-catenin signaling has been also shown for the sea urchin Wnt8 (28, 29), in which it is required for the maintenance of the vegetal Wnt signaling center during gastrulation. This conservation in the prebilaterian and bilaterian animals implies that the positive autoregulatory feedback machinery of Wnt/β-catenin signaling can be an ancient tool used for the maintenance of the Wnt signaling center during metazoan evolution.
Although the activation of HyWnt3act requires the Hyβ-catenin/HyTcf transcriptional complex binding to this element, HyWnt3act appears to act as a hub of autoregulatory signals and inputs of other transcriptional activators. A 5′ 227-bp deletion from the HyWnt3act fragment (−758/−386) resulted in loss of reporter expression, although the functional TCF binding sites remained intact (Fig. 2). We found that a number of potential transcription factor binding motifs exist in this region (Table S1). In particular, CBP/CREB, Forkhead domain factors, and LIM homeodomain transcription factors are potential candidate molecules. CBP is known to act as a transcriptional coactivator of β-catenin (30), and is activated during early head regeneration in Hydra (18, 19, 31). In Drosophila, forkhead transcription factor is required for expression of the Drosophila Wnt1 homologue wingless in the posterior end of the embryo (32). The Hydra orthologue of forkhead, budhead, is expressed in the hypostome (33), making possible a similar genetic cascade on HyWnt3 activation. The LIM homeobox gene functions in activation of vertebrate organizer genes (34), and recently a cnidarian LIM counterpart suggested its conserved role in organizer gene activation (35). It is possible that transcription factors that are expressed in Wnt signaling centers in both prebilaterians and bilaterians have played a role in establishing Wnt expression in combination with Wnt autoregulation in the evolution of the Wnt signaling center.
Given the positive autoregulation on HyWnt3 transcriptional control, how can its expression be confined to the head organizer? Also, how can HyWnt3 emerge as a restricted domain from the broad distribution of HyTcf and Hyβ-catenin (3, 15) despite their major contributions to the autoregulation? Reaction-diffusion models of pattern formation (12) predict that the autocatalytic activator produces a long-range inhibitor that restricts the activating source to the organizing center. The Wnt antagonist Hydra Dickkopf1/2/4 (HyDkk1/2/4) (36) was postulated as a potential inhibitor (37). As Dkk inhibits ligand receptor interactions, this mechanism does not explain how Wnt expression is inhibited on the transcriptional level.
We showed that transcriptional regulation is essential to restrict Wnt expression to the site of the signaling center. We discovered a HyWnt3rep repressor element that is necessary and sufficient for localizing HyWnt3 expression to the head organizer. The removal of HyWnt3rep resulted in an expansion of gene expression that was dependent on Tcf-binding sites. This suggests that HyWnt3 has the potential to be activated in a Wnt/β-catenin–dependent manner in a region broader than expected. This expansion occurs even in the context of intact HyDkk1/2/4 function. Transcriptional regulation of HyWnt3 expression is therefore independent of HyDkk1/2/4, although HyWnt3rep action can be complemented by HyDkk1/2/4. However, a high β-catenin signal can suppress HyWnt3rep function, and the signaling center can therefore emerge by local suppression of an inhibitor being uniformly present. We speculate that the repressive activity of HyWnt3rep gets locally abolished at the onset of regeneration and budding, thereby permitting local HyWnt3 transcription and new organizer formation. The activation of canonical Wnt signaling by alsterpaullone treatment in the presence of a putative repressor in the body column results in multiple small foci of Wnt activation (Fig. 5 A and B). Interestingly, these foci develop into tentacles, but not into complete heads (6, 36), suggesting the input of additional transcriptional activators (as detailed earlier) in head development.
Although the identity and expression pattern of the repressor molecule regulating HyWnt3rep are still unknown, it is expected to be absent from the head organizer or exist uniformly with its activity suppressed in the head organizer. HyWnt3rep contains putative binding sites for several transcription factors (Table S2). Among them, we found binding sites for the GATA transcription factor, which was reported to act as a transcriptional repressor (38). A Hydra GATA orthologue is expressed in the body column (Fig. S3), making it a possible repressor. Interestingly, in DNase I footprinting experiments, the cis-regulatory region of the Hydra head-specific gene ks1 were also identified to specifically bind nuclear proteins from basal tissue (39), although the identity of the corresponding repressors is unknown. As HyWnt3prox shares 21 bp with the HyWnt3act and −2149/−386 that did not drive reporter expression in the body column, the HyWnt3prox may be a bipartite element; one represses gene expression in the lower hypostome and the other blocks expression in the body column. It is thus possible that the HyWnt3rep function is controlled by several transcriptional repressors. The identification and functional analysis of the corresponding molecules governing HyWnt3rep will clarify the mechanisms underlying the localized HyWnt3 expression. In line with this strategy, it will be necessary to unravel the regulatory network of the cnidarian head and blastopore organizer that includes the other Wnt genes and yet-unidentified transcriptional repressors and activators.
The establishment of local sources of the Wnt ligand could have been important in the evolution of metazoan axis formation. We presume that the cis-regulatory control mechanism combining autoactivation and repression resulted in the establishment of a locally restricted Wnt signaling center. This regulatory mechanism can be independent of the function of extracellular Wnt antagonists. Although multiple Wnt antagonistic regulators have been identified from cnidarians to vertebrates, these molecules are absent from several metazoan lineages (e.g., sea urchin and some protostomes) (1). A combination of autoregulation and repression can therefore account for the localized Wnt expression in other animals as well. The presence of this transcriptional control in a basal metazoan suggests possible conservation in other phyla and a key role in the evolution of the spatially localized Wnt signaling centers.
Materials and Methods
Animals.
The Hydra magnipapillata 105 strain (40) and Hydra vulgaris AEP strain (41) were used. Animals were cultured as described previously (42). To induce gametogenesis, the AEP animals were starved for 1 wk and then fed twice per week.
Cloning and Constructs.
For the HyWnt3FL-EGFP construct, a 2,149-bp HyWnt3 promoter fragment (HyWnt3FL) was amplified by PCR from genomic DNA of the 105 animals and subcloned into the hoT G (16), which resulted in hoT G-HyWnt3FL-EGFP (HyWnt3FL-EGFP). The derivative deletion or mutation constructs were generated by PCR and/or restriction digest (SI Materials and Methods), and subsequent subcloning into Hydra transgenesis vectors, pBSSA-AR or its derivative containing the Hydra hsp70 minimal promoter (pBSSA-AR-hsp.mini-EGFP; SI Materials and Methods), which contain the Hydra actin promoter and the RFP gene. Additional information on the constructs is provided in SI Materials and Methods and Tables S3−S7.
Generation of Transgenic Hydra Polyps.
Generation of transgenic Hydra polyps was carried out as previously described (16). To ensure whether transgenic cells have a transgene, we constructed and used a vector system for Hydra transgenesis described in Fig. S1 and SI Materials and Methods. Each transgene construct was injected into 100 to 300 embryos of the AEP strain. The resultant hatched polyps were collected for 1 to 2 mo after injection and maintained individually. Polyps ubiquitously expressing the transgene were generated by clonal propagation, asexual budding. For each transgene, two to 30 independent transgenic lines were obtained, and at least two lines were analyzed. Details of generation of transgenic experiments are described in Table S3.
In Situ Hybridization.
Whole-mount in situ hybridization was performed as described previously (33, 43).
Immunohistochemistry.
Immunohistochemistry was performed by using GFP antibody (1:1,000; Abcam), anti-nuclear Hyβ-catenin antibody (1:100) (14), Alexa Fluor 488 goat anti-chicken IgG (1:500; Invitrogen), and Alexa Fluor 568 goat anti-guinea pig IgG (1:150; Invitrogen). DAPI staining was done with 1:5,000 dilution. Hydra polyps were relaxed with 2% urethane/hydra medium for 2 min, fixed with 4% formaldehyde (Sigma) for 30 min at room temperature, and then permeabilized for 30 min in PBS solution with 0.1% Triton X-100. Incubation with primary antibodies was done overnight at 4 °C in PBS with 0.1% Tween-20 with 5% donkey serum (Sigma). The anti-nuclear Hyβ-catenin antibody was produced by immunizing guinea pigs with a synthetic peptide (YQDIQRRGPGAQNMQD) encompassing amino acid region 603 to 618 of Hyβ-catenin. The antibody was affinity-purified by using the antigenic peptide.
Alsterpaullone Treatment.
Animals were incubated in 5 μM alsterpaullone (Calbiochem) in Hydra medium for 24 h (6). Thereafter, they were rinsed with Hydra medium several times and cultured in Hydra medium.
Regeneration Experiments.
Budless polyps were bisected at 80% body length.
Microscopy.
Fluorescent micrographs were acquired on a Nikon 80i upright microscope equipped with a 10× PlanApo objective, NA 0.45, onto a Nikon DS-1QM cooled digital camera. Alternatively, animals were documented with a MonoZoom microscope (AZ100; Nikon) equipped with a DS-Qi1Mc cooled digital camera.
EMSA.
Recombinant His-tagged XTcf-3 was expressed in Escherichia coli (44). EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer's instructions. Glycerol (5%), MgCl2 (5mM), poly(dI-dC) (50 ng/μL), and Nonidet P-40 (0.05%) were included in the binding reaction. WT or mutant probes were present at 2 nM final concentration. The purified His-XTcf-3 was present at 63 ng/μL final concentration in the binding reaction. In competition assays, DNA oligonucleotides containing WT or mutant Tcf sites from the X. siamois promoter were used as competitors. Oligonucleotide sequences used in EMSA are provided in Table S4.
ChIP Assay.
Approximately 180 Hyβ-catenin-EGFP transgenic animals were treated overnight with 5 μM alsterpaullone or with DMSO, and then washed with Hydra medium. The animals were fixed with 1% formaldehyde for 10 min and then processed using the ChIP Assay Kit (USB) according to the manufacturer's instructions. The antibody used was rabbit anti-GFP polyclonal (Abcam). The chromatin DNA was fragmented with an S-4000 sonicator (Misonix) to an average of 100 bp. The ChIP and input was compared with real time PCR by using the Chromo4 RT PCR detector attached on a DNA engine thermal cycler (Bio-Rad). Primers were designed to amplify approximately 80-bp sequences containing site 1 and sites 2 and 3, as well as a part of the HyActin promoter (sequences are provided in Table S5). The analysis was done by normalizing against input and actin promoter sequence using the −ΔΔC(t) method (45).
Supplementary Material
Acknowledgments
We thank Thomas C. G. Bosch for the hoTG construct, Robert E. Steele for the pHyVec5 construct, Dietmar Gradl for the XTcf-3 construct, Jutta Tennigkeit for helping with XTcf-3 production, and the Nikon Imaging Center at the Heidelberg University (Ulrike Engel). We thank Jan Lohmann, Hans Meinhardt, and Mihaela Žigman for critically reading the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grants DFG-SFB 488 (to T.W.H.) and DFG-FOR 942 (to T.W.H. and S.Ö.) and by the Cluster of Excellence CellNetworks (C.D.T. and T.W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018109108/-/DCSupplemental.
References
- 1.Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 2.Niehrs C. On growth and form: A Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development. 2010;137:845–857. doi: 10.1242/dev.039651. [DOI] [PubMed] [Google Scholar]
- 3.Meinhardt H. Models for the generation of the embryonic body axes: Ontogenetic and evolutionary aspects. Curr Opin Genet Dev. 2004;14:446–454. doi: 10.1016/j.gde.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Adamska M, et al. Wnt and TGF-beta expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE. 2007;2:e1031. doi: 10.1371/journal.pone.0001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobmayer B, et al. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 6.Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development. 2005;132:2907–2916. doi: 10.1242/dev.01848. [DOI] [PubMed] [Google Scholar]
- 7.Lengfeld T, et al. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol. 2009;330:186–199. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Philipp I, et al. Wnt/beta-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc Natl Acad Sci USA. 2009;106:4290–4295. doi: 10.1073/pnas.0812847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gee L, et al. beta-catenin plays a central role in setting up the head organizer in hydra. Dev Biol. 2010;340:116–124. doi: 10.1016/j.ydbio.2009.12.036. [DOI] [PubMed] [Google Scholar]
- 10.Browne EN. The production of new hydranths in hydra by the insertion of small grafts. J Exp Zool. 1909;7:1–37. [Google Scholar]
- 11.Broun M, Bode HR. Characterization of the head organizer in hydra. Development. 2002;129:875–884. doi: 10.1242/dev.129.4.875. [DOI] [PubMed] [Google Scholar]
- 12.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 13.Meinhardt H. The radial-symmetric hydra and the evolution of the bilateral body plan: an old body became a young brain. Bioessays. 2002;24:185–191. doi: 10.1002/bies.10045. [DOI] [PubMed] [Google Scholar]
- 14.Guder C, et al. The Wnt code: Cnidarians signal the way. Oncogene. 2006;25:7450–7460. doi: 10.1038/sj.onc.1210052. [DOI] [PubMed] [Google Scholar]
- 15.Holstein TW, Hobmayer E, Technau U. Cnidarians: An evolutionarily conserved model system for regeneration? Dev Dyn. 2003;226:257–267. doi: 10.1002/dvdy.10227. [DOI] [PubMed] [Google Scholar]
- 16.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA. 2006;103:6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galliot B, Welschof M, Schuckert O, Hoffmeister S, Schaller HC. The cAMP response element binding protein is involved in hydra regeneration. Development. 1995;121:1205–1216. doi: 10.1242/dev.121.4.1205. [DOI] [PubMed] [Google Scholar]
- 19.Kaloulis K, Chera S, Hassel M, Gauchat D, Galliot B. Reactivation of developmental programs: The cAMP-response element-binding protein pathway is involved in hydra head regeneration. Proc Natl Acad Sci USA. 2004;101:2363–2368. doi: 10.1073/pnas.0306512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chera S, et al. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev Cell. 2009;17:279–289. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Kusserow A, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 22.Broun M, Sokol S, Bode HR. Cngsc, a homologue of goosecoid, participates in the patterning of the head, and is expressed in the organizer region of Hydra. Development. 1999;126:5245–5254. doi: 10.1242/dev.126.23.5245. [DOI] [PubMed] [Google Scholar]
- 23.Technau U, Bode HR. HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development. 1999;126:999–1010. doi: 10.1242/dev.126.5.999. [DOI] [PubMed] [Google Scholar]
- 24.Martin BL, Kimelman D. Wnt signaling and the evolution of embryonic posterior development. Curr Biol. 2009;19:R215–R219. doi: 10.1016/j.cub.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin BL, Kimelman D. Regulation of canonical Wnt signaling by Brachyury is essential for posterior mesoderm formation. Dev Cell. 2008;15:121–133. doi: 10.1016/j.devcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berns N, Kusch T, Schröder R, Reuter R. Expression, function and regulation of Brachyenteron in the short germband insect Tribolium castaneum. Dev Genes Evol. 2008;218:169–179. doi: 10.1007/s00427-008-0210-7. [DOI] [PubMed] [Google Scholar]
- 27.Shinmyo Y, et al. brachyenteron is necessary for morphogenesis of the posterior gut but not for anteroposterior axial elongation from the posterior growth zone in the intermediate-germband cricket Gryllus bimaculatus. Development. 2006;133:4539–4547. doi: 10.1242/dev.02646. [DOI] [PubMed] [Google Scholar]
- 28.Minokawa T, Wikramanayake AH, Davidson EH. cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 29.Wikramanayake AH, et al. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 30.Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chera S, Buzgariu W, Ghila L, Galliot B. Autophagy in Hydra: A response to starvation and stress in early animal evolution. Biochim Biophys Acta. 2009;1793:1432–1443. doi: 10.1016/j.bbamcr.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Hoch M, Pankratz MJ. Control of gut development by fork head and cell signaling molecules in Drosophila. Mech Dev. 1996;58:3–14. doi: 10.1016/s0925-4773(96)00541-2. [DOI] [PubMed] [Google Scholar]
- 33.Martinez DE, et al. Budhead, a fork head/HNF-3 homologue, is expressed during axis formation and head specification in hydra. Dev Biol. 1997;192:523–536. doi: 10.1006/dbio.1997.8715. [DOI] [PubMed] [Google Scholar]
- 34.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 35.Yasuoka Y, et al. Evolutionary origins of blastoporal expression and organizer activity of the vertebrate gastrula organizer gene lhx1 and its ancient metazoan paralog lhx3. Development. 2009;136:2005–2014. doi: 10.1242/dev.028530. [DOI] [PubMed] [Google Scholar]
- 36.Guder C, et al. An ancient Wnt-Dickkopf antagonism in Hydra. Development. 2006;133:901–911. doi: 10.1242/dev.02265. [DOI] [PubMed] [Google Scholar]
- 37.Kondo S, Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- 38.Okumura T, Matsumoto A, Tanimura T, Murakami R. An endoderm-specific GATA factor gene, dGATAe, is required for the terminal differentiation of the Drosophila endoderm. Dev Biol. 2005;278:576–586. doi: 10.1016/j.ydbio.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 39.Endl I, Lohmann JU, Bosch TC. Head-specific gene expression in Hydra: Complexity of DNA- protein interactions at the promoter of ks1 is inversely correlated to the head activation potential. Proc Natl Acad Sci USA. 1999;96:1445–1450. doi: 10.1073/pnas.96.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugiyama T, Fujisawa T. Genetic analysis of developmental mechanisms in hydra. I. Sexual reproduction of Hydra magnipapillata and isolation of mutants. Dev Growth Differ. 1977;19:187–200. doi: 10.1111/j.1440-169X.1977.00187.x. [DOI] [PubMed] [Google Scholar]
- 41.Martin VJ, Littlefield CL, Archer WE, Bode HR. Embryogenesis in hydra. Biol Bull. 1997;192:345–363. doi: 10.2307/1542745. [DOI] [PubMed] [Google Scholar]
- 42.Hobmayer B, Rentzsch F, Holstein TW. Identification and expression of HySmad1, a member of the R-Smad family of TGFbeta signal transducers, in the diploblastic metazoan Hydra. Dev Genes Evol. 2001;211:597–602. doi: 10.1007/s00427-001-0198-8. [DOI] [PubMed] [Google Scholar]
- 43.Grens A, Gee L, Fisher DA, Bode HR. CnNK-2, an NK-2 homeobox gene, has a role in patterning the basal end of the axis in hydra. Dev Biol. 1996;180:473–488. doi: 10.1006/dbio.1996.0321. [DOI] [PubMed] [Google Scholar]
- 44.Pukrop T, et al. Identification of two regulatory elements within the high mobility group box transcription factor XTCF-4. J Biol Chem. 2001;276:8968–8978. doi: 10.1074/jbc.M007533200. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.