Abstract
Several yeast and mammalian peroxisomal membrane proteins (PMPs) are delivered to peroxisomes via the endoplasmic reticulum (ER). Fluorescence microscopy showed a focused assembly of PMPs in a specialized domain of the ER, referred to as the preperoxisomal ER. It is proposed that preperoxisomal vesicles containing PMPs bud from this domain to either fuse with preexisting peroxisomes or to mature into functional peroxisomes by uptake of peroxisomal membrane and matrix proteins. However, such vesicular entities are not identified nor are the biochemical requirements for the budding process known. We developed an in vitro cell-free ER-budding assay using Pichia pastoris and followed two endogenous PMPs, Pex11p and Pex3p during their ER exit. Both the PMPs were copackaged in the ER-budded vesicles that float on a Nycodenz gradient. PMP budding from the ER was dependent on ATP, temperature, cytosol, and Pex19p and generated preperoxisomal vesicles with an incomplete complement of PMPs. Surprisingly, Pex11p budding was independent of Pex3p; however, the budded vesicles were devoid of most of the PMPs otherwise present in the wild-type vesicles and might represent peroxisomal remnants. Our findings provide a biochemical platform to uncover the mechanism of PMP budding from the ER.
Keywords: biogenesis, endoplasmic reticulum-to-peroxisome trafficking
Peroxisomes are ubiquitous organelles present in almost all eukaryotic cell types and house oxidative enzymes involved in lipid metabolic pathways. Although great advances have been made in the biogenesis of peroxisomal matrix proteins, the origin of PMPs has been the subject of much debate. Earlier it had been suggested that peroxisomes grow in size by the sequential uptake of membrane and matrix proteins and then divide to produce new peroxisomes (1, 2). In yeast, at the time of cell division, some peroxisomes are transferred to the growing bud, whereas others are retained in the mother cell (3). These observations led to the growth and division model wherein preexisting peroxisomes and their division maintain the peroxisome number (4, 5). However, in certain pex mutants where peroxisome biogenesis is completely blocked, new peroxisomes appear upon reintroduction of the missing genes (5–12). A similar observation was made in yeast cells lacking peroxisomes as a result of an inheritance defect (5, 13). To account for these results, which are incompatible with the growth and division model, a de novo pathway for peroxisome biogenesis was postulated.
Several studies suggest that peroxisomes could be derived de novo from the endoplasmic reticulum (ER). This assumption was based on the following observations: (i) the presence of certain phospholipids in the peroxisomal membrane, which are exclusively synthesized on the ER (14) and (ii) electron microscopy data indicating close proximity between peroxisomes and the ER lamellae (15, 16). Further evidence came from pulse-chase experiments demonstrating that Yarrowia lipolytica PMPs, such as Pex2p and Pex16p, undergo core N-glycosylation in the ER before appearing on peroxisomes (17). Similarly, in mammalian cells, Pex13p was localized in subdomains of the ER (16). More recent in vivo studies in yeast demonstrated more than 16 fluorescently labeled PMPs, including Pex3p, trafficking to peroxisomes via the ER (6, 9, 12, 18). These data suggest that PMPs and peroxisomes can indeed be derived de novo from the ER.
The involvement of the ER in the secretory pathway is well characterized. Secretory proteins are processed in the ER lumen, packaged into a vesicular compartment, and transported to the Golgi apparatus (requiring the COPI and COPII families of proteins) for further processing and targeting to membranes of the secretory compartments, or for eventual secretion. A similar pathway could be hypothesized for the transport of PMPs from the ER to peroxisomes; however, the proteins necessary for assembly of COPI and COPII vesicles were unnecessary for peroxisome biogenesis (19, 20), suggesting the requirement of distinct components for ER-to-peroxisome trafficking. Previously, immature preperoxisomal vesicles, containing partially overlapping sets of peroxins and distinct from the mature peroxisomes, were defined in Y. lipolytica (21). Pex19p might be one of these components required for ER-to-peroxisome protein trafficking because in vivo studies suggest its essential role in the exit of PMPs from the ER (6, 18).
To unravel the mechanism by which PMPs exit from the ER and to elucidate the biochemical requirements for the budding process, we dissected the early events of PMP biogenesis from the ER. We followed the trafficking of two endogenous PMPs, an HA-tagged Pex11p and a GFP-tagged Pex3p, as markers for vesicular carriers emerging from the ER using a cell-free in vitro ER-budding assay. We identified a vesicular carrier for the trafficking of these PMPs emerging from the ER and the biochemical requirements for the budding of these carriers. We show that both these PMPs are incorporated selectively into preperoxisomal vesicles in an ATP-, temperature-, cytosol-, and Pex19p-dependent manner.
Results
Pex11p and Pex3p Are Mislocalized in Δpex19 Cells.
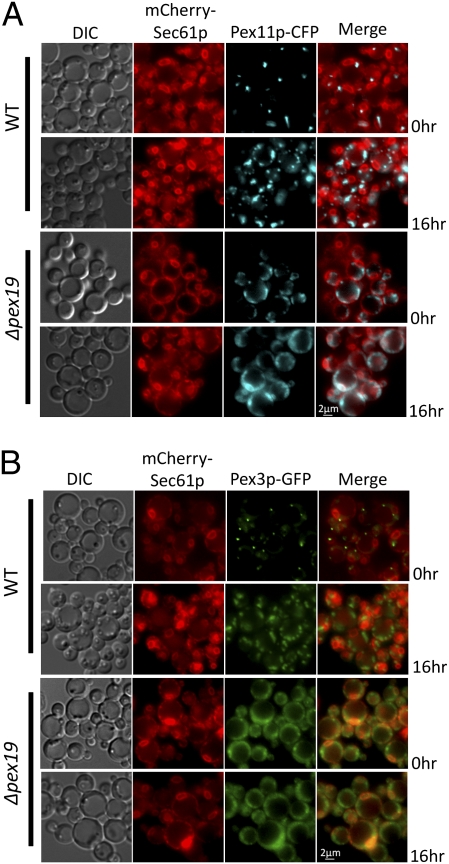
Several PMPs transit via the ER en route to peroxisomes (9). To understand ER-to-peroxisome trafficking of endogenous PMPs, Pex11p and Pex3p were used as markers because their transit via the ER was previously established (9, 22). CFP-tagged Pex11p and GFP-tagged Pex3p were expressed and colocalized with mCherry–Sec61p in WT and Δpex19 cells. For cells just shifted to oleate medium, at 0 h, Pex11p–CFP and Pex3p–GFP were localized in a single dot per cell, partially colocalized with mCherry–Sec61p in punctate structures at the cell cortex, perhaps representing a subdomain of the ER in the WT cells. When cells were transferred to oleate medium for 16 h, Pex11p–CFP and Pex3p–GFP were localized to typical punctate clusters representing mature peroxisomes, well segregated from the mCherry–Sec61p (Fig. 1 A and B). In contrast, in Δpex19 cells, Pex11p–CFP and Pex3p–GFP were mislocalized to punctate structures close to the mCherry–Sec61p-labeled peripheral ER, even after shifting the cells to oleate medium for 16 h (Fig. 1 A and B). Interestingly, when Pex19p was reintroduced in these cells, Pex11p–CFP was relocalized to the newly formed peroxisomes within 3 h (Fig. S1A). These observations suggest that Pex11p and Pex3p might transit the ER en route to the peroxisomes and that in the absence of Pex19p, both Pex11p and Pex3p are mislocalized partially with Sec61p near the cell periphery.
Fig. 1.
Pex11p localization in WT and Δpex19 cells. Fluorescence microscopy analysis of oleate-grown WT and Δpex19 cells coexpressing the relevant proteins from PGAP–PEX11–CFP or PPEX3–PEX3–GFP and PSEC61–mCherry–SEC61. Cells were grown on YPD and switched during exponential phase to oleate medium for 0 or 16 h. mCherry–Sec61p (ER marker) localizes to punctate structures at the peripheral and nuclear ER. (A) In WT cells, Pex11p–CFP or Pex3p–GFP was partially localized with the Sec61p-labeled ER (0 h) and subsequently were found on the mature peroxisome cluster (16 h in oleate medium). (B) However, in Δpex19 cells Pex11p–CFP or Pex3p–GFP was mislocalized near the cell periphery partially associated with mCherry–Sec61p in the peripheral ER. No nonlinear adjustments or changes to gamma settings were made in the images.
Pex11p and Pex3p Traffic from the ER to a Vesicular Carrier.
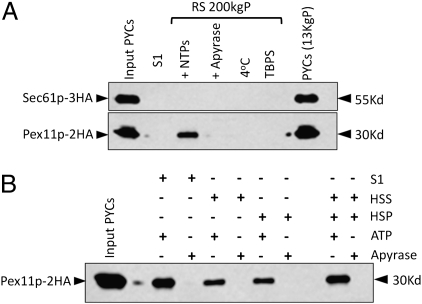
We modified the classical cell-free in vitro ER-budding assay to identify the vesicular carriers delivering PMPs to the peroxisomes. The assay was originally developed to identify the components of the secretory pathway involved in ER to Golgi trafficking (23, 24). Pex11p served as a marker for peroxisome-specific vesicular traffic emanating from the ER. For all of the biochemical assays, Pex11p was expressed as a HA-tagged fusion protein (Pex11p–2HA) and, in certain experiments, was coexpressed with Sec61p that was tagged genomically with 3HA (Sec61p–3HA). The in vitro ER-budding assay has three major components: a donor membrane compartment, soluble budding factors, and an ATP-regenerating system. The donor compartment was provided by permeabilized yeast cells (PYCs), prepared from the cells that were induced for peroxisome biogenesis. The S1 fraction (crude cytosol), which is added exogenously to the PYCs, supplies soluble factors necessary for budding in the presence of an ATP-regenerating system. Apyrase was added in place of an ATP-regenerating system as a control. The reactions were incubated at 20 °C for 90 min. The donor PYCs were separated from the released vesicles by a brief centrifugation step, the supernatant was removed and spun at 200,000 × g, and the pellet fraction was analyzed by SDS/PAGE and immunoblotting for the presence of Pex11p–2HA. We detected Pex11p–2HA in the 200,000 × g pellet in the reaction when WT cytosol was used with an ATP-regenerating system. However, in the control with apyrase, the Pex11p–2HA signal was dramatically decreased (Fig. 2A). In other controls, where cytosol was substituted by TBPS buffer, or when the reaction was incubated at 4 °C, the signal was abolished.
Fig. 2.
Cell-free in vitro assay for Pex11p–2HA budding from the ER. (A) PYCs prepared from WT PPY12 cells expressing Pex11p–2HA and Sec61p–3HA were incubated with the WT S1 fraction for 90 min at 20 °C (lanes 3, 4, and 6) or 4 °C (lane 5) in the presence of an ATP-regenerating system (lanes 3, 5, and 6) or apyrase (lane 4). At the end of the budding reaction, samples were centrifuged at 13,000 rpm for 1 min to separate supernatants from the PYC pellet (lane 7), which still had the majority of the Pex11p–2HA and Sec61p–3HA. The reaction supernatant (RS) was centrifuged again at 200,000 × g and the pellet (RS 200 KgP) was resuspended in TBPS and analyzed here. PYCs in lane 1 represents nearly 3% load of the starting PYCs. (B) Soluble cytoplasmic (HSS) and the high-speed pelletable (HSP) membrane fractions obtained after further fractionating S1 were compared independently and together for their potential to support budding of Pex11p–2HA from the ER.
Vesicle budding from the ER is expected to be selective, excluding ER-resident proteins from the vesicles. Therefore, if the Pex11p–2HA-containing budded vesicles are real transport vesicles, then the yeast ER membrane protein, Sec61p, should be retained in donor cells during vesicle formation. We observed that Sec61p resided in the pelleted donor PYC and no detectable amounts were released during the assay (Fig. 2A).
When the S1 fraction was further subfractionated into a high-speed supernatant (HSS) and pellet (HSP), the ER budding of Pex11p–2HA vesicles was reduced about twofold with each fraction alone, but could be restored when HSS and HSP were added together (Fig. 2B), indicating that the components from both fractions are required for optimal budding of Pex11p–2HA vesicles.
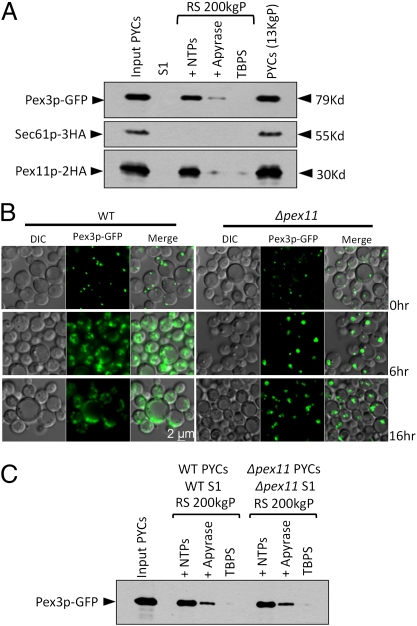
Previous studies have demonstrated the contribution of the ER to peroxisome biogenesis by investigating the trafficking of Pex3p in yeast (6, 12) and mammalian cells (25). We performed similar ER-budding experiments with a GFP-tagged Pex3p, coexpressed with Pex11p–2HA and Sec61p–3HA. Pex3p–GFP was detected with Pex11p–2HA, but not Sec61p–3HA, in the supernatant fraction (Fig. 3A). The budding was inhibited with apyrase and abolished when cytosol was replaced with TBPS. These observations indicate that our system recapitulates selective Pex11p and Pex3p budding from the ER, while excluding an ER-localized protein.
Fig. 3.
Division of preexisting peroxisomes is not the primary source of budded vesicles. (A) ER-budding assay was performed with WT cells coexpressing Pex11p–2HA, Sec61p–3HA, and Pex3p–GFP. Lane 1 represents nearly 3% load of the starting PYCs. (B) Fluorescence microscopy analysis of oleate-grown WT cells and Δpex11 cells expressing Pex3p–GFP. Cells were grown on YPD and switched during exponential phase to oleate medium. In WT cells, Pex3p–GFP labeled proliferating peroxisomes forming numerous punctate structures. The Δpex11 cells showed a severe block in peroxisome division even after growth for 16 h on oleate medium, resulting in bigger peroxisomes compared with those in WT cells. (C) ER-budding assay performed with WT cells and Δpex11 cells expressing Pex3p–GFP. Lane 1 represents nearly 3% load of the starting PYCs.
Preperoxisomal Vesicle Budding Is Independent of Division of Preexisting Peroxisomes.
To exclude the possibility that the preperoxisomal vesicles bud from the preexisting peroxisomes, we performed the budding assay with a Δpex11 mutant in which peroxisome division is blocked. As observed using fluorescence microscopy, peroxisomes labeled with Pex3p–GFP in Δpex11 cells grew bigger in size compared with the WT cells with time, but as expected, exhibited a severe block in division (Fig. 3B). However, the budding assay showed normal budding of Pex3p–GFP vesicles, suggesting that preexisting peroxisomes are not the source of these vesicles (Fig. 3C).
ER Exit of Pex11p–2HA Requires Pex19p but Not Pex3p.
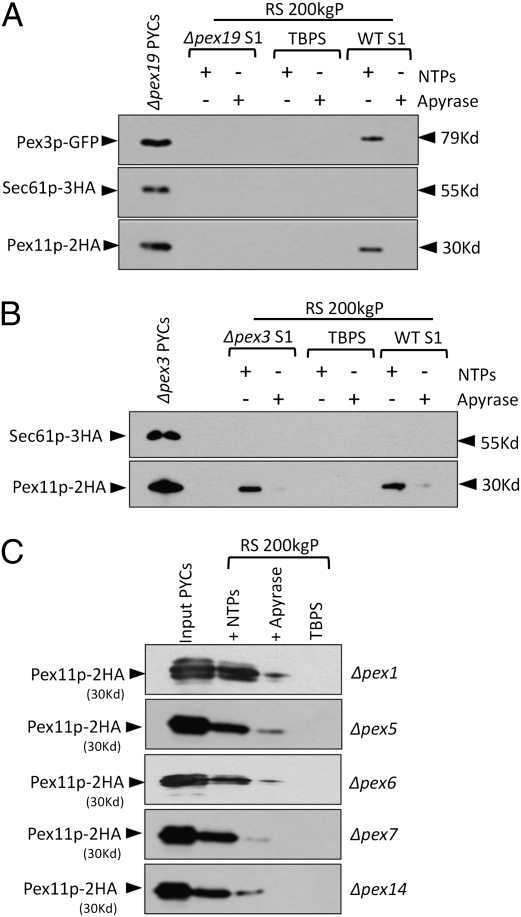
Recent in vivo studies indicate that Pex19p plays a critical role in the ER exit of PMPs as judged by their ER accumulation in Δpex19 cells. Moreover, the induction of PEX19 restored peroxisomes in Δpex19 cells (6, 18, 26, 27). To directly test the requirement of Pex19p in the ER exit of Pex11p–2HA and Pex3p–GFP in the in vitro ER-budding assay, we expressed these proteins in Δpex19 cells. Because peroxisome biogenesis was blocked in these mutants, we found the expression of Pex11p–2HA was notably low, thus to obtain comparable signals, we pooled and analyzed the supernatant of five ER-budding reactions. Interestingly, with Δpex19 cells we did not detect either Pex11p–2HA or Pex3p–GFP in the supernatant of the budding reaction when Δpex19 cytosol was used (Fig. 4A). However, the budding of Pex11p–2HA and Pex3p–GFP was restored when WT cytosol (S1) was added to the budding reaction (Fig. 4A). Because Pex19p is a predominantly cytosolic protein (28), it is very likely that the WT cytosol supplied Pex19p to restore budding. These observations provide evidence for the role of Pex19p in exit of PMPs from the ER.
Fig. 4.
Pex19p is required for the budding of peroxisomal vesicles from the ER. (A) ER-budding assay was performed with Δpex19 cells coexpressing Pex11p–2HA, Sec61p–3HA, and Pex3p–GFP with either Δpex19 or WT cytosol. The expression of Pex11p–2HA was markedly low in Δpex19 and Δpex3 cells, so RS fractions of five budding reactions were pooled and analyzed together to obtain comparable levels of the protein. Neither Pex11p–2HA nor Pex3p–GFP was detected in the RS 200 KgP when Δpex19 cytosol was used. The budding of Pex11p–2HA was restored with WT cytosol (WT S1) in an ATP-dependent manner. Likewise, Pex3p–GFP budding too was restored with the WT cytosol (WT S1). PYCs in lane 1 represent nearly 3% load of the starting PYCs. (B) A similar ER-budding assay was performed with Δpex3 cells coexpressing Pex11p–2HA and Sec61p–3HA with Δpex3 or WT cytosol. Pex11p–2HA was detected in the RS 200-KgP fraction, indicating that Pex3p is not required for budding of preperoxisomal vesicles. Further addition of WT cytosol did not increase the budding of Pex11p–2HA vesicles. (C) The ER-budding assay was performed with Δpex1, Δpex5, Δpex6, Δpex7, and Δpex14 cells expressing Pex11p–2HA with their respective cytosols. Pex11p–2HA was detected in the RS 200 KgP in all of the mutants.
We also addressed the requirement of coat protein complex II (COPII), an indispensable component for the budding of ER-derived vesicles en route to the Golgi. In a similar budding assay, we selectively blocked COPII-mediated budding using a dominant negative mutant form Sar1p(T34N) of the Sar1p GTPase, which blocks COPII vesicle formation (29). As a control, we confirmed the phenotype of the Sar1p(T34N) mutant in blocking maturation of the vacuolar protein, carboxypeptidase Y (CPY), as well as pexophagy (Fig. S2 A and B). First, the budding of Pex11p–2HA was unaffected in cells expressing Sar1p(T34N) (Fig. S2C). Second, when coexpressed with Sar1p(T34N), the peroxisomal localization of Pex11p–CFP was unaffected (Fig. S3). Additionally, when Pex19p expression was induced in Δpex19 cells, the relocalization of Pex11p–CFP to the newly formed peroxisomes remained unaffected when Sar1p(T34N) was coexpressed (Fig. S1B). These observations suggest distinct COPII-independent requirements for the budding of Pex11p–2HA vesicles.
In view of data showing that Pex3p is the docking protein necessary to anchor cytosolic Pex19p on the peroxisome membrane (11), we evaluated the requirement of Pex3p in the budding of Pex11p–2HA. Interestingly, Δpex3 cytosol supported the budding of Pex11p–2HA vesicles from Δpex3 cells (Fig. 4B). Moreover, the addition of the WT cytosol did not increase the budding of Pex11p–2HA from Δpex3 cells (Fig. 4B). As described in the next section, Pex11p–2HA vesicles budded from the Δpex3 cells were affinity captured with an HA affinity matrix and analyzed for the presence of various membrane and matrix peroxins. Supporting the requirement of Pex3p and Pex19p for the recruitment of PMPs to the peroxisomal membrane, the Δpex3-budded vesicles were devoid of most of the membrane peroxins compared with the vesicles purified from the WT cells (shown later in Fig. 5B).
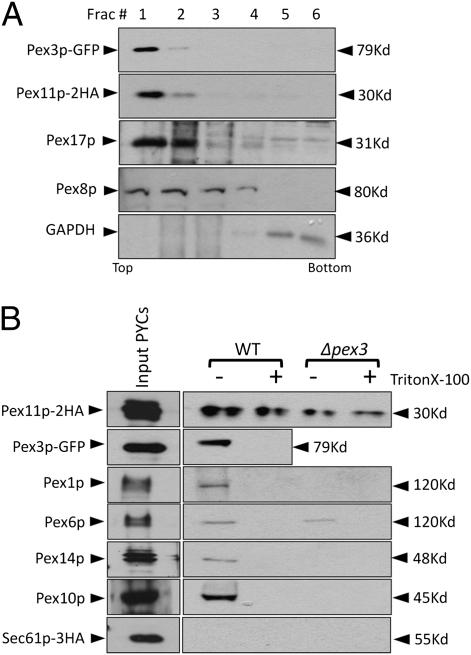
Fig. 5.
Pex3p and Pex11p budded from PYCs are membrane associated and are copackaged in the same vesicles. (A) The supernatant (RS) of the in vitro budding reaction was analyzed using a flotation gradient as described in Materials and Methods. Fractions (50 μL each) were collected from the top and analyzed by immunoblotting. Both the peroxins were found associated with buoyant membranes. (B) The RS fractions of 10 in vitro budding assays performed with WT and Δpex3 PYCs were pooled (900 μL) and spun at 200,000 × g. The pellet was resuspended in TBPS with or without Triton X-100 (1% vol/vol) and incubated with 80 μL of HA-affinity matrix to capture Pex11p–2HA-associated membranes. The incubation was for 6 h at 4 °C. The beads were washed three times, resuspended in 100 μL of TBPS, and analyzed.
We also analyzed other mutants defective in cytosolic and peroxisomal membrane peroxins for their effect on ER budding of Pex11p–2HA. All of the analyzed mutants (Δpex1, Δpex5, Δpex6, Δpex7, and Δpex14) affecting components of the peroxisomal matrix protein import pathways, were proficient in budding of Pex11p–2HA vesicles (Fig. 4C).
Pex11p–2HA and Pex3p–GFP Are Associated with Membranous Vesicles.
We performed flotation gradient centrifugation to ensure the membrane association of Pex11p–2HA and Pex3p–GFP. The supernatant of the budding reaction was mixed with Nycodenz to a final concentration of 35% (wt/vol) and layered at the bottom of a Nycodenz flotation step gradient with decreasing densities of Nycodenz layered on top. The membranes were floated to light density by ultracentrifugation at 50,000 rpm for 2 h at 4 °C. Both Pex11p–2HA and Pex3p–GFP were at the top of the gradient, colocalized with peroxisomal membrane (Pex17p) and matrix (Pex8p) markers and well resolved from the cytosolic marker, GAPDH, present in the bottom fractions (Fig. 5A). This observation, together with the fact that these PMPs are in the supernatant fraction after pelleting PYCs, indicates that the PMPs are in membranous vesicles.
Pex3p and Pex11p Are Captured in the Same Vesicles.
It has been shown that nonidentical populations of preperoxisomal vesicles fuse to form mature peroxisomes (30). Are Pex3p and Pex11p packaged in the same budded vesicles or in different vesicles that fuse after budding? We addressed this question by performing an affinity capture of Pex11p–2HA in presence or absence of Triton X-100 with HA affinity matrix and the beads were analyzed after thorough washing. We detected Pex11p–2HA in the presence as well as absence of Triton X-100, whereas Pex3p–GFP was coeluted only in the absence of Triton X-100 (Fig. 5B, lanes 1 and 2). These results suggest that Pex3p–GFP and Pex11p–2HA are assembled in the same vesicles. We also observed various other PMPs in the affinity captured vesicles including Pex1p, Pex6p, Pex10p, and Pex14p but were unable to detect several other matrix and membrane proteins including Pex2p, Pex5p, Pex8p, Pex12p, Pex13p, Pex17p, thiolase, and catalase. In contrast, the Pex11p–2HA vesicles budded from Δpex3 cells, further cementing our earlier conclusion that the vesicles could not be of peroxisomal origin, but they lacked all peroxins tested, except a small amount of Pex6p.
Discussion
Several PMPs, tracked using microscopic visualization, transit via the ER en route to peroxisomes (6, 9, 12, 18). However, a vesicular carrier for the trafficking of these proteins emerging from the ER has not been identified, nor have the biochemical requirements for the budding of such vesicular carriers been defined. Understanding these early events of peroxisome biogenesis is critical to clarify the origin and fate of preperoxisomal vesicles and will provide a deeper understanding of the molecular mechanisms involved in peroxisome assembly and maintenance.
We modified the classical cell-free in vitro ER-budding assay to identify the vesicular carriers delivering PMPs to the peroxisomes. We tracked Pex11p–2HA as a marker for the vesicular carriers because it is known to be the most abundant endogenous PMP (31). Additionally, recent reports in Saccharomyces cerevisiae indicate that Pex11p might be channeled from the ER to peroxisomes, although concrete evidence is still lacking, because the translocation of an ER-localized pool of Pex11p to peroxisomes was not shown conclusively (9, 22). Pex11p–2HA was expressed from its endogenous promoter to avoid overexpression-associated artifacts as observed with the newly synthesized Pex15p that accumulates in the ER when overexpressed (32). Upon incubating the freshly prepared permeabilized cells with crude cytosol and an ATP-regenerating system, Pex11p was present in the supernatant fraction of the budding reaction that typically contains ER-derived vesicles. We also found Pex3p–GFP, an essential PMP well characterized for its peroxisomal trafficking via the ER (6), incorporated in the budded vesicles, as a further validation of the assay. Furthermore, Sec61p, an integral ER marker was not detected in the supernatant, indicating a selective translocation of peroxins to the budded vesicles. The budding reaction was cytosol, temperature, and ATP dependent.
We ruled the possibility that the division of preexisting peroxisomes might be the primary source of budded vesicles by carrying out a similar budding assay with a Δpex11 mutant incapable of peroxisome division. The budding of Pex3p–GFP vesicles was found to be unaffected, indicating that the division of preexisting peroxisomes did not contribute to budded vesicles (Fig. 3C). Furthermore, the budding of Pex11p–2HA vesicles even in Δpex3 cells (Fig. 5B), known to lack peroxisomes, shows that the source of membranes for the vesicles is likely to be the ER, rather than peroxisomes.
Our results show that Pex3p–GFP and Pex11p–2HA were copackaged in the same vesicles, suggesting a common carrier that traffics these PMPs from the ER to the peroxisomes. Furthermore, the vesicles obtained in our reactions are preperoxisomal vesicles in that they have not yet acquired their full complement of peroxisomal membrane and matrix contents (Fig. 5B).
Previous studies in S. cerevisiae showed the requirement of Pex19p for the exit of PMPs including Pex3p from the ER (6, 18). In our in vitro budding assay, we observed a complete block in ER budding in the absence of Pex19p. Interestingly, the block was removed when cytosol containing Pex19p was added to reaction mixture from Δpex19 cells. These observations provide evidence for the role of Pex19p in exit of PMPs from the ER, in agreement with the in vivo studies where PMPs were found to accumulate in the ER in Δpex19 cells. Previously, we have argued that in Pichia pastoris, Pex19p is required for some step in peroxisome biogenesis after PMP insertion into membranes (33). The demonstration here of the requirement of Pex19p for the budding of PMPs, inserted into the ER even in the absence of Pex19p, shows clearly that this function of Pex19p is in facilitating the budding of ER-derived preperoxisomal vesicles.
Surprisingly, Δpex3 cells retained budding of Pex11p–2HA vesicles (Fig. 4B). It is an interesting result because Pex3p is believed to be the docking factor for Pex19p on membranes (11), but these observations suggest a Pex3p-independent role of Pex19p in facilitating the budding of PMP vesicles from the ER. Further, it was found that the vesicles budded from the Δpex3 cells lack most of the PMPs otherwise present on the WT vesicles (Fig. 5B). We speculate that the Pex11p–2HA vesicles budded from the Δpex3 cells might represent the peroxisomal remnants (“ghosts”) as often observed in the Δpex3 mutant (34). One possibility, worth exploring in the future, is whether the interaction of Pex19p with proteins other than Pex3p in the ER is sufficient to drive budding of vesicles in a Pex3p-independent fashion.
In summary, the availability of a cytosol-, temperature-, ATP-, and Pex19-dependent, but Pex3p-independent system for the selective budding of multiple PMPs into preperoxisomal vesicles represents a major advance in the field, making the system amenable to further fractionation and mechanistic studies.
While this manuscript was being assembled, the Schekman laboratory reported a similar assay, using S. cerevisiae, for the ATP and Pex19p-dependent budding of a different PMP, Pex15p, from the ER (35). We are pleased to note that the major results from the two laboratories are in significant agreement.
Materials and Methods
Yeast Strains and Growth Conditions.
Yeast used for the preparation of permeabilized cells and S1 fractions were grown at 30 °C on a shaker set at 250 rpm, in YPD medium (1% yeast extract, 2% peptone, 2% glucose) to an OD of 1.2–2.0 and were transferred to oleate medium [0.67% yeast nitrogen base w/o amino acids, 0.02 g L-histidine/L, 0.02 g L-arginine/L, 0.1% yeast extract, 0.2% (vol/vol) oleate, and 0.02% (vol/vol) Tween-40] for 6 h and used to prepare PYC, S1, or HSS and HSP fractions. Plasmids pJCF515 and pJCF 533 were kind gifts of J. C. Farré in this laboratory. Yeast strains and plasmids used in this study are listed in Table S1.
Fluorescence Microscopy.
Cells were grown on YPD and switched to oleate medium during exponential phase. Images were captured using a Plan Apochromat 100× 1.40 NA oil immersion objective on a motorized fluorescence microscope (Axioskop 2 MOT plus; Carl Zeiss) coupled to a monochrome digital camera (AxioCam MRm; Carl Zeiss) and processed using AxioVision software (version 4.5; Carl Zeiss).
Subcellular Fractionation.
Oleate grown cells were harvested (3,000 rpm, 5 min) at room temperature and resuspended in low glucose medium (YP medium with 0.1% glucose) and incubated for 30 min at 25 °C (50 mL per 75 OD600 units). Cells were pelleted again and spheroplasting was performed using a described procedure (36). The regenerated spheroplasts were used to prepare permeabilized cells (75 OD600 units) and S1 (1,500 OD600 units) or HSS and HSP (3,000 OD600 units), and all of the subsequent steps were carried out at 4 °C. To prepare the permeabilized cells, the regenerated spheroplasts were harvested, resuspended in 5 mL of permeabilization buffer (0.1 M potassium acetate, 0.2 M sorbitol, 2 mM magnesium chloride, and 20 mM Hepes, pH 7.2) and centrifuged at 3,000 rpm for 5 min. The supernatant was carefully removed and the pellet was resuspended in 50 μL of CB + DTT buffer (250 mM sucrose, 4 mM DTT, 1 mM EGTA, 20 mM Hepes, pH 7.4) with 1× protease inhibitor mixture [PIC; Sigma; P8215 with NaF (50 mM), leupeptin (12.5 μg/mL), aprotinin (50 μg/mL), PMSF (10 mM)]. To prepare the S1 fraction, the regenerated spheroplasts (1,500 OD600 units) were resuspended (with gentle vortexing) in 3.36 mL of 20 mM Hepes, pH 7.2, and then centrifuged at 1,000 rpm for 10 min. The supernatant was carefully removed and used to prepare HSS and HSP fractions. The S1 fraction was centrifuged at 100,000 × g for 1 h, the pellet (HSP) was resuspended in an equal volume of 20 mM Hepes, pH 7.2, or at 170,000 × g for 4 h and the supernatant (HSS) was collected and aliquots were frozen at −80 °C. Before the budding assay, the final concentration of buffer in each fraction was adjusted to: 115 mM potassium acetate, 2.5 mM magnesium chloride, 0.2 M sorbitol, 1× PIC, and 35 mM Hepes (pH 7.2). The protein concentration of each fraction was estimated using Bradford assay (37) with BSA as standard.
In Vitro ER-Budding Assay.
Permeabilized cells were washed twice with TBPS (115 mM potassium acetate, 2.5 mM magnesium acetate, 0.25 M sorbitol, 1× PIC, and 25 mM Hepes, pH 7.2) and then resuspended in the same buffer (4.5 OD600/25 μL per reaction). The budding reaction contained 4.5 OD600/25 μL PYCs, 1 mg S1 fraction, or 1 mg HSS fraction, or 1 mg HSP fraction and ATP-regenerating system (1 mM ATP, 0.1 mM GTP, 20 mM creatine phosphate, 0.2 mg/mL creatine phosphate kinase) in a 100-μL total reaction volume. The reaction mixture was incubated at 20 °C for 90 min and the reaction was terminated by chilling the samples on ice. To deplete samples of ATP, apyrase (Sigma; A6410) was added instead of the ATP-regenerating system. After the reaction, PYCs were pelleted by spinning the reaction at 13,000 rpm for 1 min. The supernatant (RS) of two reactions was pooled and spun at 200,000 × g for 1 h. The pellet (RS 200 kgP) was resuspended in SDS sample buffer, heated and analyzed on 12.5% SDS/PAGE, and immunoblotting was performed with appropriate antibodies.
Affinity Capture of Pex11p–2HA-Associated Membranes.
After separating the PYCs from the supernatant at the end of in vitro budding assay, RS of 10 reactions were pooled (900 μL) and spun at 200,000 × g. The pellet was resuspended in TBPS with or without Triton X-100 (1% vol/vol) and incubated with 80 μL of EZview Red affinity matrix (Sigma; E6779), which was washed and equilibrated with the TBPS. The incubation was carried out for 6 h at 4 °C on a rotating shaker. The beads were washed three times and finally resuspended in 100 μL of TBPS and analyzed on SDS/PAGE and immunoblotted with rat anti-HA monoclonal antibody (Roche; 11802600) to ascertain affinity capture of Pex11–2HA-containing vesicles.
Membrane Flotation Assay.
The supernatant (RS) of the in vitro budding reaction, after separating from the PYCs, was mixed with Nycodenz to a final concentration of 35% (150 μL) and placed at the bottom of a SW50.1 centrifuge tube and was layered with decreasing densities of Nycodenz (100 μL each of 35, 25, and 10% in TBPS). Tubes were centrifuged for 50,000 rpm for 2 h (Beckman; SW50.1) and 50 μL fractions were collected from the top followed by SDS/PAGE and immunoblot analysis with respective antibodies.
Supplementary Material
Acknowledgments
We thank Dr. Susan Ferro-Novick and Christopher Lord in the Department of Cellular and Molecular Medicine at University of California at San Diego for help and valuable advice in establishing the assay described here. We thank Dr. J. C. Farré for providing various strains and plasmids. This work is supported by National Institutes of Health MERIT Award DK41737 (to S.S.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018749108/-/DCSupplemental.
References
- 1.Fujiki Y, Rachubinski RA, Lazarow PB. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc Natl Acad Sci USA. 1984;81:7127–7131. doi: 10.1073/pnas.81.22.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rachubinski RA, Fujiki Y, Mortensen RM, Lazarow PB. Acyl-Coa oxidase and hydratase-dehydrogenase, two enzymes of the peroxisomal β-oxidation system, are synthesized on free polysomes of clofibrate-treated rat liver. J Cell Biol. 1984;99:2241–2246. doi: 10.1083/jcb.99.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fagarasanu A, Mast FD, Knoblach B, Rachubinski RA. Molecular mechanisms of organelle inheritance: Lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol. 2010;11:644–654. doi: 10.1038/nrm2960. [DOI] [PubMed] [Google Scholar]
- 4.Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: A story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 5.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry RJ, Mast FD, Rachubinski RA. Endoplasmic reticulum-associated secretory proteins Sec20p, Sec39p, and Dsl1p are involved in peroxisome biogenesis. Eukaryot Cell. 2009;8:830–843. doi: 10.1128/EC.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Zand A, Braakman I, Tabak HF. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell. 2010;21:2057–2065. doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elgersma Y, et al. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y, Morrell JC, Jones JM, Gould SJ. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol. 2004;164:863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam YY, Fagarasanu A, Fagarasanu M, Rachubinski RA. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- 13.Chang J, et al. Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J Cell Biol. 2009;187:233–246. doi: 10.1083/jcb.200902117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 15.Novikoff AB, Novikoff PM, Davis C, Quintana N. Studies on microperoxisomes. II. A cytochemical method for light and electron microscopy. J Histochem Cytochem. 1972;20:1006–1023. doi: 10.1177/20.12.1006. [DOI] [PubMed] [Google Scholar]
- 16.Geuze HJ, et al. Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell. 2003;14:2900–2907. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Titorenko VI, Ogrydziak DM, Rachubinski RA. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol. 1997;17:5210–5226. doi: 10.1128/mcb.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan M, Rachubinski DA, Joshi S, Rachubinski RA, Subramani S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol Biol Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voorn-Brouwer T, Kragt A, Tabak HF, Distel B. Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J Cell Sci. 2001;114:2199–2204. doi: 10.1242/jcs.114.11.2199. [DOI] [PubMed] [Google Scholar]
- 20.South ST, Sacksteder KA, Li X, Liu Y, Gould SJ. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol. 2000;149:1345–1360. doi: 10.1083/jcb.149.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titorenko VI, Rachubinski RA. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol. 2000;150:881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knoblach B, Rachubinski RA. Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem. 2010;285:6670–6680. doi: 10.1074/jbc.M109.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groesch ME, Ruohola H, Bacon R, Rossi G, Ferro-Novick S. Isolation of a functional vesicular intermediate that mediates ER to Golgi transport in yeast. J Cell Biol. 1990;111:45–53. doi: 10.1083/jcb.111.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruohola H, Kabcenell AK, Ferro-Novick S. Reconstitution of protein transport from the endoplasmic reticulum to the Golgi complex in yeast: The acceptor Golgi compartment is defective in the sec23 mutant. J Cell Biol. 1988;107:1465–1476. doi: 10.1083/jcb.107.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuzaki T, Fujiki Y. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J Cell Biol. 2008;183:1275–1286. doi: 10.1083/jcb.200806062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuzono Y, Fujiki Y. In vitro transport of membrane proteins to peroxisomes by shuttling receptor Pex19p. J Biol Chem. 2006;281:36–42. doi: 10.1074/jbc.M509819200. [DOI] [PubMed] [Google Scholar]
- 28.Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol. 2004;164:57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titorenko VI, Mullen RT. Peroxisome biogenesis: The peroxisomal endomembrane system and the role of the ER. J Cell Biol. 2006;174:11–17. doi: 10.1083/jcb.200604036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCammon MT, Veenhuis M, Trapp SB, Goodman JM. Association of glyoxylate and β-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J Bacteriol. 1990;172:5816–5827. doi: 10.1128/jb.172.10.5816-5827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder WB, Koller A, Choy AJ, Subramani S. The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J Cell Biol. 2000;149:1171–1178. doi: 10.1083/jcb.149.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazra PP, Suriapranata I, Snyder WB, Subramani S. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3:560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- 35.Lam SK, Yoda N, Schekman R. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2010;107:21523–21528. doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groesch ME, Rossi G, Ferro-Novick S. Reconstitution of endoplasmic reticulum to Golgi transport in yeast: In vitro assay to characterize secretory mutants and functional transport vesicles. Methods Enzymol. 1992;219:137–152. doi: 10.1016/0076-6879(92)19016-y. [DOI] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.