Abstract
Acetylation of histones triggers association with bromodomain-containing proteins that regulate diverse chromatin-related processes. Although acetylation of transcription factors has been appreciated for some time, the mechanistic consequences are less well understood. The hematopoietic transcription factor GATA1 is acetylated at conserved lysines that are required for its stable association with chromatin. We show that the BET family protein Brd3 binds via its first bromodomain (BD1) to GATA1 in an acetylation-dependent manner in vitro and in vivo. Mutation of a single residue in BD1 that is involved in acetyl-lysine binding abrogated recruitment of Brd3 by GATA1, demonstrating that acetylation of GATA1 is essential for Brd3 association with chromatin. Notably, Brd3 is recruited by GATA1 to both active and repressed target genes in a fashion seemingly independent of histone acetylation. Anti-Brd3 ChIP followed by massively parallel sequencing in GATA1-deficient erythroid precursor cells and those that are GATA1 replete revealed that GATA1 is a major determinant of Brd3 recruitment to genomic targets within chromatin. A pharmacologic compound that occupies the acetyl-lysine binding pockets of Brd3 bromodomains disrupts the Brd3-GATA1 interaction, diminishes the chromatin occupancy of both proteins, and inhibits erythroid maturation. Together these findings provide a mechanism for GATA1 acetylation and suggest that Brd3 “reads” acetyl marks on nuclear factors to promote their stable association with chromatin.
Keywords: hematopoiesis, posttranslational modifications, gene regulation
Posttranslational acetylation of lysine residues is a widely used cellular signaling mechanism involving histone and nonhistone proteins. Typically, acetylated lysines (acK) serve as docking sites for bromodomains that are found in numerous chromatin-related proteins (1, 2). Thus, bromodomains serve as “readers” of acK marks in a cascade of signaling events that influence diverse processes within chromatin. The hematopoietic zinc finger protein GATA1 was one of the earliest reported acetylated transcription factors (3, 4). GATA1 is a pivotal regulator of the erythroid, megakaryocyte, and mast cell lineages (5–9). GATA1 activates all known erythroid-specific genes, and mice lacking GATA1 succumb to anemia due to failed maturation and apoptosis of erythroid precursor cells (5, 10, 11). GATA1 also functions as a repressor, down-regulating genes associated with the proliferative, immature state. Mutations in the zinc finger region of GATA1 underlie congenital anemias and thrombocytopenias (12), whereas alterations at the N terminus of GATA1 are associated with megakaryoblastic leukemias in patients with Down syndrome (13). Previously, we found that GATA1 interacts with the acetyltransferase CREB binding protein (CBP) (14) to stimulate histone acetylation at GATA1 target genes (15, 16). CBP and its paralog p300 also acetylate GATA1 at two lysine-rich clusters C-terminal to each of its zinc fingers (Fig. 1A) (3, 4). CBP-mediated acetylation of GATA1 can be antagonized by the myeloid transcription factor PU.1 (17) that promotes myeloid differentiation at the expense of erythroid development in part by inhibiting GATA1 (18). The oncogenic fusion protein AML1-ETO also inhibits acetylation of GATA1, which might account for the inhibition of erythroid development associated with t(8;21) myeloid leukemias (19).
Fig. 1.
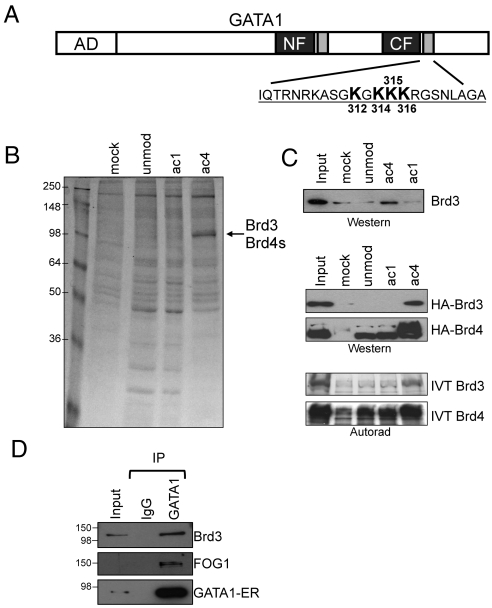
Brd3 is an acetyl-GATA1 associated protein. (A) Schematic of GATA1. AD, activation domain; gray boxes near the N-terminal (NF) and C-terminal (CF) zinc fingers represent conserved acetylated motifs. The sequence of the C-terminal acetylation motif is shown with four major acetylatable lysines in bold. (B) Colloidal blue stained gel of proteins bound to unmod, ac1, ac4 peptides, or to beads alone (mock). (C) Endogenous Brd3 (Top), stably expressed HA-Brd3 or HA-Brd4 (Middle), or IVT Brd3 and Brd4 bound to indicated (Bottom) GATA1 peptides were detected by Western blot or autoradiography. (D) Immunoprecipitation of GATA1-ER from G1E-ER4 cells followed by Western blot for GATA1, Brd3, and FOG1. Input equals 2.5%.
Despite the growing body of data supporting the importance of GATA1 acetylation, the functional and mechanistic consequences of this modification have remained unclear. Although acetylation of chicken GATA1 has been reported to enhance its affinity for DNA in vitro, little to no effect on DNA binding was observed with mouse or zebrafish GATA1 (3, 4, 20, 21). It has been suggested that acetylation of GATA1 promotes its turnover (22) perhaps involving an interaction with HSP27 (23). However, when expressed in erythroid cells, mutations of the relevant lysine residues in GATA1 did not alter steady-state protein levels nor did they impair nuclear localization of GATA1 or binding to naked DNA templates in vitro (20). Notably, although acetylation-defective versions of GATA1 can activate transiently transfected reporter genes, they fail to stably associate with their chromatinized endogenous targets in vivo (20). This suggests that acetylation functions specifically in the context of native chromatin.
The mammalian BET (Bromodomain Extra Terminal) family of proteins includes Brd2, Brd3, Brd4, and Brdt, and is characterized by the presence of two N-terminal bromodomains (BD1 and BD2) and an extraterminal protein interaction domain. Bromodomains recognize acK in the context of flanking amino acids that mediate binding selectivity. Brd4 binds to histone H4 in its di- (K5, K12) or tetraacetylated (K5, K8, K12, K16) forms (24, 25) although binding to monoacetylated H4K16 and dually acetylated histone H3K9 and H3K14 has also been observed (24, 26). In addition to binding acetylated histones, Brd4 recruits the positive transcription elongation factor b (P-TEFb) to the promoters of active genes (27, 28). Current models suggest that signal-dependent augmentation in H4 acetylation at inducible promoters leads to Brd4-mediated P-TEFb recruitment, followed by phosphorylation of the C-terminal domain of RNA polymerase 2 (pol II) and transitioning to productive transcription elongation (25, 26).
Brd2 and Brd3 associate with acetylated H3K14, H4K5, and H4K12 (29). These marks are found in the transcribed portion of active genes and are thought to contribute to Brd2/3 recruitment in vivo. In vitro, Brd2 and Brd3 facilitate the passage of pol II through nucleosomal templates in a histone acetylation-dependent manner (29). Although a single bromodomain usually interacts with one acK (30), a distinct binding mode was recently described by which the first bromodomain of Brdt binds to two acetylated lysines (H4K5 and H4K8) (31, 32). This expands the way acK marks are interpreted while providing an additional layer of specificity.
A complete understanding of the in vivo function of BET proteins was hindered by the early embryonic lethality of Brd2 and Brd4 nullizygous mice (33, 34). A Brd3 knockout mouse has not been reported. Very recent studies described highly selective compounds that specifically interfere with the function of BET bromodomains by blocking their interaction with acK (35–37). Interest in these compounds is high given their great potential as immunomodulatory drugs (35) and anticancer therapeutics (36).
Here, we report the identification of Brd3 as an acetylated GATA1 interacting partner. An intact acetyl-lysine binding site within BD1 is essential for the physical interaction with acetylated GATA1 and its recruitment to GATA1-occupied sites (GATA1 OSs) in vivo. Genome-wide ChIP followed by massively parallel sequencing (ChIP-seq) analysis revealed that Brd3 is enriched at GATA1 bound enhancers and promoters but not in the transcribed portion of target genes. Dominant negative forms of Brd3 or pharmacologic inhibition of GATA1’s interaction with Brd3 reduce the association of both proteins at key erythroid genes and impair GATA1-mediated erythroid maturation, closely phenocopying the erythroid defects observed with GATA1 acetylation site mutants. In concert, these results reveal that Brd3 serves as reader of acK marks on a key hematopoietic transcription factor to promote its chromatin binding. Moreover, our studies reveal that although compounds targeting BET proteins might be used to manipulate hematopoietic development for exploratory or therapeutic purposes, their inhibitory effects on erythroid maturation highlight the need for further improvement of target specificities.
Results
Brd3 Binds to Acetylated GATA1.
GATA1 bearing mutations at the lysine-rich motifs C-terminal to the zinc fingers fails to stably bind endogenous chromatinized targets in vivo and trigger erythroid maturation (3, 20). However, acetylation-defective GATA1 is functional in transient transfection assays, suggesting that acetylation conveys an activity involving native chromatin. Previously, in vitro acetylation of murine GATA1 peptides coupled with Edman degradation identified four acetylatable lysines at positions 312, 314, 315, and 316 in the C-terminal acetylation motif (Fig. 1A) with K312 being the strongest and K316 the weakest (3). However, this approach did not address whether multiple residues could be acetylated simultaneously. Therefore, we investigated by mass spectrometry the CBP-mediated acetylation of GATA1 using the entire zinc finger domain as substrate. The spectra obtained from trypsin-digested material (Fig. S1) revealed that GATA1 can be multiply acetylated at lysines 312, 314, 315, and 316. In addition, lysine 308 was acetylated, consistent with results obtained at the homologous site in chicken GATA1 (4).
To identify proteins that preferentially interact with acetylated GATA1, we performed peptide affinity purification using nuclear extract from mouse erythroleukemia (MEL) cells. We generated three 24 amino acid peptides corresponding to the C-terminal acetylation motif of GATA1. These were unacetylated (unmod), monoacetylated (ac1), or acetylated at lysines 312, 314, 315, and 316 (ac4) residues that were previously identified as important for GATA1 function (3). The ac4 peptide specifically retained proteins of 110 kDa (Fig. 1B), which were identified by mass spectrometry as Brd3 (22 peptides) and lesser amounts of the short isoform of Brd4 (Brd4s, 6 peptides).
For our studies of endogenous Brd3 and Brd4, we raised antisera and confirmed their specificity in Western blots and immunoprecipitation experiments (see SI Materials and Methods and Fig. S2). The enrichment of Brd3 in the ac4 peptide bound fraction was confirmed by Western blot. In addition, HA-tagged Brd3 expressed in G1E-ER4 erythroid cells specifically bound the ac4 peptide (Fig. 1C). In contrast, although HA-tagged Brd4 bound to the ac4 peptide, it also interacted with unmodified and ac1 peptides (Fig. 1C). Similar binding profiles were observed when using in vitro translated (IVT) Brd3 and Brd4 (Fig. 1C), suggesting that the interaction is direct.
To examine whether the Brd3-GATA1 interaction occurs in vivo, we performed coimmunoprecipitations (co-IP) using nuclear extracts from estradiol-treated G1E-ER4 cells. G1E-ER4 cells are derived from G1E cells, GATA1-null murine progenitors (38, 39) that stably express GATA1 fused to the ligand-binding domain of the estrogen receptor. Estradiol treatment leads to synchronous erythroid differentiation and expression of all known GATA1 target genes. This powerful system enables assaying the function of GATA1 in its natural context. Whole genome expression studies revealed that differentiating G1E-ER4 cells mimic the normal erythroid maturation program with high fidelity (39–42). As shown in Fig. 1D, Brd3 coprecipitated with GATA1, suggesting that GATA1 and Brd3 associate in vivo. FOG1, a known GATA1 binding protein, served as positive control.
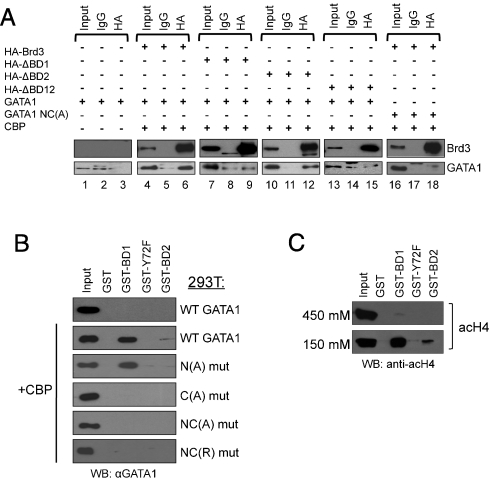
To determine whether GATA1 and Brd3 interact in vivo in an acetylation-dependent manner, we carried out coIP experiments with nuclear extracts from 293T cells transfected with vectors expressing GATA1, the acetyltransferase CBP, and HA-Brd3. GATA1 coprecipitated with wild-type Brd3 (Fig. 2A, lanes 3 and 6), but deletion of BD1(ΔBD1) or both bromodomains (ΔBD12) reduced GATA1 binding to background (IgG) levels (compare lanes 6, 9, 12, and 15). Deletion of BD2 (ΔBD2) had little effect. An acetylation-defective mutant of GATA1 [GATA1 NC(A)] failed to bind Brd3. In summary, the acetylation dependence of the interaction is supported by the requirement for intact acetylation sites in GATA1 and an intact first bromodomain of Brd3.
Fig. 2.
Acetylation-dependent interaction of BD1 with the C-terminal acetylation motif of GATA1. (A) Extracts from 293T cells transfected with indicated plasmids were immunoprecipitated with antibodies against HA or control IgG, followed by Western blotting with HA or GATA1 antibodies. Input equals 2.5%. (B) Immobilized GST-BD1, -BD2, and -Y72F were exposed to nuclear extracts from 293T cells expressing wild-type GATA1 or indicated GATA1 mutants with or without CBP, followed by anti-GATA1 Western blotting. Input equals 2.5%. (C) Immobilized GST-BD1, -BD2, and -Y72F were exposed to bulk chromatin from 293T cells, washed with 150 mM NaCl or 450 mM NaCl, followed by anti-acH4 Western blotting. Input equals 2.5%.
BD1 Binds to the C-Terminal Acetylation Motif of GATA1.
To examine whether BD1 is sufficient for binding full-length, mammalian-cell expressed GATA1, immobilized GST-BD1 was incubated with extracts from 293T cells transfected with GATA1 alone or together with CBP (Fig. 2B). Western blots of bound proteins showed that CBP coexpression was required for GATA1 binding to GST-BD1. We also assayed binding of GATA1 to GST-BD2 as well as GST-BD1 bearing a phenylalanine substitution at a conserved tyrosine residue (Y72) critical for acetyl-lysine binding (43). Neither GST-BD2 nor mutant GST-BD1(Y72F) showed any interaction. Interestingly, GATA2, which retains the N- and C-terminal acetylation motifs, also bound specifically to GST-BD1 when coexpressed with CBP (Fig. S3). Hence, Brd3 may be a widely used partner for acetylated GATA factors in diverse lineages. Additional evidence supporting the acetylation dependence of the GATA1-BD1 interaction derives from the observation that alanine (A) or arginine (R) substitutions at the C-terminal acetylation motif [C(A) mut] or both motifs [NC(A) mut or NC(R) mut] disrupted binding, whereas alanine substitutions at the N-terminal acetylation motif (N(A) mut) bound to GST-BD1 similar to wild-type GATA1 (Fig. 2B).
Because Brd3 has been reported to bind acetylated histone H4 (29), we examined the association of GST-BD1, -BD2, or -BD1(Y72F) with acetylated histone H4 from nuclear extracts. Using an antibody against tetraacetyl H4 (K5, 8, 12, 16) we detected binding to BD1 but not BD1(Y72F) (Fig. 2C). Binding to BD2 was less avid. Because the GATA1-Brd3 binding studies were carried out under stringent washing conditions (450 mM NaCl), we tested acH4 binding under high ionic strength and found BD1 binding to be dramatically diminished (Fig. 2C). These results suggest that acH4, like acGATA1, can be bound by BD1, but that the acH4-BD1 interaction might be less robust. In summary, BD1 binds the C-terminal acetylation motif of GATA1, whereas BD2 likely associates with acetylated lysines present in other transcription factors or histones.
The first bromodomain of Brdt binds to histone H4 when it is acetylated simultaneously at two lysine residues separated by two amino acids (31, 32). Therefore, we surmised that BD1 of Brd3 might employ a similar binding mode in the context of acetylated GATA1. In fact, a GATA1 peptide diacetylated at K312 and K315 bound GST-BD1 almost as avidly as the ac4 peptide, whereas none of the GATA1 peptides with singly acetylated lysines bound with high avidity. This suggests that the binding mechanism of Brd3 BD1 to acetyl-GATA1 is likely similar to that between Brdt and acetyl-H4. However, this does not preclude other possible combinations of acK-BD1 interactions. These data and a detailed structural and biochemical analysis of the interaction between acetylated GATA1 and Brd3 are published elsewhere.
Brd3 and GATA1 Cooccupy Chromatin in Vivo.
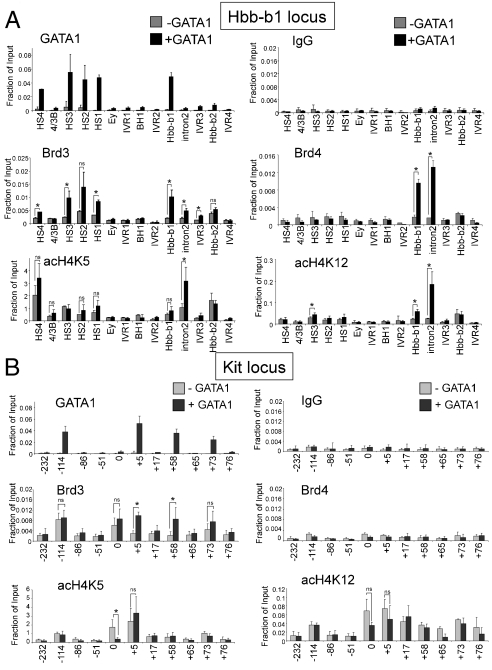
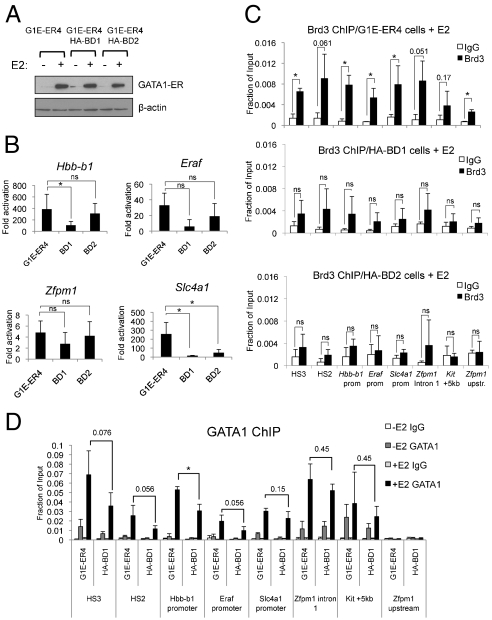
Anti-GATA1 and anti-Brd3 ChIP experiments were carried out in GIE and GIE-ER4 cells following GATA1 activation. At the β-globin locus, GATA1 bound to DNase I hypersensitive sites (HS) 1, 2, 3, and 4 of the locus control region (LCR) and the Hbb-b1 promoter (Fig. 3A), consistent with previous studies (44, 45). Notably, Brd3 occupancy increased at all GATA1 OSs but not at sites lacking GATA1, such as intervening regions (IVR) 1, 2, 3, and 4. Brd3 was also detected at all other examined GATA1 OSs near erythroid genes, including Hba-a1, Klf1, Eraf, Zfpm1, and Slc4a1 (Fig. S4) but not in the transcribed portion of the Eraf gene. Total Brd3 levels did not change significantly upon erythroid differentiation, indicating that recruitment instead of increased production is the cause for Brd3 occupancy (Fig. S5). Although Brd4 bound to acetyl-GATA1 peptides in vitro (Fig. 1) its presence at the β-globin locus correlated poorly with that of GATA1 (Fig. 3A). Significant amounts of Brd4 were observed only at the Hbb-b1 promoter and Hbb-b1 intron 2. At the remaining GATA1-regulated genes, Brd4 was enriched at the Eraf and Zfpm1 genes but not the Hba-a1, Klf1, and Slc4a1 genes (Fig. S4). Because expression of Brd2 and Brdt is low/absent in these cells (39), Brd3 is likely the most relevant GATA1-binding BET protein. The anti-Brd3 and anti-Brd4 ChIP experiments were confirmed by anti-HA ChIP experiments in cell lines stably expressing HA-Brd3 or HA-Brd4 (Fig. S6A).
Fig. 3.
Cooccupancy of Brd3 and GATA1 at active and repressed target genes. (A) ChIP-qPCR with indicated antibodies or control (IgG) in G1E (−GATA1) and estradiol-treated G1E-ER4 (+GATA1) using primers specific for the β-globin locus. G1E-ER4 cells are definitive erythroid cells expressing adult (Hbb-b1) but not embryonic (Ey, bh1) globin genes. (B) ChIP-qPCR as in A using primers specific for the Kit locus. The data shown are the average of 3–6 experiments. Asterisks indicate p < 0.05. ns, not significant. Error bars represent standard deviation.
To examine the generality of the combined presence of Brd3 and GATA1 on cellular chromatin, we performed ChIP in the erythroid cell line MEL before and after DMSO-induced erythroid differentiation. Again, a tight correlation was found between Brd3 and GATA1 at the β-globin locus as well as at the Eraf promoter (Fig. S6B). Brd3 and GATA1 also cooccupied GATA1 target genes in the megakaryocytic cell line L8057 (Fig. S6C), indicating that the Brd3-GATA1 interaction occurs in distinct cell lineages. To confirm our findings in primary cells, ChIP was carried out in E14.5 fetal liver erythroblasts. Brd3 was detected at high levels at four GATA1 OSs (HS2, HS2, and the promoters of Hbb-b1 and Slc4a1) but not a control region 1kb upstream of the Hbb-b1 promoter (Fig. S6D). Taken together, these results demonstrate that GATA1 and Brd3 cooccupy chromatin in vivo.
Brd3 Recruitment Correlates with GATA1 but Poorly with Histone Acetylation.
The activation of GATA1 target genes is associated with increased histone acetylation (15, 16). Because Brd3 has been reported to bind acetylated histones, in particular acH4K5 and acH4K12 (29), its recruitment could be an indirect consequence of GATA1-induced transcription. Therefore, we examined acH4K5 and acH4K12 levels by ChIP at the β-globin locus. In general, both the pattern and dynamics of these histone marks correlated poorly with those of Brd3 at the locus (Fig. 3A). For instance, although Brd3 was high at the Hbb-b1 promoter and low at Hbb-b1 intron 2, the converse was found for acH4K5. Almost opposite patterns of Brd3 and acH4K5 were observed across the LCR (Fig. 3A). Additional results obtained with di-acH3 (K9/14) and tetra-acH4 (K5/8/12/16) antibodies confirmed the relatively poor correlation between histone acetylation and Brd3 occupancy after GATA1 activation. (Fig. S6E). Finally, acH4K5 and acH4K12 also displayed profiles distinct from Brd3 at the Hba-a1, Klf1, Eraf, Zfpm1, and Slc4a1 genes (Fig. S4).
GATA1 functions as a direct repressor of genes associated with the immature, proliferative state, such as the Kit cytokine receptor gene (46, 47). Repression is associated with reduced histone acetylation, thus providing an opportunity to distinguish whether Brd3 recruitment is controlled by acetylated GATA1 or histones. We measured Brd3 and GATA1 occupancy across the Kit locus. GATA1 was detected at sites -114, +5, +58, and +73 kb with respect to the transcription start site, consistent with previous observations (47). Notably, following GATA1 induction, Brd3 occupancy increased at the +5, +58, and +73 sites and remained high at -114 (Fig. 3B). The levels of acH4K5 and acH4K12 tended to decline, albeit not dramatically, following GATA1 activation. Importantly, the overall patterns of these modifications were distinct from that of Brd3. Interestingly, acH4K12 not only marked the distal -114-kb enhancer but also the transcribed portion of the gene that extends up to approximately 80 kb (Fig. 3B) (47). acH4K12 might thus play a role in transcription elongation (48). Additional points deserve comment. First, Brd3 was found at the -114-kb region prior to GATA1 induction. This could be due to the presence of substantial amounts of GATA2 at this site (47). Second, Brd3 was also found at the Kit promoter that lacks high levels of GATA1. This clearly indicates that, not surprisingly, Brd3 can be recruited by additional, GATA1-independent mechanisms. Third, although Brd3 has been implicated in transcription elongation (29) there was a trend toward increased Brd3 in the body of the Kit gene during repression. Fourth, Brd4 was undetectable at the Kit locus even under conditions of high Kit expression. Thus, the presence of Brd4 is neither correlated with GATA1 nor does it appear to be universally associated with active genes. Finally, we also examined Brd3 and Brd4 recruitment at two additional GATA1 repressed genes, Lyl1 (39, 49) and Gata2 (50). Consistent with the results at the Kit locus, Brd3 but not Brd4 was inducibly recruited by GATA1 while acH4K5 and acH4K12 declined (Fig. S6F). In summary, Brd3 recruitment at GATA1 OSs appears to be largely independent of the gene expression state and histone acetylation.
BD1 of Brd3 Is Necessary and Sufficient for Recruitment to GATA1 Targets.
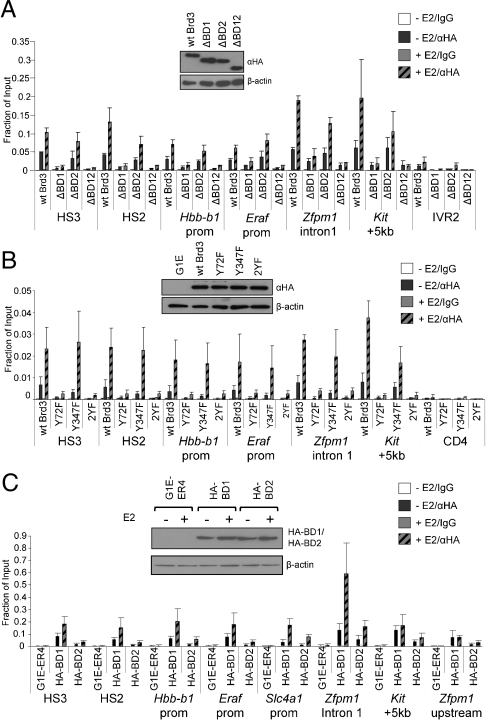
To determine whether GATA1 acetylation is required for Brd3 recruitment in vivo, we performed ChIP of HA-tagged, stably expressed Brd3 proteins with mutations in the bromodomains in G1E-ER4 cells before (−E2) or after activation of GATA1 (+E2). All constructs were expressed comparably (Fig. 4A, Inset). Deletion of BD1 (ΔBD1) or both bromodomains (ΔBD12) almost completely abrogated Brd3 recruitment to all examined GATA1 OSs, including those where GATA1 functions as an activator (HS2, HS3, Zfpm1, Hbb-b1, and Eraf) and repressor (Kit) (Fig. 4A). Importantly, the effects of the Y72F substitution were tantamount to the loss of the entire BD1 (Fig. 4B). In contrast, deletion (ΔBD2) or mutation (Y347F) of BD2 had no effect. Combining both point mutations in Brd3 (2YF) produced the same effects as Y72F or ΔBD1.
Fig. 4.
Association of Brd3 with GATA1 target sites requires acK binding by BD1. (A) Anti-HA ChIP of G1E-ER4 cells expressing HA-Brd3 or versions lacking BD1, BD2, or both bromodomains before or after estradiol (E2) treatment. (B) ChIP as in A in G1E-ER4 cells expressing HA-Brd3 with Y72F and/or Y347F substitutions. (C) BD1 is sufficient for recruitment by GATA1. HA-BD1 or HA-BD2 expressing G1E-ER4 cells were analyzed by anti-HA ChIP before and after treatment with E2. All constructs were expressed at comparable levels (Insets). Negative controls include 1 kb upstream of Hbb-b1 (IVR2), the 5′ transcribed region of CD4, or upstream of the Zfpm1 promoter. The data shown are the average of three to five experiments. All error bars represent standard deviation.
To test whether BD1 is sufficient for recruitment to GATA1 targets, we expressed HA-tagged BD1 or BD2 in G1E-ER4 cells (Fig. 4C). Anti-HA ChIP revealed that HA-BD1 is recruited to GATA1 targets. Interestingly, HA-BD2 was also found at GATA1 OSs, albeit at lower levels. Because BD2 does not bind acetylated GATA1, this association might be mediated by acetylated histones or other nuclear factors in the vicinity of GATA1. Thus, BD1 is the major binding determinant, whereas BD2 might confer some target specificity. Because it is difficult to directly compare the recruitment levels of HA-BD1 and full-length Brd3, these experiments do not rule out a role for other domains of Brd3 in stabilizing the interaction with chromatin. Nevertheless, these results are in agreement with the binding experiments above and strongly suggest that in vivo, acetylation of GATA1 contributes to the normal recruitment of Brd3 via BD1.
Brd3 Occupies GATA1 Targets Genome-Wide.
To measure Brd3 chromatin occupancy in an unbiased fashion, we carried out anti-Brd3 ChIP-seq in G1E cells with or without active GATA1. ChIP material was analyzed by single molecule deep sequencing using the Helicos platform, which yielded approximately 21 × 10e6 and approximately 11 × 10e6 aligned reads each in GATA1 null and GATA1 replete conditions, respectively.
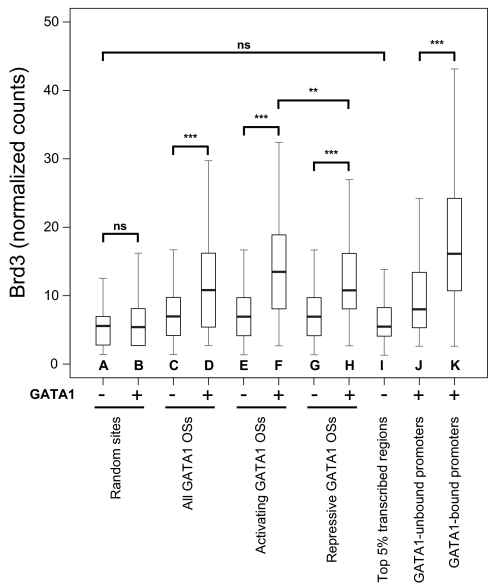
The correlation between GATA1 and Brd3 occupancy was examined by placing the number of raw Brd3 sequencing reads into 500-bp genomic bins and multiplying the number placed into each bin by a constant to account for the differences in total number of sequence reads between the two datasets (with or without GATA1). This enabled us to quantify Brd3 binding at roughly 15,000 GATA1 OSs (41) and compare counts to the same sites before and after GATA1 occupancy. We observed a highly significant increase of Brd3 at GATA1 OSs in induced G1E-ER4 cells when compared to G1E cells (Fig. 5, compare Fig. 5 C and D) or at randomly chosen sites (compare Fig. 5 A and B). We next separated the GATA1 OSs into those that are near active or repressed genes. Because the latter generally display lower histone acetylation levels (15, 47, 50), it allowed us to evaluate the contribution of histone acetylation in Brd3 recruitment. At both activating (Fig. 5 E and F) and repressive (Fig. 5 G and H) GATA1 OSs we found a highly significant increase of Brd3 with a slight, albeit significant, preference for activating sites (median of 13 vs. 11 normalized counts). This supports the results obtained by ChIP-qPCR showing that GATA1 is a critical recruiter of Brd3 regardless of the state of transcription. We stress that although the large majority of GATA1 OSs had elevated read counts for Brd3, a substantial fraction of Brd3-occupied sites were devoid of GATA1, consistent with GATA1-independent modes of recruitment.
Fig. 5.
GATA1 occupancy is a major determinant for Brd3 occupancy genome-wide. The distributions of levels of Brd3 occupancy as determined by genome-wide ChIP in cells with or without active GATA1 are shown as box plots. Black bars denote the median number of normalized read counts, and boxes denote interquartile ranges (IQR). Whiskers demarcate the range of values excluding outliers as conventionally defined by > 1.5 × IQR. Statistical significance of difference was estimated by applying the Mann–Whitney U test on a random subset of data points (n = 400). “ns” indicates not significant; * indicates p < 0.05; ** indicates p < 10-5; *** indicates p < 10-10.
It has been suggested that Brd3 broadly “coats” the transcribed portion of active genes (29). Therefore, we determined the number of Brd3 reads in the center of the transcribed portions of the top 5% most highly expressed genes in the absence of GATA1. Notably, there was no statistically significant enrichment of Brd3 at these regions when compared to random genomic sites (compare Fig. 5 A and I). To further examine the distribution of Brd3 along the bodies of transcribed genes, we generated composite profiles of Brd3 binding along the top 1%, 5%, or 25% expressed genes extending one gene length upstream and downstream in cells with or without active GATA1. Strikingly, Brd3 was enriched around the transcription start site (TSS) but relatively depleted throughout the gene body (Fig. S7).
To rule out the possibility that Brd3 binds near the TSS indirectly, for example via acetylated histones or other transcription factors, we divided active promoters into those bound by GATA1 and those that are not. Brd3 occupancy was markedly higher at the former (compare Fig. 5 J and K). In summary, these data strongly suggest that GATA1, instead of acetylated histones, is a major determinant of Brd3 occupancy on a genome-wide scale. However, this does not rule out a partial contribution of acetylated histones for Brd3 binding to chromatin.
Brd3 Is Required for GATA1 Chromatin Occupancy and Normal Erythroid Maturation.
Several attempts to stably knock down Brd3 in erythroid cells were unsuccessful. Knockdown of Brd3 has previously been reported to impair growth and viability of some cell types (29, 51). Therefore, we aimed to more specifically target the GATA1-dependent functions of Brd3, which might be less detrimental than global loss of Brd3. To interfere with the GATA1-Brd3 interaction in vivo we stably expressed HA-BD1 and HA-BD2 in G1E-ER4 cells (Fig. 4C). HA-BD1, and to a lesser extent HA-BD2, impaired the transcriptional up-regulation of several GATA1 target genes, including Hbb-b1, Eraf, and Slc4a1 (Fig. 6B). Zfpm1 was affected to a lesser extent. Overexpression of either bromodomain did not impact steady-state levels of GATA1-ER (Fig. 6A). HA-BD1 expression reduced Brd3 chromatin occupancy by 2- to 3-fold at GATA1 target sites (Fig. 6C). HA-BD2 expression also reduced the amounts of Brd3 on chromatin, suggesting that BD2-interacting partners contribute to stable Brd3 chromatin association. If acetylation facilitates GATA1 chromatin binding via Brd3, HA-BD1 is expected to reduce chromatin occupancy of GATA1. Indeed, ChIP showed reduced association of GATA1 with its targets in cells expressing HA-BD1 (Fig. 6D). The effects were modest likely because high BD1 expression levels could not be achieved. Because HA-BD1 affected GATA1 chromatin occupancy to a lesser degree than GATA1 target gene expression (Fig. 6B), this suggests additional roles for Brd3 in GATA1-dependent gene expression. BD2 overexpression also led to reduced erythroid maturation. Because BD2 does not strongly interact with acetylated GATA1, this is likely an indirect effect of reduced Brd3 occupancy. As additional control we attempted to express HA-BD1 carrying the Y72F mutation. However, this protein could not be expressed at detectable levels likely as a result of instability.
Fig. 6.
Effects of BD1 or BD2 on GATA1 function. (A) Control Western blot showing that expression of HA-BD1 or HA-BD2 does not affect GATA1-ER levels. (B) qRT-PCR measuring fold induction of indicated GATA1 target genes in G1E-ER4 cells expressing HA-BD1 or HA-BD2 before and after E2 treatment. Data are the average of five independent experiments. (C) Anti-Brd3 ChIP in parental G1E-ER4 cells (Top) or cells expressing HA-BD1 (Middle), HA-BD2 (Bottom). Data are the average of three independent experiments. (D) Anti-GATA1 ChIP comparing GATA1 occupancy in parental G1E-ER4 cells and HA-BD1 expressing cells. Data are the average of three to four independent experiments. Asterisks indicate p < 0.05. ns, not significant. All error bars denote standard deviation.
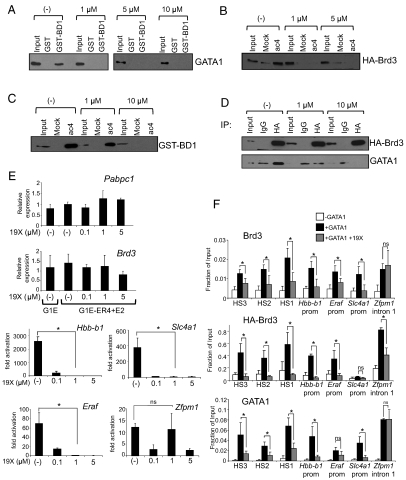
To avoid possible toxicities due to prolonged disruption of Brd3 function, we used the pharmacologic compound GW841819X (hereafter called 19X), which is virtually identical to the recently described compound I-BET in its binding profile and affinity toward BET proteins (35, 37). 19X specifically targets both bromodomains of BET family proteins with affinities ranging from approximately 50 nM to approximately 60 nM, but does not bind to bromodomains from other classes of molecules, including p300/CBP and P/CAF (35, 37). 19X is also extremely similar in structure and function to the compound JQ1, which also targets BET proteins in vitro and whole animals (36). At concentrations ranging between 1 and 10 μM, 19X inhibited binding between full-length GATA1 and GST-BD1 (Fig. 7A), HA-Brd3 and the ac4 GATA1 peptide (Fig. 7B), GST-BD1 and the ac4 GATA1 peptide (Fig. 7C), and full-length GATA1 and Brd3 in co-IP experiments (Fig. 7D).
Fig. 7.
19X disrupts the interaction between Brd3 and acetylated GATA1, blocks erythroid maturation, and impairs GATA1 binding to chromatin. (A) GST-BD1 was exposed to nuclear extracts from 293T cells expressing GATA1 and CBP in the presence of the indicated concentrations of 19X. Bound material was analyzed by anti-GATA1 Western blot. Input equals 2.5%. (B) GATA1 peptides were incubated with nuclear extracts from 293T cells expressing full-length HA-Brd3 in the presence of the indicated concentrations of 19X. Retained material was probed by anti-HA Western blotting. Input equals 2.5%. (C) GATA1 peptide binding assays with GST-BD1 in the presence of varying concentrations of 19X. Bound material was analyzed by Western blotting using GST antibodies. Input equals 4%. (D) Anti-HA immunoprecipitation of nuclear extracts from 293T cells expressing GATA1 and HA-Brd3 in the presence of 19X, followed by Western blotting with anti-GATA1 or anti-HA antibodies. Input equals 2.5%. (E) qRT-PCR measuring fold changes in mRNA levels of GATA1 target genes Hbb-b1, Eraf, Slc4a1, and Zfpm1 (Right) and control genes Pabpc1 and Brd3 (Left) in G1E and estradiol-treated G1E-ER4 cells. Prior to harvesting, cells were grown in the presence of the indicated concentrations of 19X for 24 h. The data shown are the average of two to four independent experiments. (F) ChIP-qPCR analysis of G1E and estradiol-treated G1E-ER4 grown in the presence or absence of 1 μM 19X for 24 h. The data shown are the average of three to four independent experiments. Asterisks indicate p < 0.05. ns, not significant. All error bars denote standard deviation.
We next determined the effects of 19X on erythroid differentiation and GATA1 target gene transcription by exposing G1E-ER4 cells to the compound for 7 and 24 h. At both time points, 19X inhibited in a dose-dependent fashion the expression of Hbb-b1, Eraf, and Slc4a1. However, 19X did not measurably alter Zfpm1 induction by GATA1, nor did it affect the mRNA levels of the housekeeping gene Pabpc1 or Brd3 itself (Fig. 7E and Fig. S8A). Importantly, Brd3 and GATA1 protein levels were unaffected by the presence of 19X (Fig. S8B).
Using ChIP, we examined the chromatin occupancy pattern of Brd3 and GATA1 in G1E-ER4 cells in the presence or absence of 19X. 19X diminished both endogenous Brd3 and HA-tagged Brd3 from positive-acting GATA1 sites, including the Hbb-b1 promoter, HS3, HS2, HS1, Slc4a1, and Eraf (Fig. 7F). Remarkably, the reduction in Brd3 occupancy was accompanied by a substantial reduction of GATA1 at these sites. 19X also disrupted Brd4 binding to the promoters of Hbb-b1 and Eraf, perhaps by inhibiting its interaction with acetylated histones (Fig. S8C). Importantly, although 19X targets Brd2, Brd3, and Brd4, its effects on GATA1 occupancy can be largely attributed to Brd3 because several GATA1 sites (e.g., HS1, HS2, and HS3 of the LCR) are not occupied by Brd4, and Brdt as well as Brd2 are absent or expressed at low levels in G1E cells (39). Thus, Brd3 is the major BET protein that is required for optimal GATA1 chromatin binding in erythroid cells.
19X treatment did not affect all GATA1 targets equally. The induction of Zfpm1 was normal in the presence of the compound, and repression of Kit was only affected moderately or not at all (Fig. 7E and Fig. S8D). This response profile largely reflected the effects of 19X on chromatin occupancy by GATA1 and Brd3 (Fig. 7F and Fig. S8D). Brd4 occupancy was also refractory to 19X at Zfpm1 but not at Hbb-b1 (Fig. S8C). What explains the differential sensitivity to 19X? First, it is possible that the strength of the Brd3-GATA1 interaction varies between sites. Second, it has been reported that genes that are already primed for activity are refractory to I-BET (35). Zfpm1, Kit, and housekeeping genes are expressed at significant levels prior to GATA1 activation and thus are bound by transcription factor complexes and have high levels of activating histone marks, such as acetylation. It is possible that following their initial recruitment, Brd3 and Brd4 are retained by mechanisms unrelated to GATA1 or GATA2. In summary, the use of 19X and dominant interfering versions of Brd3 establish a role for Brd3 in chromatin occupancy of acetylated GATA1, which is critical for execution of the transcriptional program of erythroid maturation.
Discussion
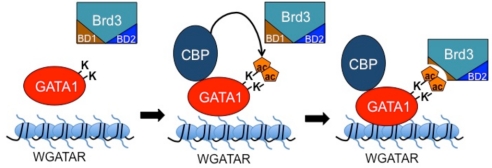
The discovery that nonhistone nuclear factors can be acetylated has revealed a distinct layer of nuclear signaling and has been likened in its impact to phosphorylation (52). Yet our knowledge of the mechanistic consequences is still relatively sparse. Acetylation of GATA1 was described over 10 years ago, but the role of this modification has remained unclear. In prior studies, we found that acetylation of GATA1 is required for its stable association with chromatin in vivo but not purified DNA templates in vitro (20). In search of a mechanism, we found that acetylated lysine residues near the C-terminal zinc finger serve as a docking site for Brd3 and that this interaction is crucial for GATA1’s stable association with chromatin. The specificity of the interaction is underscored by our observation that other bromodomain-containing proteins such as p300 and Brg1 were not bound by acetylated GATA1 peptides. Brd4s was also enriched in the GATA1 peptide affinity purification, which is not surprising given the similarity between their bromodomains. Nevertheless, the in vivo genomic distributions of Brd4 and Brd3 are remarkably distinct, indicating that domains outside BD1 contribute to binding selectivity in vivo. In contrast to Brd4, Brd3 was found by ChIP-qPCR at all examined GATA1 OSs. Likewise, in genome-wide studies GATA1 emerged as a key determinant of Brd3 binding, indicating that Brd3 is the most relevant BET family protein with regard to GATA1 function.
A major unmet challenge has been the identification of the exact acetylated lysine residues of endogenous GATA1 in vivo. However, there is strong evidence supporting the importance of GATA1 acetylation in vivo: First, binding of GATA1 to BD1 is stimulated by CBP. Second, co-IP of GATA1 and Brd3 requires intact GATA1 acetylation sites. Third, the interaction between GATA1 and Brd3, as well as the recruitment of Brd3 by GATA1 in vivo, depends on an intact BD1, specifically the conserved acK-binding residue Y72. Of note, BD1 binding to GATA1 requires at least two acetylated residues, K312 and K315. This is only the second example of a single bromodomain contacting more than one AcK residue in cis (31, 32). However, it is possible that other combinations of diacetylated forms of GATA1 might also be capable of binding to BD1, especially because in vivo, protein–protein interactions are influenced by their molecular surroundings, such as the presence of cofactor complexes. Detailed biochemical and structural studies on this interaction are reported elsewhere.
An open question is the mechanism by which the other major acetylation motif near the N-terminal zinc finger of GATA1 contributes to GATA1 activity (3). The spacing of the acetylated lysines would suggest a different mode of interaction with bromodomain-containing proteins. For example, in the case of Brd4, both bromodomains contribute to the binding of a single acetylated residue of the NFκB subunit RelA (53) and histone H4 (54). Thus, despite the similarities between Brd3 and Brd4, distinct binding mechanisms might govern the selectivity of interactions with acetylated nuclear factors or histones. More generally, it is becoming clear that there exists a significant degree of ligand selectivity among BET family proteins and their bromodomains with regard to the combination and spacing of acetyl-lysine residues (55).
Our genome-wide analysis revealed several important aspects of Brd3 recruitment. First, Brd3 is enriched at GATA1-occupied genomic segments when compared to random sequences, confirming ChIP-qPCR experiments. Second, Brd3 recruitment occurred at GATA1 sites near both active and repressed genes, supporting a model in which acetylated GATA1, but not necessarily histone acetylation, is critical for Brd3 recruitment. This also suggests that Brd3 is versatile in nature, occupying both transcriptionally active and inactive euchromatin. Third, Brd3 is found at sites that are not occupied by GATA1. This agrees with ChIP-qPCR results showing that Brd3 can be found at the Kit promoter, which is devoid of GATA1. A meaningful estimate of the number of Brd3 peaks was not obtainable because the signal-to-noise ratio of the Brd3 ChIP was not as high as that of GATA1 (41). This is likely due to multiple factors, including a broader genomic distribution, the indirect association of Brd3 with DNA, which reduces cross-linking efficiency, and possible differences in antibody affinities. As a result, depending on the threshold settings, the number of called peaks varied significantly. Thus, the exact number of Brd3 occupied sites devoid of GATA1 is not provided here. However, we were able to bypass this obstacle and obtain solid datasets of Brd3 and GATA1 occupancy by assessing the number of Brd3 reads at GATA1-occupied and -depleted sites. It remains to be determined what additional factors and mechanisms specify Brd3 enrichment at GATA1-depleted sites. Fourth, on a genome-wide scale, Brd3 is significantly enriched at promoters favoring those occupied by GATA1 over those devoid of it. This clearly indicates that Brd3 recruitment is not simply the consequence of high-level transcription. Fifth, despite a recent report that Brd3 coats the transcribed portion of active genes (29), our analysis did not reveal a substantial Brd3 enrichment throughout the bodies of highly transcribed genes despite the presence of acetylated histones. Even at the top 5% most highly expressed genes Brd3 enrichment in the middle of the transcribed domain was only minimally elevated over random sequences and much lower when compared to GATA1 occupied sequences. The reasons for this discrepancy include possible differences between cell lines and genes analyzed. In addition, the results from LeRoy et al. (29) are based on the overexpression of Brd3 in 293T cells. We noticed that forced expression of HA-Brd4 produced a broader genomic occupancy pattern when compared to endogenous Brd4, highlighting the importance of appropriate expression levels when examining the distribution of BET proteins.
The biological relevance of Brd3 binding to GATA1 is underscored by experiments in which the interaction has been disrupted by forced expression of BD1 or 19X. Both approaches produced similar results, including impaired erythroid maturation, lack of activation of GATA1 target genes, and reduced chromatin occupancy by GATA1 and Brd3. Although it remains possible that off-target effects involving other BET proteins contributed to the observed phenotypes, we studied genomic sites where Brd3 but not Brd4 is present together with GATA1. This permits the conclusion that at these sites, the BD1- or 19X-induced loss of GATA1 binding to chromatin specifically results from perturbation of Brd3 (expression of Brdt and Brd2 are absent/low in these cells). Because compounds such as 19X and JQ1 bind to both bromodomains of BET proteins, their phenotypic effects should be interpreted with caution. In light of the growing appreciation of the distinct acK-binding modes of the BET family bromodomains, more selective ligands with diminished pleiotropism may be developed. This holds the promise of improved therapeutic compounds for the treatment of diseases in which chromatin-related pathways are perturbed (35–37). Our study underscores the need to further improve the binding site selectivity of pharmacologic compounds to increase their therapeutic benefit and lower the potential for unwanted side effects, such as the potential impairment of erythroid maturation. Structural analysis of the acetyl-GATA1-BD1 interaction will be important in this regard.
Acetylation site mutations or disruption of its interaction with Brd3 diminish GATA1’s ability to associate with chromatin. What is the mechanism by which Brd3 facilitates the binding of GATA1 to chromatinized targets? One highly speculative model invokes the ability of BET proteins to bind to mitotic chromatin (24, 56). This contrasts with most transcription factors and coregulators, which are dispersed into the cytoplasm after mitosis (57, 58). The reassembly of transcription factor complexes at the appropriate time and location upon mitotic exit thus presents a challenge to the newly emerging daughter nuclei. Retention on condensed mitotic chromosomes of “bookmarking” proteins might facilitate this process by directing nuclear factors to the genes they control to stably transmit transcriptional programs through mitosis. For example, Brd4 is required to recruit the transcription elongation factor P-TEFb to sites of transcription in anaphase (59). Brd3 might perform a similar function for GATA1. Loss of Brd3 function would result in diminished chromatin occupancy in stable cell lines with cells continuously dividing and failure of GATA1 to efficiently reassociate with chromatin. This might also explain why in transient transfection assays using episomal plasmids and nondividing cells this function might be dispensable. Future experiments are aimed at testing this model by determining the mitotic occupancy of GATA1 and Brd3. In addition, Brd3 might stabilize chromatin binding of GATA1 by mechanisms unrelated to the cell cycle. Because transcription factors typically nucleate large multiprotein complexes, disruption of any protein interaction could lead to reduced association with chromatin. Indeed, numerous mutations in GATA1 that impair distinct functions are associated with reduced chromatin occupancy (60–62).
Of course, Brd3 might serve additional functions related to transcription and chromatin remodeling as has been proposed for Brd4 (55). In one scenario, BD1 might recruit Brd3 to GATA1 OSs, whereas BD2 or other domains of Brd3 might connect to components of the transcription machinery.
In summary, our work reveals that a pivotal hematopoietic transcription factor involved in tissue specification associates with Brd3 in an acetylation-dependent manner to facilitate stable association with chromatin. The interaction occurs via recognition of multiply acetylated lysine residues, which sets it apart from the great majority of acK-bromodomain interactions. In light of the great therapeutic promise afforded by pharmacologic compounds that target BET proteins (35, 36), this work together with future studies on the structural and biochemical features of this type of interaction might aid in the production of more selective compounds with more targeted therapeutic effects.
Experimental Procedures
Protein Binding Studies.
Details on the conditions for all binding studies can be found in SI Text.
Chromatin Immunoprecipitation.
ChIP was performed as described (15). Primer sequences are listed in SI Text.
ChIP-Seq Analysis.
Anti-Brd3 ChIP material from G1E or induced G1E-ER4 cells was sequenced on a HeliScope Single Molecule Sequencer (Helicos). Details of the analysis are described in SI Text.
qRT-PCR.
RNA was extracted with Trizol reagent (Invitrogen). Reverse transcription reactions were performed using Superscript II (Invitrogen). Results were quantified by real-time PCR on an ABI Prism 7900 system using SYBR Green dye.
Supplemental Data
Supplemental experimental procedures and eight figures are available online.
Supplementary Material
Acknowledgments.
We thank Drs. David Steger and Mitch Lazar for helpful suggestions. We are grateful to Ross Tomaino at the Taplin Mass Spectrometry facility at Harvard for help in the data interpretation. We thank Rena Zheng, Nancy L. Maas, Rachel Kadzik, and Thomas Kadauke for technical assistance. This work was supported by National Institutes of Health (NIH) Grant DK054937 (to G.A.B.), NIH training grants T32 HL007971-07 (to J.M.L.) and T32 DK07780 (to A.E.C.), and American Heart Association training Grant 09PRE2300139 (to S.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 8927.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102140108/-/DCSupplemental.
References
- 1.Zeng L, Zhou MM. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 2.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migliaccio AR, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiyama C, et al. GATA-1 is required for expression of FcϵRI on mast cells: Analysis of mast cells derived from GATA-1 knockdown mouse bone marrow. Int Immunol. 2005;17:847–856. doi: 10.1093/intimm/dxh278. [DOI] [PubMed] [Google Scholar]
- 10.Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 11.Weiss MJ, Orkin SH. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols KE, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurbuxani S, Vyas P, Crispino JD. Recent insights into the mechanisms of myeloid leukemogenesis in Down syndrome. Blood. 2004;103:399–406. doi: 10.1182/blood-2003-05-1556. [DOI] [PubMed] [Google Scholar]
- 14.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg EC, et al. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong W, et al. Inhibition of CBP-mediated protein acetylation by the Ets family oncoprotein PU.1. Mol Cell Biol. 2002;22:3729–3743. doi: 10.1128/MCB.22.11.3729-3743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- 19.Choi Y, Elagib KE, Delehanty LL, Goldfarb AN. Erythroid inhibition by the leukemic fusion AML1-ETO is associated with impaired acetylation of the major erythroid transcription factor GATA-1. Cancer Res. 2006;66:2990–2996. doi: 10.1158/0008-5472.CAN-05-2944. [DOI] [PubMed] [Google Scholar]
- 20.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishikawa K, et al. Self-association of Gata1 enhances transcriptional activity in vivo in zebra fish embryos. Mol Cell Biol. 2003;23:8295–8305. doi: 10.1128/MCB.23.22.8295-8305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Hernandez A, et al. Acetylation and MAPK phosphorylation cooperate to regulate the degradation of active GATA-1. EMBO J. 2006;25:3264–3274. doi: 10.1038/sj.emboj.7601228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Thonel A, et al. HSP27 controls GATA-1 protein level during erythroid cell differentiation. Blood. 2010;116:85–96. doi: 10.1182/blood-2009-09-241778. [DOI] [PubMed] [Google Scholar]
- 24.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 28.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 29.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 31.Moriniere J, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci USA. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung C, et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem. 2011 doi: 10.1021/jm200108t. in press. [DOI] [PubMed] [Google Scholar]
- 38.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welch JJ, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Primary sequence and epigenetic determinants of in vivo occupancy of genomic DNA by GATA1. Nucleic Acids Res. 2009;37:7024–7038. doi: 10.1093/nar/gkp747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanno T, et al. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 44.Im H, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson KD, et al. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munugalavadla V, et al. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jing H, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson KD, et al. Friend of GATA-1-independent transcriptional repression: A novel mode of GATA-1 function. Blood. 2007;109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grass JA, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishii H, Mimori K, Mori M, Vecchione A. Differentially expressed genes in endothelial differentiation. DNA Cell Biol. 2005;24:432–437. doi: 10.1089/dna.2005.24.432. [DOI] [PubMed] [Google Scholar]
- 52.Kouzarides T. Acetylation: A regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, et al. Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry. 2008;47:6403–6417. doi: 10.1021/bi8001659. [DOI] [PubMed] [Google Scholar]
- 55.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 56.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 58.Delcuve GP, He S, Davie JR. Mitotic partitioning of transcription factors. J Cell Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci USA. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pal S, et al. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee HY, et al. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol Cell. 2009;36:984–995. doi: 10.1016/j.molcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]