Abstract
The in vitro cloning of DNA molecules traditionally uses PCR amplification or site-specific restriction endonucleases to generate linear DNA inserts with defined termini and requires DNA ligase to covalently join those inserts to vectors with the corresponding ends. We have used the properties of Vaccinia DNA topoisomerase I to develop a ligase-free technology for the covalent joining of DNA fragments to suitable plasmid vectors. This system is much more efficient than cloning methods that require ligase because the rapid DNA rejoining activity of Vaccinia topoisomerase I allows ligation in only 5 min at room temperature, whereas the enzyme’s high substrate specificity ensures a low rate of vector-alone transformants. We have used this topoisomerase I-mediated cloning technology to develop a process for accelerated cloning and expression of individual ORFs. Its suitability for genome-scale molecular cloning and expression is demonstrated in this report.
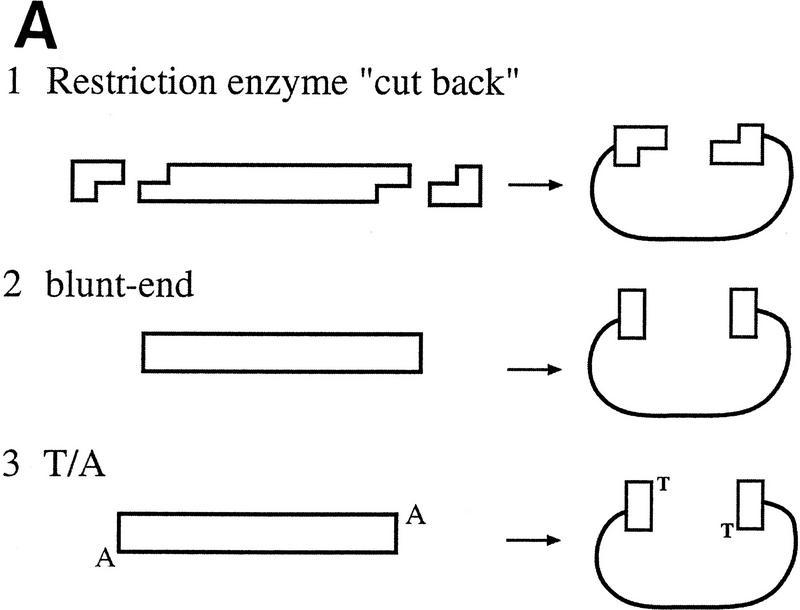
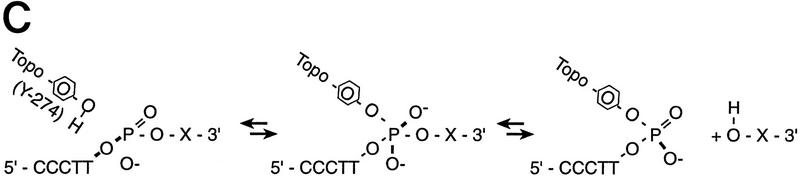
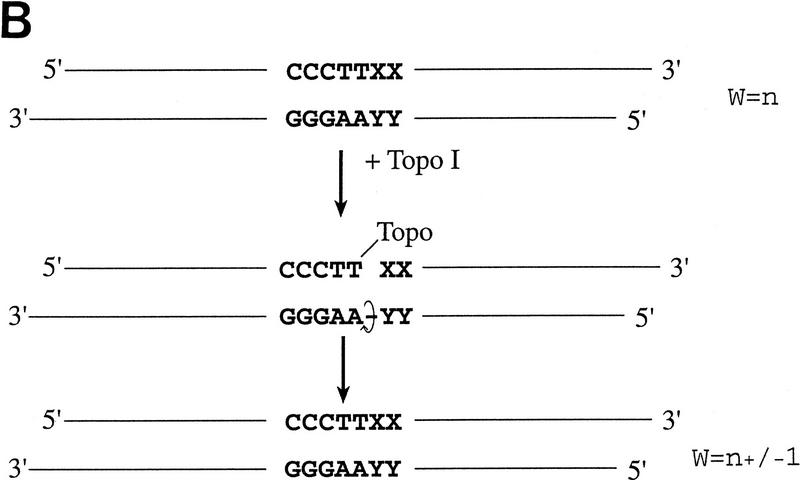
With conventional cloning methods, linear DNA inserts to be cloned are generated by either PCR amplification or by the cleaving action of restriction endonucleases that leave the DNA fragments with blunt ends or specific overhangs. In a second step, the corresponding ends of a DNA insert are covalently joined to the appropriately prepared complementary ends of a plasmid vector by the action of DNA ligase (Fig. 1A). Here, we present a new approach to molecular cloning that exploits the unique activity of a single enzyme, Vaccinia DNA topoisomerase I, to both cleave and rejoin DNA strands with a high sequence specificity. The enzyme, a 314-amino-acid virus-encoded eukaryotic type I topoisomerase (Shuman and Moss 1987), binds to duplex DNA and cleaves the phosphodiester backbone of one strand at a consensus pentapyrimidine element 5′-(C/T)CCTT in the scissile strand (Shuman and Prescott 1990; Shuman 1991a,b). In the cleavage reaction, bond energy is conserved by formation of a covalent adduct between the 3′ phosphate of the incised strand and a tyrosyl residue (Tyr-274) of the protein (Fig. 1B,C). The covalent complex can reclose across the same bond originally cleaved (as occurs during DNA relaxation) or it can combine with a heterologous acceptor DNA that has a 5′ hydroxyl tail complementary to that of the adduct, and thereby create a recombinant molecule, as first described by Shuman (Shuman 1992a,b).
Figure 1.
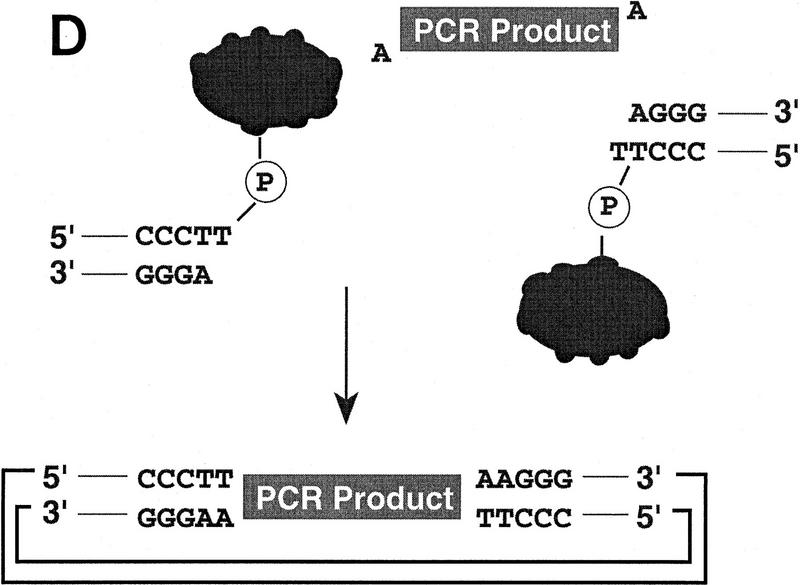
(A) Conventional techniques used to clone PCR products include (1) restriction-cutback methods, in which a restriction endonuclease is used to cleave both DNA insert and vector, leaving complementary overhangs; (2) blunt-end cloning, in which both insert and vector are prepared to have blunt ends; (3) TA Cloning uses the thermophilic TaqI polymerase to add a single 3′A overhang to each end of a PCR product, which can then be joined to a TA Cloning vector with single 3′T overhangs. All three methods involve a second step in which the PCR product and the vector are joined by the action of DNA ligase. (B) During DNA relaxation, the enzyme Vaccinia DNA topoisomerase I cleaves the phosphodiester backbone of one strand at a consensus pentapyrimidine element 5′-(C/T)CCTT in the scissile strand, allowing the DNA to unwind and reduce its winding (W) number, n, to n+1 or n−1 (for DNA that was negatively or positively supercoiled, respectively). (C) In the cleavage reaction, bond energy is conserved by formation of a covalent adduct between the 3′ phosphate of the incised strand and a tyrosyl residue (Tyr-274) of the protein. The covalent complex can reclose across the same bond originally cleaved or it can combine with a heterologous acceptor DNA that has a 5′ hydroxyl tail complementary to that of the adduct, and thereby create a recombinant molecule. (D) Topoisomerase I-mediated cloning uses the reaction mediated by Vaccinia DNA topoisomerase I to join PCR-amplified DNA fragments into plasmid vectors. PCR fragments have 5′ hydroxyl residues from the primers used for the amplification reaction, and therefore are an ideal substrate for the topoisomerase ligation reaction. The topoisomerase I (solid black shape) is shown linked to the vector through the 3′ phosphate (P) of the incised strand. The PCR product has single 3′ A overhangs (A).
Topoisomerase I-mediated cloning uses the above reaction to join DNA fragments containing 5′ hydroxyl groups to acceptor plasmid vectors. PCR fragments are well-suited for this topoisomerase-mediated ligation step because they generally have 5′ hydroxyl residues from the primers used for the amplification reaction (Fig. 1D). In fact, only DNA that has a 5′ hydroxyl group can serve as a substrate for the topoisomerase-mediated ligation and this contributes to the low rate of vector-only transformants (Shuman 1994). In addition, because low-melt agarose and slightly elevated temperatures (22–42°C) do not interfere with topoisomerase I activity, it is possible to purify desired DNA fragments by electrophoresis through low-melt agarose followed by excision of the appropriate band. This DNA purification method is amenable to high-throughput and ensures that only the desired DNA fragments are included in cloning reactions. These features of topoisomerase I, as well as the speed of its DNA rejoining activity, have been exploited to develop a high-throughput cloning technique.
This high-throughput cloning technique serves as the platform in the process for accelerated cloning and expression of open reading frames (ORFs) that is described here. We have performed two feasibility studies to demonstrate the suitability of this process for genome-scale molecular cloning and expression. In one study, we attempted to clone all 6035 ORFs of the yeast Saccharomyces cerevisiae into both the yeast pYES2/GS and the mammalian pcDNA3.1/GS expression vectors, and we then tested the positive-orientation clones for their ability to direct recombinant protein synthesis in yeast and in Chinese hamster ovary (CHO) cells, respectively. In the second feasibility study, we demonstrated the power of this technology for cloning and expressing human cDNAs. In this case, primer sets for 288 human kinases were used to amplify full-length ORFs from cDNA, and these ORFs were then taken through the cloning and expression process. The results are presented below.
RESULTS
High-Throughput Cloning of Yeast ORFs
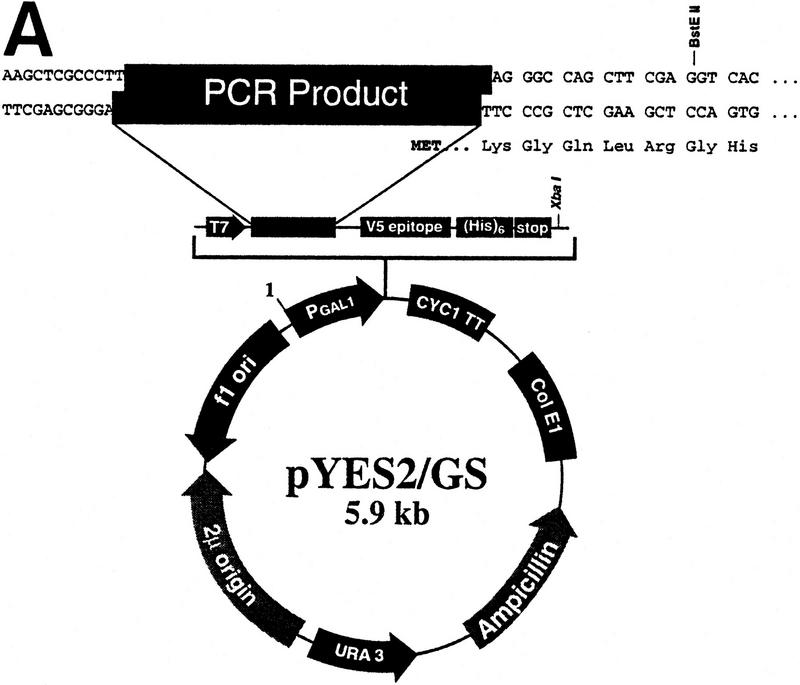
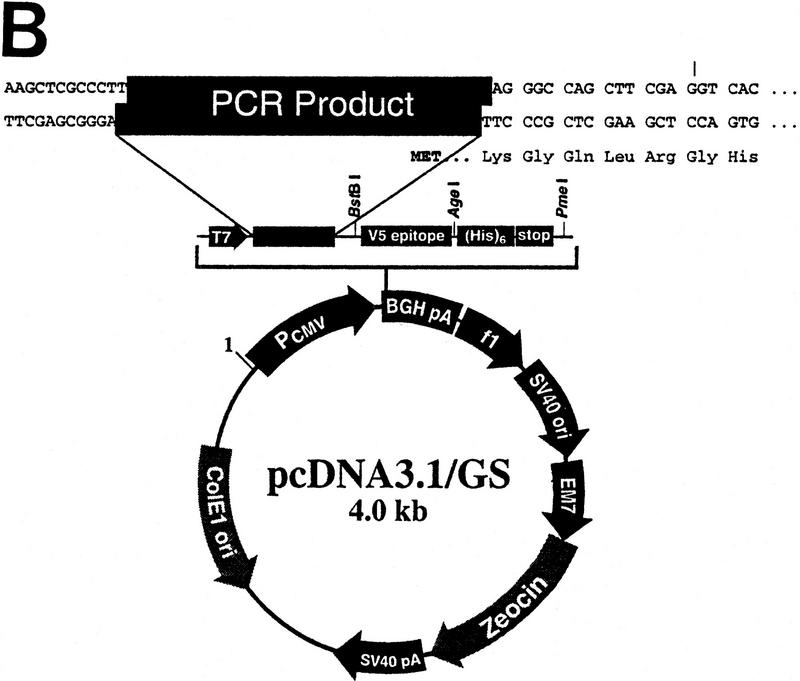
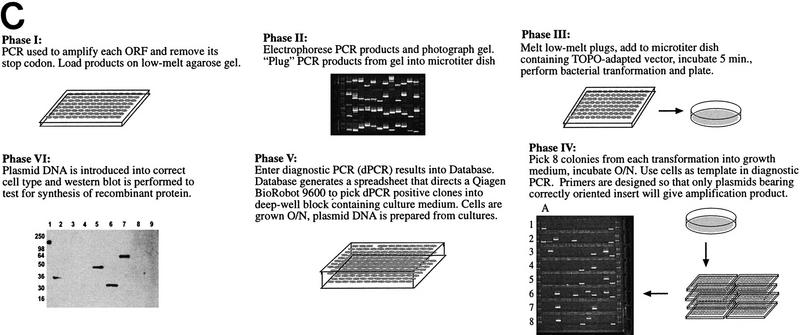
In this study 6035 ORFs from S. cerevisiae were amplified by PCR and inserted into two separate expression vectors (pYES2/GS and pcDNA3.1/GS; see Fig. 2A,B). The plasmids were tested for insert orientation, and orientation-positive plasmids were expression tested in yeast or CHO cells. There are essentially six phases to this process, which are schematically represented in Figure 2C and outlined below. Process details are given in Methods.
Figure 2.
(A) Vector pYES2/GS for expression in yeast cells includes the GAL1 promoter for inducible expression in appropriate S. cerevisiae strains; the 2 μ origin of replication for maintenance of high copy number and improved expression; the URA3 gene for stable selection of transformants in ura3 S. cerevisiae strains. (B) Vector pcDNA3.1/GS for expression in mammalian cells includes the CMV promoter and enhancer sequences for high-level transient or stable expression; and the multipurpose Zeocin resistance antibiotic for selection of bacterial transformants and of stably transfected mammalian cells. Both vectors also include the V5 epitope tag after the insert to allow detection of proteins for which antibodies are not available, a carboxy-terminal polyhistidine tag (His)6 to allow metal-affinity protein purification, and the T7 promoter to enable in vitro transcription and translation of the target gene. (C) Schematic representation of the topoisomerase I-mediated cloning process. Phases are described in the text.
Phase I—Amplification
PCR was performed to amplify each yeast ORF and remove its stop codon. The 6035 yeast ORFs and corresponding gene-specific primers for the 3′ end of each were provided by Research Genetics (Huntsville, AL) in a 96-well format.
Phase II—Insert Purification
PCR products were loaded onto 1% low-melt agarose gels and separated by electrophoresis. PCR products of the correct size were removed from the gel and transferred to a corresponding 96-well plate. We found this step to be essential—when we attempted to clone straight from the PCR reaction, we obtained an unacceptably high number of clones that contained primer-dimer PCR products (data not shown). Phase I and II resulted in the isolation of 5632 ORFs, for an amplification success rate of ∼93%.
Phase III—Topoisomerase I-Mediated Cloning
Low-melt agarose plugs were melted in the 96-well plates and a portion of this agarose/PCR fragment mixture was multichannel-pipetted into a 96-well tray containing the topoisomerase-adapted expression vector of choice. After a 5-min incubation at room temperature, a standard bacterial transformation was performed.
Phase IV—Diagnostic PCR
Bacterial transformants were screened by PCR to determine the size and orientation of the plasmid insert. Primers for this amplification were designed such that only plasmids with correctly oriented inserts gave amplification products. In the panel representing phase IV in Figure 2C, each column of eight wells (i.e., A1–A8) contains diagnostic PCR results for eight colonies from a single transformation.
Phase V—Plasmid Preparation
Positive clones identified by the diagnostic PCR were robotically organized and cultured in replicate 96-well deep block plates. After overnight growth of these cultures, one block was used to prepare glycerol stocks, whereas plasmid DNA was robotically prepared from the other.
Phase VI—Expression Testing
DNA prepared from the PCR-positive cultures was tested for its ability to direct protein synthesis in yeast or CHO cells. The DNA was transformed into yeast cells or transfected into CHO cells in 96-well format. After growth and/or induction, the cells were pelleted, lysed, and loaded onto SDS–polyacrylamide gels. Samples were then analyzed by Western blot for the presence of the V5 epitope tag that is cloned in-frame with all of the ORFs. Plasmids that directed synthesis of the recombinant protein of the correct size were considered to be expression-positive.
The first pass through the set of yeast ORFs generated 7511 vectors with correctly inserted yeast ORFs (3749 in pYES2/GS and 3762 in pcDNA3.1/GS). Therefore, the success rate for obtaining at least one positive orientation clone from eight picked colonies was 67% (number of unique ORFs correctly cloned/number of ORFs attempted). After DNA from the orientation-positive clones was prepared and introduced into the correct cell type, 1217 (or 20% of the clones tested) directed synthesis of detectable levels of recombinant protein (659 in pYES2/GS and 558 in pcDNA3.1/GS). Combined, the results for the two vectors (11% and 9%) yield a success rate of ∼10% for the overall process (from amplification PCR through positive expression). Table 1 shows the results for each of the stages in the cloning of yeast ORFs into the pYES2/GS and pcDNA3.1/GS expression vectors. This first pass was completed by eight people in 3 months.
Table 1.
Cloning and Expression Results for Yeast Genome and Human Kinase Studies
| Yeast first-pass process | No. successful | No. attempted | Success rate (%) | |||
|---|---|---|---|---|---|---|
| pYES2 | pcDNA3.1 | pYES2 | pcDNA3.1 | pYES2 | pcDNA3.1 | |
| ORF amplification | 5632 | 6035 | 93 | 93 | ||
| Topoisomerase I-mediated cloning + orientation | 3749 | 3762 | 5632 | 5632 | 67 | 67 |
| Expression QC positive | 659 | 558 | 3749 | 3762 | 18 | 15 |
| Start-to-finish summary | 659 | 558 | 6035 | 6035 | 11 | 9 |
| Yeast second-pass process | ||||||
| ORF amplification | 5000 | 5093 | 5376 | 5477 | 93 | 93 |
| Topoisomerase I-mediated cloning + orientation | 3750 | 3266 | 5000 | 5093 | 75 | 64 |
| Expression QC positive | 1553 | 1197 | 3750 | 3266 | 41 | 37 |
| Start-to-finish summary | 1553 | 1197 | 5376 | 5477 | 29 | 22 |
| Human kinase process | No. successful (pcDNA3.1) | No. attempted (pcDNA3.1) | Success rate (%) (pcDNA3.1) | |||
| RT–PCR | 179 | 288 | 62 | |||
| Topoisomerase I-mediated cloning + orientation | 140 | 179 | 78 | |||
| Expression QC positive | 108 | 140 | 77 | |||
| Start-to-finish summary | 108 | 288 | 38 | |||
Cloning and expression results for the yeast genome and human kinase studies. Success rates are expressed as percentages of successful attempts over the total number of attempts for each step of the cloning and expression process.
During the first pass, we identified several aspects of the process that could be improved. The improvements (described in the beginning of the Discussion) were applied in a second-pass cloning of the yeast ORFs into pYES2/GS and pcDNA3.1/GS, and a comparison of the results obtained in each pass reveals the effect of the process improvements (Table 1). In the first pass, we started with 6035 yeast ORFs and were able to construct 659 pYES2/GS-based and 558 pcDNA3.1/GS-based plasmids that were expression-positive (start-to-finish success rates of 11% and 9%, respectively). In the second pass for cloning into the pYES2/GS vector, we started with 5376 yeast ORFs and constructed 1553 expression-positive plasmids, whereas the cloning into the pcDNA3.1/GS vector was performed with 5477 yeast ORFs and generated 1197 expression-positive plasmids. Therefore, the second pass start-to-finish success rates were 29% for vector pYES2/GS and 22% for vector pcDNA3.1/GS, a marked improvement over the first pass (the second pass was not started with 6035 yeast ORFs because the ORFs that were successfully cloned and expressed in the first pass were removed from the second pass).
High-throughput Cloning of Human Kinase ORFs
Next, we analyzed the possibility of utilizing this cloning system, coupled with an initial high-throughput reverse transcriptase-PCR (RT-PCR) (Saiki et al. 1985; Sambrook et al. 1989) step, to obtain full-length human ORFs and insert them into the pcDNA3.1/GS vector. We focused on human ORFs that encode kinases, a family of proteins involved in signal transduction. The results described below are summarized in Table 1.
RT-PCR
To assess the feasibility of large-scale RT-PCR amplification of full-length human ORFs, we designed primer sets to amplify 288 full-length human kinases. PolyA+ mRNA was isolated from human fetal heart tissue, converted to first-strand cDNA, and used as template for PCR amplifications primed with the 288 human kinase primer sets (see Methods). A single pass with these 288 primer pairs resulted in RT-PCR generation of 179 products of the predicted size.
Topoisomerase I-mediated Cloning, Diagnostic PCR, and Plasmid Preparation
The 179 RT-PCR products were cloned into the pcDNA3.1/GS vector using the protocol described in the previous study. Diagnostic PCR on eight colonies from each of 179 transformations indicated that 140 of the 179 PCR products were cloned in the correct orientation into the expression vector. This represents a 78% success rate for this step. One or two colonies harboring these plasmids were grown overnight in deep-well 96-well blocks and plasmid DNA was prepared (in cases in which diagnostic PCR identified more than one positive-orientation clone, DNA was prepared from two clones and transfected—see below).
Expression Testing in CHO Cells; Sequence Analysis of Plasmids
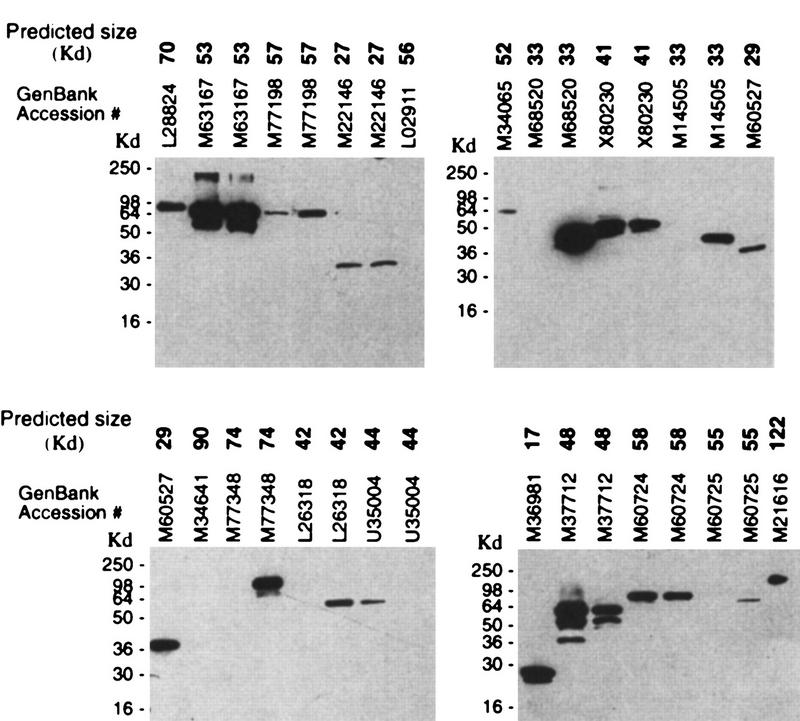
The expression plasmids bearing the 140 unique PCR products were transfected into CHO cells in 96-well, deep-well blocks. Cell lysates were made 48 hr after transfection and assayed by Western blot (Fig. 3). Analysis of positive signals and comparison to expected mobilities indicated that 115 of the transfected plasmids directed synthesis of an appropriately sized recombinant protein. So far, sequence data have been obtained for 113 of these 115 constructs: 108 (95%) contained the appropriate kinase and the predicted insert/vector junctions. Four of the incorrect plasmids contained DNA inserts of the correct size but the incorrect identity, and the fifth contained the correct insert, but in the reverse orientation. These five inserts did not contain a stop codon in the same frame as the epitope tag, resulting in expression of a fusion protein of the predicted size, but which was not the correct protein. The overall success rate of the process from RT-PCR through positive expression was 38% (108/288). Generally, in most transfections performed with DNA prepared from two unique clones bearing insert from the same RT-PCR, both DNAs directed expression of the protein of the predicted size (data not shown). This feasibility study (excluding the sequencing) took the equivalent of one person two weeks to complete.
Figure 3.
Western analysis of cell lysates from CHO cells transfected with pcDNA3.1/GS-based plasmids bearing the full-length human gene indicated by GenBank accession numbers. Predicted size of the recombinant protein is given in Kd (boldface). Cell lysates were made 48 hr after transfection and loaded on 12% Tris-glycine polyacrylamide gels. Proteins were transferred to membrane filters, and the filters were probed with the anti-V5/HRP conjugated antibody. Migration of recombinant proteins was visualized by chemiluminescence.
Success Rates—Yeast vs. Human ORFs into Vector pcDNA3.1/GS
As mentioned above, the overall success rate for cloning the yeast ORFs into pcDNA3.1/GS, starting with amplification PCR and finishing with expression-positive QC in CHO cells, was 9% for the first pass, and 22% for the second pass. In contrast, the overall success rate for cloning and expressing the human kinases was 38% in a single pass. This single pass also included an RT–PCR step (which was not needed with the yeast ORFs) to acquire the full-length template for cloning. Table 2 summarizes the efficiencies at each step of the process for the yeast clones and the human kinase clones. Possible reasons for the differing success rates are given in the Discussion.
Table 2.
Summary of Cloning and Expression Results in pcDNA3.1/GS for Yeast ORFs and Human Kinases
| Process | Success rate of yeast ORFs (%) | Success rate human kinases (%) | |
|---|---|---|---|
| first pass | second pass | ||
| RT–PCR/human kinases | N.A. | N.A. | 62 |
| ORF amplification/yeast | 93 | 93 | N.A. |
| Topoisomerase I-mediated cloning + orientation | 67 | 64 | 78 |
| Expression QC positive | 15 | 37 | 77 |
| Start-to-finish summary | 9 | 22 | 38 |
(N.A.) Not applicable.
ORF Length Correlates Negatively to Success Rate of Diagnostic PCR
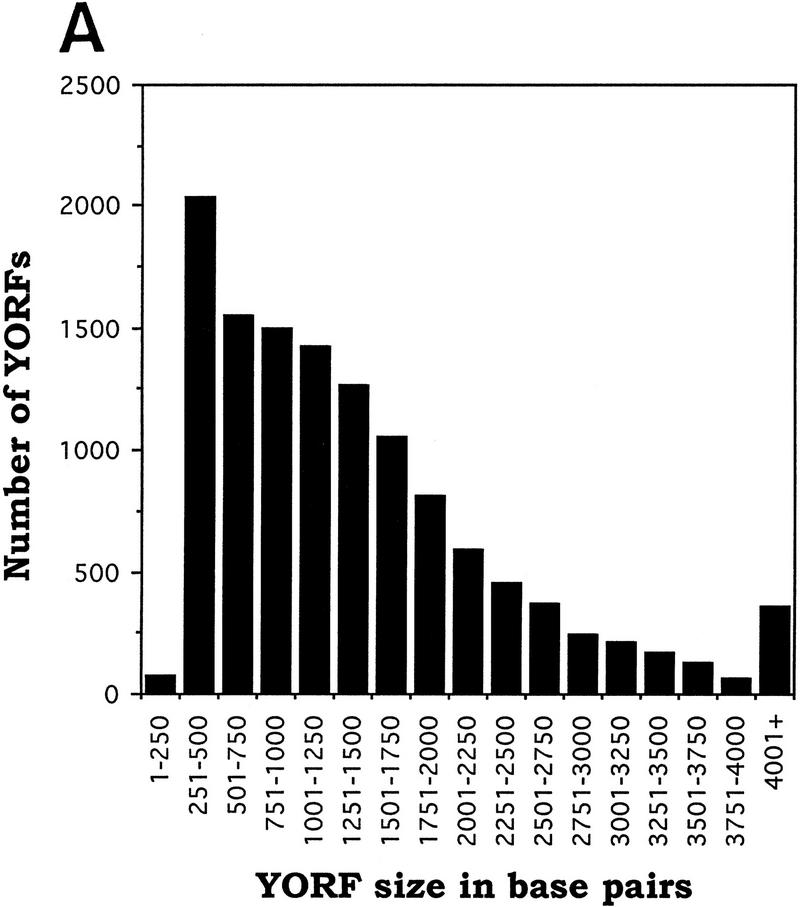
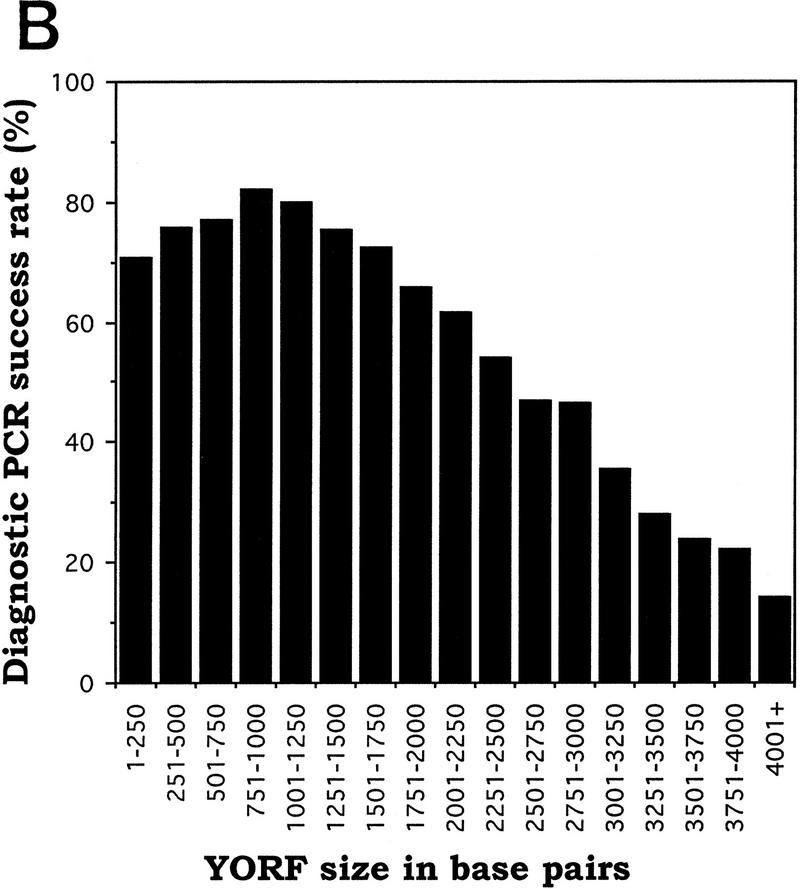
It is to be expected that success rates for several phases of the high-throughput cloning process will be influenced by the size of the ORF being processed. We have analyzed the data from the first and second pass cloning of yeast ORFs into vector pYES2/GS, as well as data from a smaller-scale topoisomerase I-mediated cloning of yeast ORFs into the same vector (data not shown), and have determined the efficiency with which we were able to identify plasmids containing positive-orientation inserts of a given length. The analysis is limited to phases III and IV of the cloning process, in which a purified yeast ORF (YORF) is mixed with topoisomerase I-adapted vector, the mixture is transformed into bacteria, and diagnostic PCR is performed on eight colonies from each transformation. Figure 4A was produced by sorting YORF PCR products by size into groups of increasing size (250 bp increments). For each group, we divided the number of YORFs for which at least one positive-orientation plasmid was identified by the total number of YORFs in that size group (Fig. 4B). These data clearly indicate that success rate of diagnostic PCR correlates negatively with increasing ORF length. For example, at least one positive-orientation plasmid was obtained for 80% of the 1001- to 1250-bp YORFs taken through phases III and IV, whereas at least one positive-orientation plasmid was obtained for only 22% of the 3751- to 4000-bp YORFs taken though the same phases. Because the data were collected from the diagnostic PCR reactions performed on colonies resulting from 12,284 separate cloning events (total from the two passes and those from the smaller-scale test cloning mentioned above), the statistical significance is high.
Figure 4.
Correlation between YORF length and success rate of diagnostic PCR (dPCR). (A) YORF PCR products were sorted by size into groups that increase in size by 250 bp increments. (B) The dPCR success rate was obtained, for each group, by dividing the number of YORFs for which dPCR on eight colonies identified ≥1 positive-orientation clones by the total number of YORFs in that size group.
DISCUSSION
During and following the first pass with the yeast ORFs, we made several improvements to the cloning and expression process. Most of these changes are process-related and are as follows: (1) low-melt agarose is now prepared fresh, to avoid generation of break-down products that interfere with the normally robust topoisomerase I-based cloning; (2) the amplification and diagnostic PCR primers have been replaced with primers of better design (see Methods); (3) the yeast induction protocol now includes short (3 hr) and long (24 hr) time points, so that yeast clones that express recombinant protein for a short window of time are identified; and (4) CHO cell culturing procedures were altered to ensure that cells were kept at lower, more transfection-competent, passage numbers. Further improvements to the process are the result of the steadily increasing skills of the personnel involved in the study.
These improvements were incorporated into a second pass through the yeast genome, and start-to-finish success rates improved significantly (see Tables 1 and 2). Because the ultimate goal of these projects was to produce reagents for the research community, only plasmids that directed Western blot-detectable synthesis of the correct recombinant protein were judged to be expression-positive. In the case of plasmids made in the pcDNA3.1/GS vector, there are several steps in the expression testing at which expression levels are likely to be reduced due to high-throughput requirements: (1) Miniprep DNA is used for the transfections, and the 96-well transfection format limits the number of cells per transfection to 3 × 105; (2) the cells are all harvested 48 hr after transfection, which might be too long if the recombinant protein is toxic, but too short if the protein accumulates slowly; and (3) transfections are done according to a fixed schedule, making it difficult to ensure that cells are at the optimal density and passage number for transfection. In the case of pYES2/GS-based plasmids, the yeast transformations, selections, and inductions are all performed in 1.4-ml cultures in 96-well blocks, conditions that are well-suited to high-throughput expression testing, but cannot be the optimal expression conditions for each plasmid. Therefore, it is likely that a percentage of both the pYES2/GS- and the pcDNA3.1/GS-based clones do direct recombinant protein synthesis, but at levels below our detection methods. We are currently assessing alternative methods, such as DNA sequencing or more sensitive protein detection techniques, to test the validity of the orientation-positive plasmids that did not direct Western blot-detectable levels of recombinant protein synthesis.
The human kinase study was undertaken after most of the high-throughput cloning and expression techniques had been optimized during the yeast ORF first pass, and this is reflected in the high rate of success of the kinase project (38% from start to finish; see Table 1). This study produced 108 unique pcDNA3.1/GS clones, each proven by Western blot to direct synthesis of the appropriately sized recombinant protein, and each partially sequenced to confirm the insert identity and to ensure that the insert/vector junctions were as predicted. This work also demonstrated that RT-PCR could be used for high-throughput generation of full-length ORFs from human mRNA.
A comparison between the human kinase and YORF projects reveals that 77% of the orientation-positive pcDNA3.1/GS human kinase clones directed synthesis of the appropriate protein (i.e., were expression-positive), whereas only 15% of the orientation-positive pcDNA3.1/GS YORF clones were expression-positive. Even in our improved second pass at the yeast genome, only 37% of the orientation-positive pcDNA3.1/GS clones tested expression-positive. The difference in success rates for yeast ORFs and human ORFs at this stage of the process might be explained by two fundamental differences between the amplified yeast ORFs and the amplified human ORFs. First, each amplified human ORF contains only a Kozak consensus sequence (CACC) (Kozak 1987) appended upstream of the start ATG, whereas each yeast ORF contains a palindromic sequence (5′-GCAGTCGTGGAATTCCAGCTGACCACC) appended immediately upstream of its ATG. It is possible that the extra palindromic sequence in each yeast ORF interferes with transcription and/or translation of the ORF. A second difference between the human and yeast ORFs is that the human ORFs were amplified once, with first-strand cDNA used as template, whereas the yeast ORFs were PCR-amplified twice—the first time by Research Genetics, using a 250:1 TaqI to PfuI polymerase (nonproofreading and proofreading polymerases, respectively) mixture and employing genomic DNA as the template; and a second time by us, in reactions that used the Research Genetics amplification products as templates and used a higher-fidelity 50:1 TaqI to PfuI mixture (Barnes 1992, 1994). Therefore the twice-amplified yeast ORFs may contain a higher number of nonsense mutations than the human ORF amplification products. We are currently conducting a variety of experiments, including the sequencing of positive-orientation clones, to determine the reasons why some correctly oriented clones do not direct Western blot-detectable levels of recombinant protein synthesis.
Analysis of the YORF cloning/expression results from the first pass revealed a strong inverse correlation between the length of an ORF and the likelihood of identifying a plasmid containing that ORF in the positive orientation. The stage analyzed encompasses phases III and IV of the cloning process, and these phases consist of three manipulations that might be influenced by YORF length: topoisomerase-mediated ligation of amplified ORFs contained in low-melt agarose plugs, bacterial transformation, and diagnostic PCR. Experiments are under way to determine which of these steps is most adversely affected by clone length. Our analysis also suggests that it may be advantageous to group PCR primers in plates according to the size of the expected amplification product. This will make it possible to pick additional transformants from the plates that contain the longer ORFs, therefore increasing the probability of obtaining orientation-positive clones.
Whereas the above studies used topoisomerase I to create recombinant plasmids, there exist numerous methods for high-throughput cloning of PCR products. For example, ligase could have been used to join the PCR products to expression vectors. This method, however, requires that PCR products be phosphorylated, either through the use of phosphorylated primers or by phosphorylation of the PCR product. Also, measures must be taken to ensure a low rate of vector-alone (containing no insert) transformants. Vector dephosphorylation is commonly employed, but this treatment often results in vectors that accept inserts with poor efficiency. Alternatively, vectors can be created so that the cloning site is located within a gene that encodes a lethal protein (Bernard et al. 1994). These vectors cannot give rise to bacterial transformants unless the lethal gene is interrupted by a cloned DNA. A drawback of this method is that the lethal gene sequences constrain the sequences surrounding the cloning site, thereby making it difficult to include desired elements in the vector (promoters, terminators, epitope tags, etc). Another method to improve ligase-mediated cloning efficiency, TA cloning (described in Fig. 1A), relies on vectors prepared with single 3′ T overhangs to limit vector-alone transformants. This method has been shown to be very effective for cloning PCR products and could certainly be adapted to high-throughput applications. Because ligase activity is reduced by increased temperatures and the presence of low-melt agarose, however, TA-cloning is not compatible with the streamlined method of PCR product purification that we used with topoisomerase I-mediated cloning.
There also exist recombination-mediated cloning strategies that use yeast (Muhlrad et al. 1992; Oldenburg et al. 1997) or Escherichia coli cells (Zhang et al. 1998). These methods generally do not suffer from problems associated with vector-alone transformants. Also, yeast-based recombination schemes offer a unique advantage in the generation of yeast expression plasmids because the recombinant plasmids can be functionally tested in the cells in which they were created. These methods, however, require that the PCR primers each have 30–40 bp of homology to the recombination target, which increases primer cost by over twofold. In addition, these methods involve transformation of PCR products, which precludes the use of the low-melt agarose method for DNA purification. Finally, it is not trivial to retrieve recombinant plasmids from yeast cells, a necessary step if the plasmids are to be sequenced or transferred into a different host.
Our studies have demonstrated that topoisomerase I-mediated cloning is a robust method to create recombinant plasmids and that it is particularly well-suited to large-scale cloning efforts. We have also shown that high-throughput RT–PCR can be used in conjunction with this cloning technique to greatly facilitate high-speed cloning and expression of ORFs from organisms whose genomic DNA contains introns.
METHODS
Preparation of Topoisomerase-Adapted Vectors
This protocol is used to prepare both the pcDNA3.1/GS and the pYES2/GS vectors. The vector is cut with HindIII, extracted with phenol/chloroform, and ethanol precipitated. TOPO-H (5′P-AGCTCGCCCTTATTCCGATAGTG) and TOPO-4 (5′-AGGGCG) oligos are ligated onto the HindIII-cut vector, the vector is phenol/chloroform extracted, ethanol precipitated, cut again with HindIII to remove re-circularized vector, and again phenol/chloroform extracted and ethanol precipitated. Purified Vaccinia topoisomerase I and TOPO-5 oligonucleotide (5′-CAACACTATCGGAATA) are added to the vector and the mixture is incubated for 15 min at 37°C (buffer is 1× NEB restriction buffer 1; New England Biolabs, Beverly, MA). During this step, the topoisomerase I cleaves after, and remains covalently attached to, the second T in the CCCTT sequence in ligated oligonucleotide TOPO-H. This leaves a vector with topoisomerase I bound to a 3′ overhanging T. The reaction is stopped by addition of 1/10 volume of TOPO-10× stop buffer. Free oligonucleotides and unbound topoisomerase I are purified away from the topoisomerase-adapted vector by agarose gel electrophoresis.
High-Throughput Cloning of Yeast ORFs
Phase I—PCR Amplification
The 6035 yeast ORFs and a corresponding gene-specific primer for the 3′ end of each were provided in 96-well format by Research Genetics (Huntsville, AL). Each gene-specific primer was designed to exclude the gene’s stop codon and to have a melting temperature of ∼62°C. Because all of the templates from Research Genetics contain the sequence 5′-GGAATTCCAGCTGACCACC immediately 5′ of the start ATG, we were able to amplify each template with the common primer YO8ATG: (5′-GCAGTCGTGGAATTCCAGCTGACCACC) and the appropriate gene-specific 3′ primer. The YO8ATG primer was designed to add bases GCAGTCGT to the 5′ end of each template so that subsequent PCR could be performed to distinguish between a nonamplified template and an amplified product (see section below on Diagnostic PCR). The reaction conditions were 1 cycle at 94°C for 4 min; 25 cycles at 94°C for 30 sec, 56°C for 45 sec, and 72°C for 3 min; followed by 1 cycle at 72°C for 4 min. Each initial amplification was performed in a 30 μl total volume with 2 units of a 50:1 (Unit/Unit) mixture of TaqI (Sigma, St. Louis, MO) and PfuI (Stratagene, San Diego, CA) polymerases, 0.4 μl of 50 mm dNTPs, 100 ng of each primer, in 1× PCR buffer J (Invitrogen, Carlsbad, CA) (final concentrations of 60 mm Tris-Cl, 15 mm (NH4)2SO4, 2.0 mm Mg2+ at pH 9.5). During the first pass for the yeast ORFs, primer YO8ATG was replaced after ∼84% of the yeast ORFs had been taken through the amplification step. A new primer, YAMP1 (5′-GCAGTCGTGGAATTCCAGCTGACCA), was used for the remainder of the first pass initial amplifications and for all of the second-pass initial amplifications.
Phase II—Insert Purification
The amplified ORF products were loaded onto 1% low-melt agarose gels and separated by electrophoresis. To excise the amplified yeast ORFs from the agarose gels, a photograph of the ethidium bromide-stained amplification products was taken, and the bands of correct size were marked. The photo with marked bands is used as a guide to ensure that only PCR products of the correct size are removed from the gel during band isolation. Disposable transfer pipettes were used to plug the amplification products from the low-melt agarose gel and to place them into a 96-well plate. The plugs were transferred so that the well location of an amplification product corresponds to the well location of the yeast ORF-specific primer and template that were used to create the product.
Phase III—Topoisomerase I-Mediated Cloning, Bacterial Transformation, and Plating
The low-melt plugs were liquified in 96-well plates on a 96-well heat block at 75°C and diluted with 50 μl of TE buffer. Three microliters of this melted agarose and PCR fragment mixture was multi-channel pipetted into a 96-well tray containing 2.5 μl of topoisomerase-adapted pcDNA3.1/GS or pYES2/GS expression vector (Invitrogen, Carlsbad, CA). After a 5-min room-temperature incubation, a multichannel pipettor was used to add 50 μl of competent E. coli (TOP10) cells to each vector/yeast ORF mixture. The tray was then placed on ice for 20 min, moved to a 42°C temperature block and subjected to a 1-min heat shock, and returned to ice for 1 min. Growth medium (140 μl) was then added to each well and the tray was placed at 37°C for 1.5 hr. The entire content of each well was then plated onto a selective plate.
Phase IV—Colony Picking and Diagnostic PCR on Bacterial Cells
Eight colonies from each selection plate were picked into a row on a 96-well tray filled with selective growth medium. These cultures were covered and grown overnight at 37°C. A 2-μl aliquot from each culture was then transferred to a corresponding well in a diagnostic PCR plate. The primers (described below) used in this reaction are designed so that only cells bearing a plasmid which contains an amplified yeast ORF in the correct orientation will give rise to an amplification product. H6STOPREVU (5′-AAACTCAATGGTGATGGTGATGATGACC) was selected as the reverse diagnostic PCR primer because it anneals to the sequence that encodes the polyhistidine tract, and therefore functions as a reverse primer for the pcDNA3.1/GS or pYES2/GS vectors. YO8DIAG (5′-CTCGCCCTTGCACTCGTGGA) was selected as the forward diagnostic primer because it anneals only to the 5′ end of an amplified yeast ORF. This is because the YO8ATG sequence is derived substantially from the sequence that is added to the yeast ORFs during the initial amplification with forward primer YO8ATG (see above for YO8ATG sequence). PCR cycling conditions were: 1 cycle of 94°C for 10 min; 25 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 3 min; followed by 1 cycle of 72°C for 4 min. Each initial amplification was performed in a 30 μl total volume with 2 units of TaqI (Sigma, St. Louis, MO) polymerase, 0.4 μl of 50 mm dNTPs, 100 ng of each primer, in 1× PCR buffer J (Invitrogen, Carlsbad, CA) (final concentrations of 60 mm Tris-Cl, 15 mm (NH4)2SO4, 2.0 mm Mg2+, pH 9.5). After ∼84% of the yeast ORF first pass was completed, YO8DIAG was replaced by 5′DIAGY1 (5′-CTTGCAGTCGTGGAATTCC), which gave a lower rate of mis-priming. 5′DIAGY1 was used for the entire second pass.
Phase V—Plasmid Preparation
The plate and well location of each PCR-positive bacterial culture were recorded in the yeast ORF database and a spreadsheet of positives was generated. This spreadsheet was downloaded to disk and then loaded on a Qiagen Biorobot 9600 (Qiagen Inc, Valencia, CA). The spreadsheet directs the robot to re-rack positive clones from the eight culture plates into two replicate 96-deep-well blocks containing growth medium. These blocks were grown overnight, then one was used for a glycerol stock plate and the other for a Qiaprep Turbo 96-well miniprep (Qiagen, Valencia, CA) by the Biorobot. The DNA prep is used for expression testing.
Phase VI—Expression Testing
Expression testing of pcDNA3.1/GS plasmids in CHO cells
In each well of a deep-well 96-well block, a mixture of 24 μg of PerFect Lipids (pFx-6) (Invitrogen, Carlsbad, CA) and 5 μg of plasmid DNA was added to 488 μl of Opti-Mem reduced serum medium (GIBCO Life Technologies, Baltimore, MD). This mixture was shaken for 5 min at room temperature, then 3 × 105 CHO suspension cells (in 500 μl Opti-Mem) were added to each well. The deep-well block was shaken for an additional 5 min and placed in a 37°C humidified incubator. After 4 hr, the cells were pelleted and the medium was replaced with 1.5 ml of CHO–S–SFM medium (GIBCO Life Technologies). The cells were incubated for 42–48 hr at 37°C, then pelleted and lysed in the 96-well blocks, and the lysates were loaded by eight-channel pipettor onto nine-well Bio-Rad 12% Tris-glycine polyacrylamide gels. Five microliters of Novex (San Diego, CA) Sea Blue markers, along with 50 ng of a control protein for the anti-V5/HRP conjugated antibody (Invitrogen, Carlsbad, CA), was loaded into lane one of each gel. Proteins were transferred to Schleicher & Schuell Optitran membrane filters, and the filters probed with the anti-V5 antibody (Invitrogen, Carlsbad, CA). Immunolocalized antibody was detected by incubation with Pierce (Rockford, IL) SuperSignal Ultra chemiluminescence and subsequent exposure to film. Transfected plasmids that directed synthesis of the correctly sized fusion protein were marked as Western positive.
Expression testing of pYES2/GS plasmids in yeast cells
Approximately 4 μg of each plasmid DNA was transformed into competent INVSc1 S. cerevisiae cells (his3Δ1 leu2 trp1-289 ura3-52) in a 96-deep-well block [transformation was performed essentially as described in S.c. EasyComp kit from Invitrogen (Carlsbad, CA), with the exception that 25 μl of competent cells were used for each transformation]. The cells were cultured in selective growth medium (1.3% yeast nitrogen base, 2% glucose, 20 μg/ml histidine, 20 μg/ml tryptophan, and 30 μg/ml leucine) for 3–4 days at 30°C, then pelleted (3000g, 10 min), and the medium was replaced with induction medium (1.1% yeast nitrogen base, 2% galactose, 1% raffinose, 20 μg/ml histidine, 20 μg/ml tryptophan, 30 μg/ml leucine). After overnight induction, the cells were pelleted, the medium decanted, and each pellet resuspended in 15 μl 1× DNase buffer [50 mm Tris-Cl at pH 7.4, 5 mm MgCl2, 0.1 mg/ml of DNase I—grade II (Boehringer-Mannheim, Chicago, IL) 1 mm PMSF, and 5% glycerol]. Fifteen microliters of 2× sample buffer [0.5 m Tris-Cl at pH 6.8, 20% glycerol, 10% (wt/vol) SDS, 0.1% Bromophenol Blue, 700 mm β-mercaptoethanol] was added to each well, the entire 96-well block was placed in boiling water for 5 min, then the block was placed on ice. Samples were analyzed by Western blot as described above for the CHO cell lysates. During the yeast ORF first pass, it was noticed that some plasmids directed recombinant protein synthesis for a short window of time. To ensure that we detected protein synthesis in subsequent similar cases, we removed 750 μl of culture from each well of the 96-well block after 3-hr induction. These cells were pelleted, frozen and saved. Fresh induction medium (750 μl) was added to each of the wells of the 96-well block and the induction was continued overnight. These cells were then pooled with the short-induction cells, and the combined cells were processed as described above. We have noticed that the expressed recombinant proteins consistently run 2–5 kD high when compared with the Sea Blue markers. The reason for this is not known, but it could be that the loading buffer used for the Sea Blue markers is different from our lysis/loading buffer.
High-Throughput Cloning of Human Kinase ORFs
RT–PCR
Primer pairs were designed to amplify 288 full-length human kinases from the ATG to the last codon before the stop codon. These primers were designed to have a melting temperature of ∼60–64°C and each 5′ primer included a Kozak sequence (CACC) immediately preceding the ATG to increase translational efficiency (Kozak 1987). Fetal human heart tissue was obtained from the International Institute for the Advancement of Medicine (IIAM), Scranton, PA. The Micro-FastTrack 2.0 Kit (Invitrogen, Carlsbad, CA) was used to isolate polyA+ mRNA. The mRNA was converted to first-strand cDNA using the cDNA Cycle Kit from Invitrogen (Carlsbad, CA), using the oligo(dT) primer provided and the protocols suggested. Eight cDNA synthesis reactions were split into the wells of a 96-well PCR amplification plate, and PCR amplifications were performed for the 288 human kinase primer sets. Cycling parameters were 1 cycle of 94°C for 4 min; 35 cycles of 94°C for 45 sec, 55°C for 45 sec, and 72°C for 3 min; followed by 1 cycle of 72°C for 4 min. Each reaction was performed in 50 μl total volume and included 2 units of a 50:1 (unit/unit) mixture of TaqI (Sigma, St. Louis, MO) and PfuI (Stratagene, San Diego, CA) polymerases, 2.0 μl of 50 mm dNTPs, 2–20 ng first-strand cDNA, and 5 μl of TA 10× PCR buffer (Invitrogen, Carlsbad, CA). Each primer was added to a final concentration of 0.6 nm. The reaction products were separated on a 1% low-melt agarose gel and visualized by ethidium bromide staining. DNA bands that represented correctly sized amplification products were removed from the rest of the gel using disposable transfer pipettes and placed into appropriate corresponding wells of a 96-well microtiter plate. These plugs were subsequently melted and used as inserts in cloning reactions as described in the previous section.
Diagnostic PCR on Bacterial Cells
Diagnostic PCR was performed as described above for the yeast ORFs, with the exception that a 5′ gene-specific primer was used in conjunction with primer H6STOPREVU for each reaction. In this case, only cells containing the correct ORF in the correct orientation should give amplification products.
DNA Sequencing
Each expression-positive human kinase plasmid was sequenced across the cloning junctions and approximately 350 bp into each end of the insert. This was performed on the Licor IR2 system, using labeled primers that flanked the cloning site.
Expression Testing
See protocol described for the yeast ORFs.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grant 1 R01 CA80224-01. We thank Heidi Kijenski and Jim Snook for programming the QIAGEN BioRobot to perform the essential task of colony re-racking. We thank Maria Knoske, Kelly Wynne, Jill Bloom, Sandy Adams, Shannon Hattier, and Josh Uhlig for performing some of the work described in this study. We also thank Karyn Hoeffler for aid in PCR primer design, Cinzia Ellero for help in writing the manuscript, and Doreen Crawford for assistance with the figures.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL john_heyman@invitrogen.com; FAX (760) 603-7201.
REFERENCES
- Barnes WM. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- ————— PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Gabant P, Bahassi EM, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shuman S. Site-specific DNA cleavage by vaccinia virus DNA topoisomerase I. Role of nucleotide sequence and DNA secondary structure. J Biol Chem. 1991a;266:1796–1803. [PubMed] [Google Scholar]
- ————— Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro [published erratum appears in J. Biol. Chem. 1991, 266: 20576–20577] J Biol Chem. 1991b;266:11372–11379. [PubMed] [Google Scholar]
- ————— DNA strand transfer reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992a;267:8620–8627. [PubMed] [Google Scholar]
- ————— Two classes of DNA end-joining reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992b;267:16755–16758. [PubMed] [Google Scholar]
- ————— Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J Biol Chem. 1994;269:32678–32684. [PubMed] [Google Scholar]
- Shuman S, Moss B. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc Natl Acad Sci. 1987;84:7478–7482. doi: 10.1073/pnas.84.21.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]