Abstract
Exogenous enzymes are administered orally to treat several diseases, such as pancreatic insufficiency and lactose intolerance. Due to the proteinaceous nature of enzymes, they are subject to inactivation and/or digestion in the gastrointestinal (GI) tract. Here we describe a convenient fluorescence-based assay to monitor the activity of therapeutic enzymes in real time in vivo in the GI tract. To establish the proof of principle, the assay was applied to proline-specific endopeptidases (PEPs), a group of enzymes recently proposed as adjuvant therapy for celiac disease (a highly prevalent immunogenetic enteropathy). A short PEP-specific peptide sequence which is part of larger immunotoxic sequences of gluten was labeled with a fluorescent dye and a corresponding quencher. Upon enzymatic cleavage, the fluorescence emission was dequenched and detected with an in vivo imaging system. PEPs originating from Flavobacterium meningosepticum (FM) and Myxococcus xanthus (MX) were evaluated after oral administration in rats. While MX PEP could not cleave the peptide in the stomach, FM PEP showed significant gastric activity reaching 40–60% of the maximal in vivo signal intensity. However, both enzymes produced comparable fluorescence signals in the small intestine. Coadministration of an antacid drug significantly enhanced MX PEP’s gastric activity due to increased pH and/or inhibition of stomach proteases. With this simple procedure, differences in the in vivo performance of PEPs, which could not be identified under in vitro conditions, were detected. This imaging assay could be used to study other oral enzymes in vivo and therefore be instrumental in improving their therapeutic efficiency.
Keywords: celiac sprue, prolyl oligopeptidase, autoimmune disease
Exogenous enzymes are being tested or already used orally to treat several diseases, including pancreatic insufficiency, lactose intolerance, and phenylketonuria (1, 2). However, harsh pH changes and the presence of proteases in the gastrointestinal (GI) environment can strongly alter the activity of orally-administered enzymes. Strategies to prevent GI degradation of enzymes include enteric-coated formulations (3), coadministration of H2-receptor antagonists (4), and polymer-based matrices (5, 6). However, to the best of our knowledge, convenient methods to evaluate oral enzyme activity in vivo are still lacking. Here, we present a method for the real time measurement of exogenous enzyme activity in the GI tract by fluorescence dequenching using celiac disease (CD) as model application. CD is a highly prevalent (∼1%), genetically-based illness of the gut, triggered by the ingestion of cereal proteins, such as wheat gluten (7).
Generally, the digestive process of proteins in mammals is initiated in the stomach by low pH and proteases (mainly pepsin). Digestion is pursued in the small intestine (pH 5–7) by pancreatic proteases (e.g., trypsin and chymotrypsin) and brush-border membrane enzymes, leading to amino acids and oligopeptides which are then absorbed (8, 9). Owing to their exceptionally high proline and glutamine content, immunogenic peptides from gluten resist enzymatic degradation by GI enzymes (10).
The current and only treatment for CD is life-long elimination of gluten from the diet (11). This dietary restriction is a difficult experience for many patients and is often associated with decreased quality of life. Poor compliance, whether inadvertent or voluntary, to a strict gluten-free diet is frequent and predisposes patients to CD complications (e.g., nutritional deficiencies, osteoporosis, secondary autoimmune disorders, malignancies) (12). Hence, there is an urgent need for complementary nondietary therapies to help treat this common disorder because it is associated with increased morbidity and mortality (13). Various therapeutic avenues are currently being explored to manage this pathology (14–17). Among these, oral administration of prolyl endopeptidases (PEPs) is one of the most studied approaches (18). Unlike human GI enzymes, PEPs can efficiently hydrolyze proline-rich gluten peptides (9) and they have been shown to alleviate gluten immunotoxicity under simulated intestinal conditions (19, 20). Earlier studies conducted with PEPs from Flavobacterium meningosepticum (FM) and Myxococcus xanthus (MX) revealed moderate enzyme stability in vitro under artificial GI conditions (21). Aspergillus niger (AN) PEP was recently investigated in an artificial compartmental system mimicking the human GI tract (TIM model). This PEP efficiently accelerated gluten breakdown under gastric conditions in vitro (22). Another PEP from Sphingomonas capsulate (SC) was evaluated in a clinical trial in a combinatory approach (23). The enzyme reduced the gluten-specific T cell response but did not significantly diminish CD-associated GI symptoms (24). Thus, a better understanding of the behavior of PEPs in vivo after oral administration may pave the way for a more efficient optimization of CD treatment (25). In this contribution, we report an in vivo fluorescence-based assay that allows for monitoring enzymatic PEP activity in real time in the GI tract. This assay could, in principle, be adapted to other oral enzymes.
Results and Discussion
In Vitro Activity of PEPs.
The activity of FM, MX, and SC PEPs was first characterized in vitro and their low stability in artificial GI fluids (Fig. S1A) was confirmed (21). Moreover, PEPs could not reduce the abundance of the peptide QLQPFPQPQLPYPQPQPF from whole wheat gluten under conditions simulating in vitro GI digestion (Fig. S1B). This model peptide comprises the major immunogenic peptide QLQPFPQPQLPY (26, 27). Since the in vivo predictability of in vitro studies is often limited, we sought to analyze PEP activity in rats.
Design of In Vivo Fluorescence Assay.
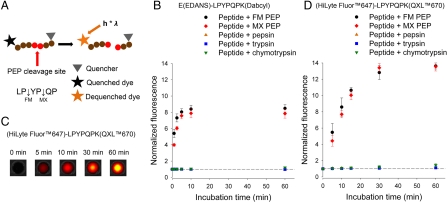
To develop the fluorescence assay, the short peptide sequence LPYPQP was selected as substrate as it constitutes the core region of both the CD-toxic peptide p62-75 from α2-gliadin (21) and from QLQPFPQPQLPYPQPQPF (Fig. 1A). To establish proof of concept, LPYPQP was first labeled with the fluorescent dye EDANS (λem = 490 nm) and corresponding quencher Dabcyl [E(EDANS)-LPYPQPK(Dabcyl)]. Fluorescence of the intact peptide was low while its proteolysis by PEPs in vitro led to its complete cleavage [as verified by liquid chromatography-mass spectrometry (LC-MS)] concurrent with an 8-fold increase in fluorescence intensity after 15 min incubation. Native enzymes of the GI tract barely hydrolyzed this peptide (Fig. 1B). For the in vivo experiments, in order to increase signal depth penetration and to reduce auto fluorescence within tissue (28), LPYPQP was labeled with the fluorescent dye HiLyte Fluor™647 (λem = 700 nm) and corresponding quencher QXL™670 [(HiLyte Fluor™647)-LPYPQPK(QXL™670)]. In vitro proteolysis of this modified peptide resulted in a > 13-fold increase of fluorescence after complete cleavage, while luminal proteases could not digest the peptide (Fig. 1 C and D). A control peptide without quencher [(HiLyte Fluor™647)-LPYPQPK] produced comparable fluorescence intensities in vitro (Fig. S2). LC-MS analysis additionally revealed that FM PEP mainly cleaved at LPYP↓QP, while MX PEP preferred the LP↓YPQP position (Fig. 1A).
Fig. 1.
In vitro cleavage of E(EDANS)-LPYPQPK(Dabcyl) and (HiLyte Fluor™647)-LPYPQPK(QXL™670). (A) Schematic representation of the dequenching assay. Arrows within the peptide sequence indicate preferential cleavage sites for FM and MX PEP. (C) In vitro cleavage of (HiLyte Fluor™647)-LPYPQPK(QXL™670) by FM PEP in phosphate buffer (pH 6.8) measured with the in vivo imaging system. (B, D) The peptides (5 and 3 μM, respectively) were incubated in the presence of FM and MX PEPs (1 μg/mL), pepsin (0.20 mg/mL), trypsin (0.22 mg/mL), or chymotrypsin (0.22 mg/mL). Mean ± SD, n = 3, fluorescence intensities in B and D were measured using plate reader and in vivo imaging system, respectively.
In Vivo Dequenching Assay.
With this tool in hand, the enzymatic activity of PEPs was examined in vivo in female rats (see Movie S1 for a live illustration of the in vivo assay). FM and MX PEP were selected for the in vivo experiments because of their comparable in vitro cleavage pattern against gliadin peptides with similar chain length (21). The animals were gavaged with PEPs, followed by the labeled peptide 5 min later. Whole wheat gliadin was coadministered to all rats to create a more realistic environment. To quantify the extent of peptide cleavage in vivo, the fully cleaved peptide (precleaved peptide) was also given to rats in a separate experiment, and its fluorescence intensity taken as 100%. After gavage, the animals were either quickly anesthetized continuously for 4 h or at selected time points for 15 min. For imaging, the rats were placed in an in vivo imaging system (Fig. S3). Images were analyzed by selecting regions of interest (ROIs) over the stomach and small intestine based on the rat’s anatomy (Fig. S4). In terms of size, form, and position on the animal’s abdominal area, ROIs were kept constant, for a given animal, throughout the duration of experiments. The fluorescence signal was not tracked in the lower GI tract because inflammation in CD occurs in the early small intestine.
In Vivo Imaging under Continuous Anesthesia.
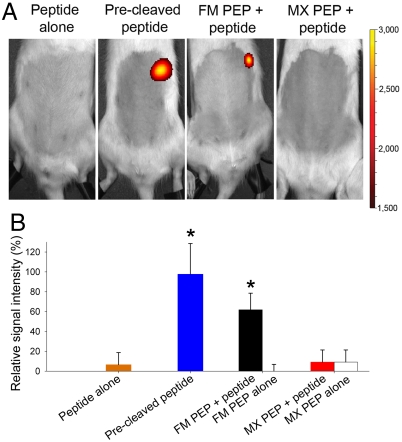
For stomach analysis experiments, rats were anesthetized 5 min after gavage. By doing so, gastric motility decreased, and the effect of prolonged exposure to gastric conditions was examined. Administration of the peptide or PEPs alone did not result in significant fluorescence in the stomach (Fig. 2, Fig. S5), while the precleaved peptide produced a strong fluorescent signal. Rats receiving FM PEP prior to the peptide displayed a rapid increase in fluorescence intensity in the stomach region (Fig. S5), reaching 60% of the signal of the precleaved peptide after 20 min (Fig. 2B). Conversely, MX PEP did not cleave the peptide efficiently in the stomach, even after 4 h. In vitro activity profiles of PEPs showed that MX PEP was not efficient at acidic pH while FM displayed very little activity at pH 4.5 (∼2% of activity) (Fig. S6A). However, in vivo, FM PEP turned out to be much more active under gastric conditions. Despite comparable inactivation rates noted in vitro in simulated GI fluids (Fig. S1, Table S1), the in vivo performance of FM and MX PEP was significantly different. In particular, this finding could have an implication for CD because in this pathology, inflammation is induced in the upper small intestine. Hence, efficient enzymatic cleavage (and thus detoxification) of gluten peptides in the early GI tract should be sought for optimal treatment efficacy.
Fig. 2.
Real time measurement of PEP activity in vivo under continuous anesthesia. (A) Fluorescence imaging and (B) relative signal intensity in the stomach region 20 min after oral administration of the labeled peptide alone, the precleaved peptide, FM, or MX PEP with or without peptide. Representative rats from each set are shown in A. Color scales are identical for all pictures. In B, signals were plotted by setting the average in vivo maximum signal (precleaved peptide after 18 min) to 100%. Mean + SD, n = 6. *Precleaved peptide and FM PEP + peptide vs. all other groups (p < 0.05).
In Vivo Imaging under Discontinuous Anesthesia.
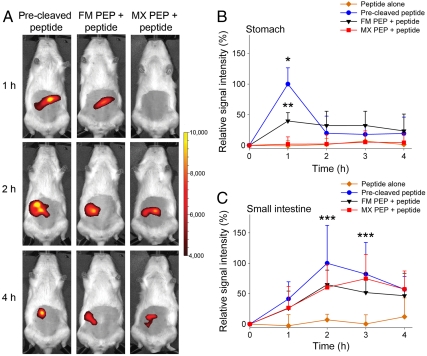
Next, PEP activity in both the stomach and small intestine, was assessed by applying discontinuous anesthesia and imaging 1, 2, 3, and 4 h after oral gavage, with recovery phases between measurements (Fig. S3). First, the impact of the fasting status on GI transit time was analyzed. Rats were deprived of provender for either 4 or 16 h and then administered the precleaved peptide. The longer fasting promoted rapid transit of the peptide from the stomach into the small intestine, with almost no peptide left in the stomach after 1 h (Fig. S7). In order to expose PEPs for a reasonable time to gastric conditions, a fasting period of 4 h was therefore applied before each experiment. Rats were fed PEPs and peptide, and anesthetized discontinuously. As seen in Fig. 3 A and B, the fluorescence intensity peaked at 1 h in the stomach and was generally lower than when continuous anesthesia was applied. In the latter case, the fluorescence signal was mainly located in the rat fore stomach while discontinuous anesthesia also led to fluorescence in the corpus stomach, indicating movement into the duodenum and small intestine (Fig. 3A, Fig. S4). Again, oral gavage of FM PEP followed by the peptide evoked a strong increase in fluorescence in the stomach region (40% of precleaved signal intensity). No peptide cleavage occurred in the stomach with MX PEP (Fig. 3B). Quantification of residual peptide in the stomach 1 h after oral administration of FM PEP with peptide showed that approximately 60% of uncleaved peptide was left in the stomach after this time (Fig. S8). In the small intestine, the signal intensity peaked between 2 h (FM PEP and the precleaved peptide) and 3 h (MX PEP) after intake. Interestingly, comparable fluorescence intensities were observed for both MX and FM PEPs in the small intestine (65 and 75% of precleaved signal intensity after 2 and 3 h, respectively) (Fig. 3 A and C). Feces collected 24 h after oral application of FM PEP with peptide contained less than 5% intact peptide. Moreover, the peptide was not absorbed systemically and could be recovered in the feces (Figs. S9 and S10).
Fig. 3.
In vivo fluorescence imaging of PEP activity in the GI tract under discontinuous anesthesia. (A) Evolution of the fluorescent signal in the abdominal region 1, 2, and 4 h after oral administration of the precleaved peptide, FM, or MX PEP followed by peptide. (B, C) Evolution of the relative signal intensity (B) in the stomach and (C) in the small intestine over 4 h. Representative rats from each set are shown in A. Color scales are identical for all pictures. At each time point, the rats were anesthetized shortly before recording a picture. In B and C, signals were plotted by setting the average in vivo maximum signal (precleaved peptide) for each region to 100%. Mean + SD, n = 6–9. *Precleaved peptide (p < 0.01) and **FM PEP + peptide (p < 0.05) vs. MX PEP + peptide and peptide alone, ***Precleaved peptide, FM, and MX PEPs + peptide vs. peptide alone (p < 0.05).
Even though it was barely active under gastric conditions, MX PEP recovered at least partially its activity upon exiting the stomach, suggesting the potential regeneration of enzyme conformation. Despite higher FM PEP gastric activity, overall, the differences in the efficacy of both PEPs were small upon reaching the intestine. Such an effect could not be readily seen under in vitro conditions.
Impact of Coadministration of Antacid Drugs and Gliadin.
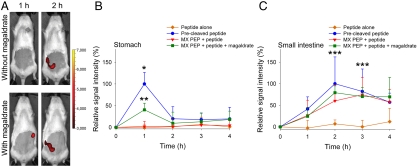
Reduced activity of PEPs in the stomach is most likely a combination of both, inactivation (by degradation or changes in conformation) and slower rate of catalysis at low pH (Fig. S6). In order to alter this environment, another series of experiments was performed with MX PEP by administering the peptide in the presence of magnesium aluminate monohydrate (magaldrate), a basic salt used to neutralize gastric acidity. The addition of magaldrate produced an increase in MX PEP activity in the stomach 1 h after oral intake and a maximum signal in the small intestine after 2 h (Fig. 4), as a result of increased stomach pH and/or inhibition of gastric protease.
Fig. 4.
Influence of magaldrate coadministration on MX PEP activity. (A) Rats were orally administered MX PEP followed by the labeled peptide with or without magaldrate. Pictures were recorded 1 and 2 h after gavage. The pictures shown are from representative rats in each set. Color scales are identical. (B, C) The graphs report the relative fluorescence signal (B) in the stomach and (C) in the small intestine with or without magaldrate treatment. Signals were plotted by setting the average in vivo maximum signal (oral administration of precleaved peptide) to 100%. Mean + SD, n = 4–9. *Precleaved peptide (p < 0.01) and **MX PEP + peptide + magaldrate (p < 0.05) vs. MX PEP + peptide and peptide alone, ***Precleaved peptide, MX PEP + peptide ± magaldrate vs. peptide alone (p < 0.05).
These data indicate that, depending on the nature of treatment, it is possible to readily visualize the increase in enzyme activity produced by the coadministration of antacid drugs.
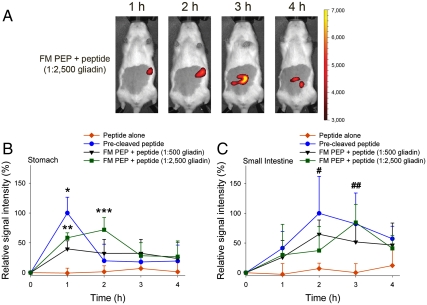
Subsequently, the influence of the amount of coadministered gliadin on the enzymatic activity was evaluated. The evolution of the fluorescence signal was analyzed after the administration of FM PEP, peptide, and different quantities of gliadin (1∶500 and 1∶2,500 PEP:gliadin, weight (w)/w). A higher amount of gliadin elicited retention of the fluorescence signal in the stomach and delayed transit into the small intestine owing to the presence of a higher quantity of dietary proteins (Fig. 5). Additionally, the presence of gliadin may, to a certain extent, have acted as competitive substrate for gastric proteases which should be at least partially responsible for PEP inactivation. Our results indicate that modification of the GI environment can considerably influence the activity of orally-administered enzymes and that a better understanding of this parameter could be instrumental in improving oral enzyme delivery.
Fig. 5.
Impact of admixed gliadin amount on FM PEP activity. Rats were administered FM PEP by gavage with peptide and gliadin addition at ratios of 1∶500 and 1∶2,500 (PEP∶gliadin, w/w), respectively. (A) Pictures depict the dynamics of the fluorescence signal during 4 h of observation in a representative animal receiving FM PEP + peptide (1∶2,500 gliadin). Color scales are identical. (B, C) Relative fluorescence signal intensity of FM PEP + peptide admixed with gliadin (1∶500 and 1∶2,500, w/w) (B) in the stomach or (C) small intestine compared to oral administration of precleaved peptide or peptide alone. Mean + SD, n = 4–9. *Precleaved peptide (p < 0.01) and **FM PEP + peptide (1∶500 and 1∶2,500 gliadin) (p < 0.05) vs. peptide alone, ***FM + peptide (1∶2,500 gliadin) vs. precleaved peptide and peptide alone, #Precleaved peptide and FM + peptide (1∶500 gliadin) vs. peptide alone, ##Precleaved peptide and FM PEP + peptide (1∶500 and 1∶2,500 gliadin) vs. peptide alone (all p < 0.05).
Conclusion
In this manuscript, a simple fluorescence dequenching assay was explored to monitor the activity of orally administered exogenous enzymes in real time in vivo. Such an assay is noninvasive and can be repeated several times on the same animals to study parameters affecting the catalytic activity of therapeutic enzymes and improve their efficiency. Furthermore, the principle could potentially be adapted to in vivo imaging of larger animals when combined with suitable hardware and computer algorithms. Fluorescence-based imaging has been used for noninvasive lymph node mapping in rabbits (29) and detection of mammary tumors in dogs (30, 31) up to 1.5 cm from tissue surface. Moreover, it has been predicted that fluorescence-assisted imaging may permit noninvasive measurements 30–40 cm deep in human tissue (32). Indocyanine green is indeed already used for angiography in humans (33).
The presented assay could also serve to monitor the degradation of peptide drugs by GI enzymes in vivo and devise new delivery strategies. The work highlighted significant discrepancies between the in vitro and in vivo properties of oral enzymes. In particular, this imaging method (potentially in combination with the TIM model) could prove useful in the optimization of PEPs and their delivery strategies for the treatment of CD.
Materials and Methods
Materials.
FM PEP (25 U/mg) was purchased from LuBioScience, MX PEP (25 U/mg) was obtained from Zedira, and SC PEP (19 U/mg) was custom-made by Prozomix Ltd., as described elsewhere (34). The labeled peptides (EDANS)-LPYPQPK(Dabcyl), (HiLyte Fluor™647)-LPYPQPK(QXL™670) and (HiLyte Fluor™647)-LPYPQPK were custom-synthesized by Peptide 2.0 and Anaspec, respectively. Whole wheat gluten, gliadin, pepsin (2,188 U/mg protein), α-chymotrypsin (96 U/mg protein), and pancreatin (USP grade) were purchased from Sigma-Aldrich. Z-Gly-Pro-pNA and trypsin (3,480 U/mg) were procured from Bachem and AppliChem, respectively. Magnesium aluminate monohydrate (magaldrate, Riopan®) was purchased in a retail pharmacy.
In Vitro Stability and Activity of PEPs.
FM, MX, and SC PEPs were incubated at 50 mU/mL in simulated gastric (pH 1.2 with pepsin) and intestinal (pH 6.8 with pancreatin) fluids United States Pharmacopeia (USP), and in USP acetate buffer (pH 4.5 with pepsin). pH-dependent activity profiles of FM and MX PEPs were determined at 50 mU/mL in HCl/KCl solution USP (pH 1.2), acetate buffer USP (pH 4.5), phosphate buffer (100 mM, pH 7.0), and trometamol/sodium chloride buffer (50/100 mM, pH 9). Activity was assessed by measuring the cleavage of Z-Gly-Pro-pNA using an Infinite M200 microplate reader (Tecan Ltd.) (20).
In vitro activity of PEPs was analyzed by incubating the enzymes directly with whole wheat gluten in 50 mM acetate buffer USP (pH 4.5). Pepsin (0.8 mg/mL) was added, and the mixture was incubated for 1 h at 37 °C. Subsequently, the pH was raised to 6.8 (using 0.2 M Na2HPO4 and 0.2 M NaOH) and trypsin and chymotrypsin (0.4 mg/mL) were added. The mixture was again incubated for 2 h at 37 °C. After heat inactivation (10 min at 95 °C), the mixture was centrifuged (14,000 × g for 10 min) and the supernatant analyzed by LC-MS.
Samples were analyzed using an LC system composed of a Rheos Allegro quaternary pump, column oven (Hot Dog 5090), C18-column (Hypersil Gold, 100 × 1 mm, 1.9 μm) and XCalibur control software (all from Thermo Fisher Scientific Inc.). Samples were injected (10 μL) at 35 °C, and a flow rate of 50 μL/ min using water (solvent A) and acetonitrile (solvent B) (both with 0.1% formic acid). The gradient consisted of: 95% solvent A as initial value, 95 - 50% A in 1–19 min, 50 - 5% A in 19–23 min, 5% A in 23–27 min, 5–95% A in 27–28 min, and 95% A in 28–31 min. The LC was directly connected to an LTQ XL linear quadrupole Ion Trap (Thermo Fisher Scientific Inc.) equipped with an electrospray ion source. Data were acquired by full MS, followed by MS2-fragmentation of the five most intense signals in an automated data-dependent scan.
Raw data were subjected to the Sequest search algorithm using Proteome Discoverer Software (Thermo Fisher Scientific Inc.). Detected peptides were searched against all sequences from the ExPASy Proteomics database (Swiss Institute of Bioinformatics) that matched the following search terms: triticum, aestivum, wheat, and gliadin. Abundance of QLQPFPQPQLPYPQPQPF was analyzed for the incubation of PEPs with whole wheat gluten. Each value was normalized to a non-PEP-treated sample. The linearity of MS quantification was determined by spiking a synthetic gluten peptide (PQPQLPYPQPQLP) into gluten-digest samples in a dose-dependent manner (R2 = 0.9945).
Dequenching Assay.
In vitro cleavage of the peptides E(EDANS)-LPYPQPK(Dabcyl) (λex = 340 nm, λem = 490 nm) and (HiLyte Fluor™647)-LPYPQPK(QXL™670) was monitored by spectrofluorometry (λex = 640 nm, λem = 700 nm) at 37 °C on an Infinite M200 microplate reader and an IVIS® Spectrum in vivo imaging system (Caliper Life Science), respectively. Complete cleavage was confirmed by LC-MS.
In Vivo Imaging.
All animal experiments were approved by the Cantonal Veterinary Office Zurich. In vivo experiments were conducted with (HiLyte Fluor™647)-LPYPQPK(QXL™670). Female Sprague-Dawley rats (4–8 weeks old, 125–200 g) on gluten-free food were fasted for 4 h, unless stated otherwise. The abdominal region of rats was shaved and a pregavage picture taken. The rats were gavaged orally with peptide alone (negative control), precleaved peptide (positive control), FM or MX PEP alone (negative control), FM or MX PEP, followed by peptide solution (peptide given 5 min after PEP; study group) (Fig. S3). Oral solutions were prepared as follows: FM and MX PEPs (0.1 μg) were diluted in 150 μL 10 mM phosphate buffer (pH 7.0). 5 μL of peptide stock solution (250 μM in 35% DMSO) was diluted in 200 μL sodium cholate [0.5%, w/volume (v)] to ensure proper solubilization. For the precleaved peptide solution, peptide and PEP were mixed and shaken at 37 °C for at least 4 h prior to each experiment. For all experiments, PEPs were admixed with gliadin, the dominant CD-toxic fraction in gluten (0.5 or 2.5 mg corresponding to a PEP:gliadin ratio of 1∶500 or 1∶2,500, w/w), 30 s prior to oral gavage. To estimate whether modified peptide was absorbed, rats were administered FM PEP with peptide and the fluorescence intensity in the saphenous vein area was imaged during 4 h of discontinuous anesthesia. In experiments aimed at assessing the impact of stomach acidity on MX PEP activity, the rats were given peptide diluted in magaldrate (Riopan®).
After oral administration, the rats were anesthetized (2% isoflurane, 2 mL/ min oxygen) for immediate imaging (continuous anesthesia) or returned to their cages for imaging at later time points (discontinuous anesthesia) (Fig. S3). Rats were placed in an IVIS® Spectrum in vivo imaging system equipped with a cone nose for anesthesia (1.5–2% isoflurane, 0.5 mL/ min oxygen) and a heated platform (37 °C) to maintain body temperature during analysis. Pictures were recorded in the following settings: λex = 640 nm, λem = 700 nm, binning 8, f/stop 2, exposure time = 2 s. The rats were assigned to groups in such a way that mean weights were equal in all groups.
For quantifying residual peptide leaving the stomach and intestine, rats were administered FM PEP with peptide and they were killed after 1 h or the feces were collected 24 h after oral gavage, respectively. Feces or stomach content were suspended in a mixture (3∶2, v/v) of phosphate buffer (10 mM, pH 7.0) and sodium cholate (0.5%) and centrifuged (3,000 × g, 5 min). 5 μL of FM PEP (0.25 mg/mL) was added to the supernatant and the extent of fluorescence dequenching (IVIS® Spectrum in vivo imaging system) was used to calculate the residual proportion of intact peptide. Then, the feces or stomach samples were extracted three times using sodium cholate dissolved in acetonitrile (0.25%, w/v). After centrifugation (20,000 × g, 10 min) supernatants were collected, combined and freeze-dried. Lyophilizates were dissolved in water and acetonitrile (1∶1, v/v) and the concentration of cleaved peptide was quantified by spectrofluorometry.
Analysis of Recorded Images.
Images were analyzed with Living Image software (Caliper Life Science). In each experiment, ROIs were defined for the stomach and small intestine (Fig. S4). At each time point, the average fluorescence intensity in each ROI was normalized to the pregavage status. The relative signal intensity was expressed in percentage of mean maximal in vivo value for each region (stomach and small intestine) obtained by administering the precleaved peptide (100%). The following equation was employed (normalized fluorescence of 1 is set to 0%)
where xt represents normalized fluorescence at a single time point and xmax is the maximal in vivo signal in the specific ROI.
Autofluorescence not related to the peptide and located outside the abdominal area was erased in all displayed pictures. All pictures were smoothed by 7 × 7 pixels to reduce background noise.
The video was created by editing the still pictures using Nero StartSmart software.
Statistical Analysis.
Mean and standard deviation (SD) are reported for all data (unless stated otherwise); n values in the legends indicate the number of independent experiments. Group values were analyzed by nonparametric Kruskal-Wallis analysis of variance, followed by Dunn’s multiple comparisons test to ascertain the significance of all paired combinations. Differences between groups were considered significant for p < 0.05.
Supplementary Material
Acknowledgments.
We thank Dr. Paola Luciani and Dr. Marc A. Gauthier for their technical help, and Dr. Hans Peter Käsermann for his help with the animal experiments. This research was supported by “IG Zöliakie der Deutschen Schweiz” and the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100285108/-/DCSupplemental.
References
- 1.Vellard M. The enzyme as drug: application of enzymes as pharmaceuticals. Curr Opin Biotechnol. 2003;14:444–450. doi: 10.1016/s0958-1669(03)00092-2. [DOI] [PubMed] [Google Scholar]
- 2.Sarkissian CN, et al. A different approach to treatment of phenylketonuria: phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proc Natl Acad Sci USA. 1999;96:2339–2344. doi: 10.1073/pnas.96.5.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stead RJ, Skypala I, Hodson ME, Batten JC. Enteric coated microspheres of pancreatin in the treatment of cystic fibrosis: comparison with a standard enteric coated preparation. Thorax. 1987;42:533–537. doi: 10.1136/thx.42.7.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regan PT, et al. Comparative effects of antacids, cimetidine and enteric coating on the therapeutic response to oral enzymes in severe pancreatic insufficiency. New Engl J Med. 1977;297:854–858. doi: 10.1056/NEJM197710202971603. [DOI] [PubMed] [Google Scholar]
- 5.Sinha VR, Trehan A. Biodegradable microspheres for protein delivery. J Controlled Release. 2003;90:261–280. doi: 10.1016/s0168-3659(03)00194-9. [DOI] [PubMed] [Google Scholar]
- 6.Morishita M, et al. Novel oral insulin delivery systems based on complexation polymer hydrogels: single and multiple administration studies in type 1 and 2 diabetic rats. J Controlled Release. 2006;110:587–594. doi: 10.1016/j.jconrel.2005.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catassi C, Fasano A. Celiac disease. Curr Opin Gastroen. 2008:687–691. doi: 10.1097/MOG.0b013e32830edc1e. [DOI] [PubMed] [Google Scholar]
- 8.Hausch F, et al. Intestinal digestive resistance of immunodominant gliadin peptides. Am J Physiol-Gastr L. 2002;283:G996–1003. doi: 10.1152/ajpgi.00136.2002. [DOI] [PubMed] [Google Scholar]
- 9.Shan L, et al. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 10.Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. 2002;2:647–655. doi: 10.1038/nri885. [DOI] [PubMed] [Google Scholar]
- 11.Tack GJ, Verbeek WHM, Schreurs MWJ, Mulder CJJ. The spectrum of celiac disease: epidemiology, clinical aspects and treatment. Nat Rev Gastroentero. 2010;7:204–213. doi: 10.1038/nrgastro.2010.23. [DOI] [PubMed] [Google Scholar]
- 12.Di Sabatino A, Corazza GR. Coeliac disease. The Lancet. 2009;373:1480–1493. doi: 10.1016/S0140-6736(09)60254-3. [DOI] [PubMed] [Google Scholar]
- 13.Biagi F, Corazza GR. Mortality in celiac disease. Nat Rev Gastroentero. 2010;7:158–162. doi: 10.1038/nrgastro.2010.2. [DOI] [PubMed] [Google Scholar]
- 14.Pinier M, Fuhrmann G, Verdu E, Verdu E, Leroux J-C. Prevention measures and exploratory pharmacological treatments of celiac disease. Am J Gastroenterol. 2010;105:2551–2561. doi: 10.1038/ajg.2010.372. [DOI] [PubMed] [Google Scholar]
- 15.Kelly CP, et al. M2048 Safety, tolerability and effects on intestinal permeability of larazotide acetate in celiac disease: results of a phase IIb 6-week gluten-challenge clinical trial. Gastroenterology. 2009;136:A–474. [Google Scholar]
- 16.Keech CL, et al. Immune tolerance induced by peptide immunotherapy in an HLA DQ2-dependent mouse model of gluten Immunity. Gastroenterology. 2009;136:A–57. [Google Scholar]
- 17.Pinier M, et al. Polymeric binders suppress gliadin-induced toxicity in the intestinal epithelium. Gastroenterology. 2009;136:288–298. doi: 10.1053/j.gastro.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Shan L, Mathews II, Khosla C. Structural and mechanistic analysis of two prolyl endopeptidases: role of interdomain dynamics in catalysis and specificity. Proc Natl Acad Sci USA. 2005;102:3599–3604. doi: 10.1073/pnas.0408286102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel M, et al. Rational design of combination enzyme therapy for celiac sprue. Chem Biol. 2006;13:649–658. doi: 10.1016/j.chembiol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Marti T, et al. Prolyl endopeptidase-mediated destruction of T cell epitopes in whole gluten: chemical and immunological characterization. J Pharmacol Exp Ther. 2005;312:19–26. doi: 10.1124/jpet.104.073312. [DOI] [PubMed] [Google Scholar]
- 21.Shan L, et al. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004;383:311–318. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitea C, et al. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for coeliac disease. Gut. 2008;57:25–32. doi: 10.1136/gut.2006.111609. [DOI] [PubMed] [Google Scholar]
- 23.Gass J, et al. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology. 2007;133:472–480. doi: 10.1053/j.gastro.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 24.Tye-Din JA, et al. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin Immunol. 2010;134:289–295. doi: 10.1016/j.clim.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Matysiak-Budnik T, et al. Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology. 2005;129:786–796. doi: 10.1053/j.gastro.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Arentz-Hansen H, et al. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med. 2000;191:603–612. doi: 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim C-Y, et al. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci USA. 2004;101:4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frangioni JV. In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Chu M, Wan Y. Sentinel lymph node mapping using near-infrared fluorescent methylene blue. J Biosci Bioeng. 2009;107:455–459. doi: 10.1016/j.jbiosc.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds JS, et al. Imaging of spontaneous canine mammary tumors using fluorescent contrast agents. Photochem Photobiol. 1999;70:87–94. [PubMed] [Google Scholar]
- 31.Gurfinkel M, et al. Pharmacokinetics of ICG and HPPH-car for the detection of normal and tumor tissue using fluorescence, near-infrared reflectance imaging: a case study. Photochem Photobiol. 2000;72:94–102. doi: 10.1562/0031-8655(2000)072<0094:poiahc>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Ntziachristos V, Ripoll J, Weissleder R. Would near-infrared fluorescence signals propagate through large human organs for clinical studies? Opt Lett. 2002;27:333–335. doi: 10.1364/ol.27.000333. [DOI] [PubMed] [Google Scholar]
- 33.Herbort CP, LeHoang P, Guex-Crosier Y. Schematic interpretation of indocyanine green angiography in posterior uveitis using a standard angiographic protocol. Ophthalmology. 1998;105:432–440. doi: 10.1016/S0161-6420(98)93024-X. [DOI] [PubMed] [Google Scholar]
- 34.Kabashima T, et al. Prolyl endopeptidase from Sphingomonas capsulata: isolation and characterization of the enzyme and nucleotide sequence of the gene. Arch Biochem Biophys. 1998;358:141–148. doi: 10.1006/abbi.1998.0836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.