Abstract
Study Design
The transition of the mouse embryonic notochord into nuclei pulposi was determined (“fate mapped”) in vivo in GDF-5 null mice using the Shhcre and R26R alleles.
Objective
To determine if abnormal nuclei pulposi formation from the embryonic notochord was responsible for defects present in adult nuclei pulposi of Gdf-5 null mice.
Summary of Background Data
The development, maintenance, and degeneration of the intervertebral disc are not understood. Previously, we demonstrated that all cells in the adult nucleus pulposus of normal mice are derived from the embryonic notochord. Gdf-5 null mice have been reported to contain intervertebral discs in which the nucleus pulposus is abnormal. It is currently unclear if disc defects in Gdf-5 null mice arise during the formation of nuclei pulposi from the notochord during embryogenesis or resulted from progressive postnatal degeneration of nuclei pulposi.
Methods
Gdf-5 mRNA expression was examined in the discs of wild-type embryos by RNA in situ hybridization to determine when and where this gene was expressed. To examine nucleus pulposus formation in Gdf-5 null mice, intervertebral discs in which embryonic notochord cells were marked were analyzed in newborn and 24 week old mice.
Results
Our Gdf-5 mRNA in situ experiments determined that this gene is localized to the annulus fibrosus and not the nucleus pulposus in mouse embryos. Notochord fate mapping experiments revealed that notochord cells in Gdf-5 null mice correctly form nuclei pulposi.
Conclusion
Our data suggest that the defects reported in the nucleus pulposus of adult Gdf-5 null mice do not result from abnormal patterning of the embryonic notochord. The use of mouse alleles to mark cells that produce all cell types that reside in the adult nucleus pulposus will allow for a detailed examination of disc formation in other mouse mutants that have been reported to contain disc defects.
Keywords: intervertebral, disc, Gdf-5, fate map, Shhcre, nucleus pulposus
Introduction
The intervertebral disc is composed of three main structures: the nucleus pulposus, annulus fibrosus and end plates. The nucleus pulposus is comprised chiefly of water and proteoglycans and is surrounded by the annulus fibrosus, which is composed of collagen fibers. This structure is sandwiched between two end plates made of hyaline cartilage to form the intervertebral disc. The disc is avascular, relying on diffusion of nutrients through the endplates to maintain disc health (1, 2).
The discs are a critical source of body support; they bear loads, and provide stability to the spinal column while allowing flexibility of the body (2, 3). During the aging process the disc undergoes several changes; it becomes less gel-like and more fibrous, loses height, and the annulus fibrosus develops fissures. As a result of these changes, the nucleus pulposus can herniate through the damaged annulus fibrosus placing pressure on the spinal nerves, resulting in back pain (2). In the United States, billions of dollars are spent on the treatment of back pain and for costs associated with back pain (for example, absenteeism and reduced productivity). Unfortunately there are few effective treatments for back pain, with most current treatments targeting the symptoms and not addressing the underlying disc degeneration that has caused pain (4).
There is relatively little known about how the disc forms, which proteins are required to maintain normal disc function or what factors contribute to its degeneration. Previously, using the mouse model system we demonstrated that all cells in the mature wild-type nucleus pulposus were derived from the embryonic notochord (5). In these experiments, the notochord was genetically marked and the fate of this structure was determined throughout embryonic and postembryonic development. This evidence plus studies on molecular markers (brachyury, others) of the notochord and nucleus pulposus indicate that the nucleus pulposus is derived entirely from notochord cells (6, 7).
Recently, Growth and Differentiation Factor 5 (GDF-5, also called BMP-14) a member of the transforming growth factor beta (TGF-β) superfamily has been implicated in disc formation (8). Mutations in TGF-β family members can lead to developmental disorders. In humans, aberrations in CDMP-1 (cartilage derived morphogenetic protein), the human homologue of GDF-5, result in Hunter-Thompson and Grebe type chondrodysplasias. Patients exhibit shortening of the long bones of the limbs and shortening of other limb elements. Both Hunter-Thompson and Grebe type chondrodysplasias are autosomal recessive mutations (9, 10). Less severe is brachydactyly type C, which results from the inactivation of one copy of CDMP-1 (11). In these patients some of the distal phalanges are shortened. In the axial skeleton of humans, premature end-plate disease was noted in the vertebral column of 4 carriers of a CDMP-1 mutation (12) suggesting that mutation of this gene in humans may cause disc defects.
In the mouse there is a naturally-occurring missense mutation in Gdf-5, called brachypodism (bp), which renders GDF-5 protein nonfunctional (13). Gdf-5 deficient mice have several skeletal abnormalities, including shorter long bones of the limb and a reduction of phalanges in the digits (8, 13, 14).
In rabbits containing damaged discs, injection of GDF-5 was reported to increase disc height (15). Studies of GDF-5 treated damaged mouse discs found increases in collagens and proteoglycans (16). GDF-5 has also been overexpressed in disc cells using adenoviruses and found to increase cell proliferation and proteoglycans in vitro (17). Nucleofection of disc cells with an expression vector containing Gdf-5 also resulted in increases of type II collagen and aggrecan (18). Though mutations in Gdf-5 are thought to affect only the appendicular skeleton, it was recently reported that Gdf-5 deficient mice also contained deformed nuclei pulposi and a decrease in type II collagen and proteoglycans in the disc (8). Another report found that some Gdf5; Gdf6 double mutants developed scoliosis post-natally and had reduced staining for cartilage matrix in vertebral processes (19).
To determine if the reported defects in nuclei pulposi of Gdf-5 null mice were due to abnormal transitioning of the embryonic notochord into the nucleus pulposus we performed a notochord fate mapping analysis in this mutant background. To do this, we employed the Shhcre allele, which can be used to determine the cellular origin of nuclei pulposi in mutant mice. Mouse Cre alleles are used to remove DNA placed between two loxP DNA sequences in a tissue-specific manner. CRE protein recognizes loxP DNA sequences and can instigate recombination between two loxP sites present as direct repeats in the genome (20). In mice containing the Shhcre allele, CRE was expressed in all cells in which Shh mRNA was observed, including the embryonic notochord. Our data shows that embryonic notochord cells normally form the nucleus pulposus of the intervertebral disc in Gdf-5 null mice. This suggests that GDF-5 is not required for the transition of embryonic notochord cells into mature nuclei pulposi. In adult Gdf-5 null mice, nuclei pulposi were found to be composed of cells derived from the notochord, paralleling the situation in normal animals. Using RNA in situ hybridization, we found that Gdf-5 mRNA expression was absent from the notochord but surprisingly present in the annulus fibrosus after discs had formed. No Gdf-5 mRNA was found within the nucleus pulposus.
Materials & Methods
Animal Model
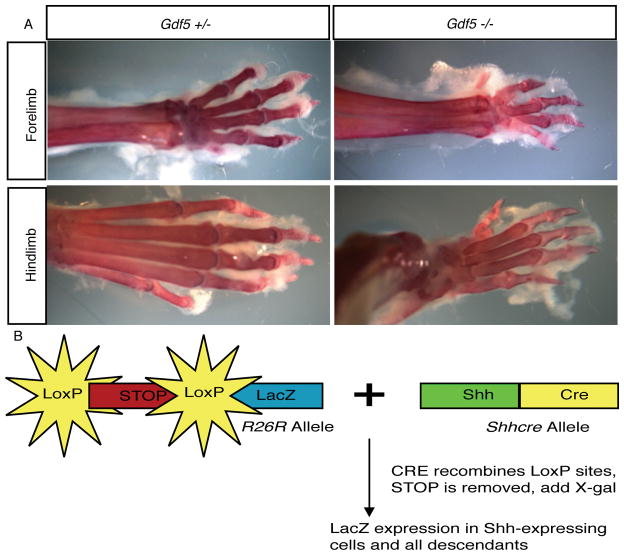
All animals were maintained at the University of XX under pathogen-free conditions. All procedures were approved by the XX IACUC. Gdf-5 (brachypodism, bp) null mice (allele Gdf5bp-J) were obtained from Jackson Laboratories (Bar Harbor, MI) and mated to animals containing both the Shhcre (21) and R26R (22) alleles. The presence of the Shhcre and R26R alleles was determined by PCR as published previously (21) and (22). Gdf-5 null mice were genotyped by examining the autopods of mutant animals (Figure 2a). The autopod is the distal-most portion of the limb and includes the bones of the carpals/tarsals and phalanges. The limb phenotype in Gdf-5 null mice is highly penetrant and observation of the limbs is frequently used to genotype these animals as opposed to PCR (8, 13). Only homozygous mutant animals will display the phenotype; wild-type and heterozygous animals are indistinguishable. The limb phenotype is identifiable at birth (13). A male of the genotype Gdf-5 +/−; Shhgfpcre/+; R26R/+ was crossed with Gdf-5-null females to obtain animals for analysis. This mating scheme maximizes the number of mutant animals with the transgenic alleles and provides heterozygotic animals to use as controls. For timed matings, animals were set up late in the afternoon and the female checked for the presence of a semen plug early the next morning. The day a plug was detected was designated E0.5. Newborn mice (P0) were harvested for β-galactosidase staining. RNA section in situ hybridization was done on wild-type animals at E10.5, E12.5, and E14.5 while other mutant mice were aged up to 24 weeks for analysis. All animals were on a mixed genetic background. For the fate-mapping and histology, 3 animals were examined for each genotype.
Figure 2. Gdf-5 deficient (bp) animals contain a reproducible and highly penetrant limb phenotype.
Gdf-5 heterozygous and null adult limbs are shown. A. Skeletal preparations of Gdf-5 adults demonstrating the presence of a smaller autopod in the mutant. B. Shhcre and R26R alleles permit the fate-mapping of cells that expressed Shh.
RNA in situ hybridization
Section in situ hybridization was performed on embryos using an antisense probe against Gdf-5 and Shh. Gdf-5 antisense probe was synthesized from a plasmid construct provided by Dr. Cliff Tabin (Harvard Medical School). Shh antisense probe was synthesized from a plasmid construct provided by Dr. Martin Cohn (University of Florida). 8μL linearized plasmid was incubated with 2μl 10x transcription buffer, 2μl DIG-labelled nucleotide mix, 0.5μL RNase inhibitor, 1μL T7 polymerase, and 6.5μl water in a 20μL reaction. All reagents were purchased from Roche Diagnostics (Indianapolis, IN). Incubation was for 2 hours at 37°C and the reaction was purified on a mini Quick Spin Column (Roche). Probes were stored at −80°C.
The procedure for RNA in situ hybridization was adapted from previously published protocols (23, 24). In brief, embryos were dissected under RNase-free conditions (DEPC-treated solutions, instruments treated with RNaseZap® Solution (Ambion, Inc.)) and put into 4% PFA made with DEPC PBS, overnight at 4°C. After fixing, tissue was put into 30% sucrose in DEPC PBS overnight at 4°C. The next day they were placed in an equal volume of 30% sucrose and OCT (Tissue Tek; Torrance, CA) until the tissue sank. The sample was then embedded in 100% OCT on dry ice. Blocks were stored at −80°C until sectioning. Cryosectioning was done on a Leica cryostat at −20°C. Sections were cut at 16μm and stored at −80°C until being used for in situ hybridization. In situs were performed on adjacent sections for each time point (1st section on one slide, next section on a second slide, 3rd section on first slide, etc).
The procedure for in situ hybridization took 4 days. On day 1, the tissue sections were treated with hydrochloric acid, proteinase K, and acetic acid before being incubated with antisense probe. All steps were done in RNase-free conditions. Day 2 involved washing off unbound probe and incubating the tissue with an antibody that recognized the DIG-label on the probe. On Day 3 excess antibody was removed, and on day 4 a color reaction was performed to detect where the probe bound. After the color reaction was completed, slides were coverslipped and photographed. Section in situs were done with E10.5 and E12.5 wild-type embryos and E14.5 vertebral columns.
LacZ-fate mapping
To generate a fate-map of the notochord, the Shhcre allele (see background) in combination with the R26R allele was used. In the R26R allele, LacZ has been inserted into the ROSA locus. Mice containing the R26R allele transcribe LacZ under control of the broadly-active ROSA promoter but no protein is made because this allele contains a stop cassette flanked by loxP sites upstream of the reporter. Cells expressing the recombinase CRE that contain the R26R allele undergo a recombination event between the two loxP sites, resulting in the permanent removal of the stop cassette and expression of the reporter (Figure 2b). Importantly, once the stop cassette is removed, reporter protein will continue to be produced in the cell in which the recombination event occurred and in all descendants of that cell throughout the life of the animal. Expression will continue irrespective of whether CRE protein is present during later development. Initial activation of the reporter only occurs when both the R26R and the Shhcre alleles are present in the same animal. Using this system, cells and their descendants in various mutant backgrounds can be irreversibly marked and the fate of these cells throughout development can be determined (20, 21, 22).
Animals positive for the Shhcre and R26R alleles (as determined by PCR genotyping of embryo tails) were sacrificed by decapitation (newborns) or cervical dislocation. LacZ staining was done as described previously (21) with the exception that tissue was fixed in 0.2% paraformaldehyde before staining. The color reaction was stopped by rinsing samples with PBS (phosphate buffered saline) 3x 10 minutes and then placed into 4% PFA to fix tissue for paraffin embedding. Decalcification, prior to sectioning, was performed for animals that were older than 1 day with Cal-EX decalcifying solution (Fisher Scientific). Vertebral columns were then dehydrated in an ethanol series, cleared through a series of xylene washes, washed in Histowax and embedded in paraffin. Sections were cut at 7μm on a microtome (Leica), and coverslipped.
Hematoxylin and eosin staining
Animals that lacked R26R and Shhcre were embedded in paraffin and sectioned as mentioned above. Slides were dewaxed in xylene, rehydrated, stained with hematoxylin and eosin, dehydrated again and coverslipped.
Skeletal Preparations
Adult Gdf-5 heterozygote and mutant animals were euthanized, limbs were removed and skinned and then fixed in 4% PFA overnight. The next morning they were washed in PBS for 30 minutes. Alcian blue (0.02%) in ethanol and acetic acid was added and allowed to stain cartilage overnight. Next, limbs were dehydrated in an ethanol series before being placed in Alizarin red overnight. Limbs were then washed in 1% KOH until tissues were cleared enough to be photographed.
Results
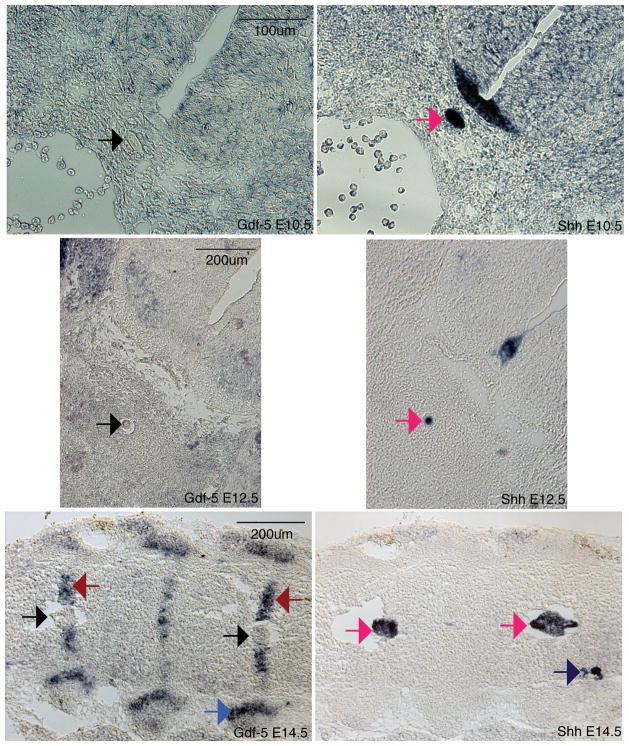
Gdf-5 is expressed in the annulus fibrosus but not the nucleus pulposus of the forming discs
Injection of GDF-5 within the disc has been reported to aid in disc repair (15, 16). However, the expression pattern of this gene during normal disc development had not been reported. To determine the in vivo localization of Gdf-5 mRNA, RNA in situ hybridization, a technique that determines mRNA localization in a tissue of interest, was performed. In E14.5 discs, Gdf-5 was found to be confined to the annulus fibrosus, which surrounds the nucleus pulposus (Figure 1, red arrow). Surprisingly, no staining was observed in the nucleus pulposus (Figure 1). Gdf-5 was also observed in the rib joints, limbs and parts of the brain, as previously reported (13) (Figure 1 and data not shown). In addition, no expression was observed in the notochord of E10.5 and E12.5 embryos (Figure 1). Gdf-5 is strongly expressed in the limbs of E12.5 embryos (data not shown). Shh in situs were performed on adjacent 16μm sections to mark either the notochord (E10.5 and E12.5) or the nucleus pulposus (E14.5). Shh was confined to the notochord, nucleus pulposus, and floorplate of the neural tube and was not expressed in the annulus fibrosus (Figure 1). Additionally, Shh was found in the dorsal root ganglia at E14.5 (Figure 1).
Figure 1. Gdf-5 and Shh expression in the intervertebral disc.
In situ hybridizations for Gdf-5 (left) and Shh (right). Gdf-5 is expressed in the annulus fibrosus of E14.5 embryos (red arrows) but not the notochord and nucleus pulposus (black arrows). Gdf5 is also present in the E14.5 rib joint (blue arrow). Shh is expressed in the notochord of E10.5 and E12.5 embryos and nucleus pulposus of E14.5 embryos (dark pink arrows). Shh is also present in the dorsal root ganglia (E14.5, dark blue arrow). Total magnification: 200× E10.5, 100× E12.5, E14.5.
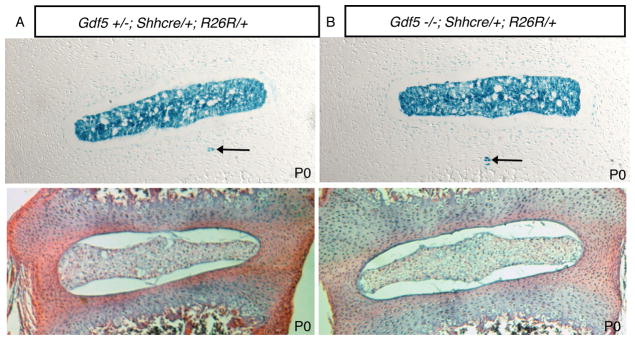
The embryonic notochord forms normal nuclei pulposi in newborn Gdf-5 null mice
GDF-5 is required for the formation of joints and recently has been demonstrated to be essential for the proper formation of adult nuclei pulposi (8). Previously, it was unknown if the disc defects reported in adult (20 week-old mice) were due to defects during embryonic development or the result of GDF-5 playing an important role during postembryonic development. To determine if GDF-5 was essential for formation of nuclei pulposi from the embryonic notochord we used the Shhcre and R26R alleles to fatemap the notochord in Gdf-5 null mice.
Newborn mutant Gdf-5 mice were identified at birth due to their characteristic small paw size (Figure 2a & reference 13). This autopod phenotype is highly penetrant and found in both the forelimbs and hindlimbs. Limbs in Gdf-5 heterozygous (Figure 2a) and wild type animals (not shown) are indistinguishable. Adults are shown here but newborns also displayed the same phenotype. The presence of Shhcre and R26R alleles was determined using PCR (see Materials and Methods). Examination of the formation of nuclei pulposi from the embryonic notochord revealed no defects in this structure in PO (newborn) mice (Figure 3). Consistent with what our laboratory previously observed in normal mice, the entire nucleus pulposus was comprised of cells that arose from the embryonic notochord (5, 6). In normal mice, rare notochord cells called notochordal remnants (blue cells outside the nucleus pulposus) were noted to reside in the annulus fibrosus (5). No increase in notochordal remnants was observed in the annulus fibrosus of Gdf-5 null mice (Figure 3).
Figure 3. Fatemap of notochord cells in P0 (newborn) Gdf-5 heterozygous and null animals.
Total magnification: 100×. Newborn Gdf-5 animals containing R26R and Shhcre alleles demonstrated that notochord cells were correctly localized to the nucleus pulposus. Both control and mutant mice contained notochordal remnants (black arrows). A. Gdf-5 heterozygote. B: Gdf-5 mutant. Lower panels: H&E-stain of newborn discs.
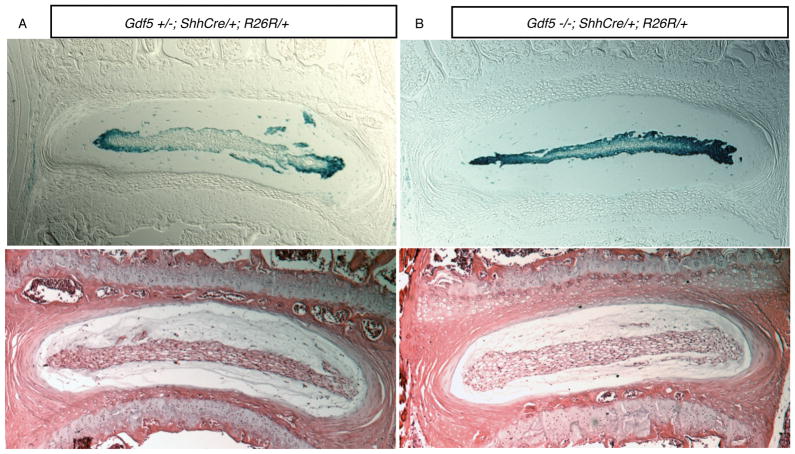
24 week-old Gdf-5 null mice contain nuclei pulposi derived from the embryonic notochord
The discs of 24 week-old mice containing the Shhcre and R26R alleles were examined to determine if nuclei pulposi were composed of notochord cells. The nuclei pulposi of both normal and Gdf-5 null mice were composed of cells derived from the embryonic notochord (Figure 4). There were no cells located in nuclei pulposi that were not β-galactosidase positive in Gdf-5 null mice indicating nuclei pulposi, similar to the discs of normal adult mice, are not composed of cells that have migrated from the surrounding annulus fibrosus or disc end plates.
Figure 4. Fatemap of notochord cells in 24 week-old Gdf-5 heterozygous and null mice.
Total magnification: 50×. Both animals have the Shhcre and R26R alleles. A. Gdf-5 heterozygote disc. B. Gdf-5 null disc. As in Figure 3, cells of the notochord were found to comprise the nucleus pulposus. Lower panels: H&E-stained discs.
Discussion
In this study we used the Shhcre and R26R alleles to determine if nuclei pulposi in Gdf-5 null mice form correctly. Our fate mapping in both PO (newborns) and animals aged to 24 weeks revealed that all cells of the nuclei pulposi were derived from the embryonic notochord. This is in agreement with published fatemaps of wild-type mice (5, 6). There were a few cells outside of the nucleus pulposus that were derived from the notochord. These cells are “notochordal remnants” and have previously been proposed in humans to form a rare type of tumor called chordoma (5, 25, 26). In Gdf-5 null animals there did not appear to be any difference in the number of notochordal remnants compared to control littermates.
Gdf-5 is expressed in many different tissues in the mouse and is known to be important for joint formation (9, 13, 27, 28). Previous reports have suggested that GDF-5 is required for formation of normal adult nuclei pulposi (8). Recently, a microarray analysis determined that Gdf-5 mRNA was highly expressed in the discs of E13.5 embryos but this study did not investigate which region of the disc Gdf-5 was expressed in (29). Our data indicates that this gene is expressed in the annulus fibrosus but not the notochord or nuclei pulposi. GDF-5 is a secreted protein and it is possible that GDF-5 protein is secreted into the nucleus pulposus from the annulus fibrosus. Supporting this hypothesis, in the adult human, GDF-5 protein has been localized to the nucleus pulposus via immunohistochemistry (30). Currently, we have only detected Gdf-5 mRNA localization during embryogenesis. It is possible that during postnatal life that Gdf-5 mRNA is expressed in nuclei pulposi and/or additional regions of the intervertebral discs.
Our studies indicate that Gdf-5 is not necessary for the transitioning of the embryonic notochord into nuclei pulposi in the mouse. The reported defect in the morphology of nuclei pulposi and decreases in collagen and proteoglycans in the Gdf-5 null mouse intervertebral discs may be due to a degenerative effect (8). On the basis of our fate mapping of the notochord, there appears to be no developmental defect of the nucleus pulposus upon loss of GDF-5 protein. We cannot rule out the possibility that a defect in the formation of the annulus fibrosus caused by Gdf-5 deficiency results in abnormal nuclei pulposi, however histological analysis suggested that the annulus fibrosus formed normally in mutant mice. We do note that our studies were performed on a mixed genetic strain background, which may influence the phenotypes produced upon removal of Gdf-5. This could explain the discrepancy between our results and that of Li and colleagues (8) in which they reported a deformity of the nucleus pulposus in Gdf-5 null. Our study used animals that were on a mixed background while Li et. al. did not. The presence of a mixed genetic background in our studies may have masked the role this gene may play in intervertebral disc development. However, the mixed genetic background of the Gdf-5 null mice had no effect on the Gdf-5 limb phenotype.
Artificial introduction of GDF-5 protein into the intervertebral discs has been proposed as a potential treatment for disc degeneration (15–17). In vivo studies were done which involved the injection of GDF-5 protein into the disc (15,16). Several in vitro studies have also been performed where cultured disc cells were treated with GDF-5 protein (15, 30). Gdf-5 null disc cells have also been treated with GDF-5 and shown to undergo upregulation of collagens and proteogylcans (8). Cultured disc cells have been transfected with expression vectors containing GDF-5 (18). Finally, GDF-5 has been overexpressed with an adenovirus in cultured disc cells (17). Our data suggests that in a normal disc, GDF-5 is produced in the annulus fibrosus, not the nucleus pulposus suggesting that GDF-5 disc-based therapies may increase their effectiveness by expressing GDF-5 in the annulus fibrosus of the intervertebral discs.
The use of a genetic-based system to follow the fate of notochord cells throughout the life of Gdf-5 null mice has allowed us to demonstrate that nuclei pulposi formation occurs normally in this mutant background. Our fate-mapping technique can be used in any mouse mutant background to determine if nuclei pulposi are formed correctly from the embryonic notochord. The ability to follow the fate of cells that normally form nuclei pulposi in various mutant backgrounds may uncover unique genetic pathways that are involved in forming the intervertebral discs.
Acknowledgments
This project was supported by NIH grants AG029353 (NIA/NIH) and AR055568 (NIAMS/NIH) to B.D.H.
References
- 1.Larson SJ, Maiman D. Surgery of the lumbar spine. New York, NY: Thieme; 1999. Lumbar Anatomy; pp. 1–12. [Google Scholar]
- 2.Raj PP. Intervertebral disc: anatomy-physiology-pathophysiology-treatment. Pain Pract. 2008 Mar;8(1):18–44. doi: 10.1111/j.1533-2500.2007.00171.x. [DOI] [PubMed] [Google Scholar]
- 3.Devereaux MW. Anatomy and examination of the spine. Neurol Clin. 2007 May;25(2):331–51. doi: 10.1016/j.ncl.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006 Apr;88( Suppl 2):21–4. doi: 10.2106/JBJS.E.01273. [DOI] [PubMed] [Google Scholar]
- 5.Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008 Dec;237(12):3953–8. doi: 10.1002/dvdy.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Risbud MV, Schaer TP, Shapiro IM. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn. 2010;239:2141–2148. doi: 10.1002/dvdy.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro IM, Risbud MV. Transcriptional profiling of the nucleus pulposus: say yes to notochord. Arthritis Res & Ther. 2010;12:117. doi: 10.1186/ar3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Leo BM, Beck G, et al. Collagen and proteoglycan abnormalities in the GDF-5-deficient mice and molecular changes when treating disk cells with recombinant growth factor. Spine (Phila Pa 1976) 2004 Oct 15;29(20):2229–34. doi: 10.1097/01.brs.0000142427.82605.fb. [DOI] [PubMed] [Google Scholar]
- 9.Thomas JT, Lin K, Nandedkar M, et al. A human chondrodysplasia due to a mutation in a TGF-beta superfamily member. Nat Genet. 1996 Mar;12(3):315–7. doi: 10.1038/ng0396-315. [DOI] [PubMed] [Google Scholar]
- 10.Thomas JT, Kilpatrick MW, Lin K, et al. Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat Genet. 1997 Sep;17(1):58–64. doi: 10.1038/ng0997-58. [DOI] [PubMed] [Google Scholar]
- 11.Polinkovsky A, Robin NH, Thomas JT, et al. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat Genet. 1997 Sep;17(1):18–9. doi: 10.1038/ng0997-18. [DOI] [PubMed] [Google Scholar]
- 12.Savarirayan R, White SM, Goodman FR, et al. Broad phenotypic spectrum caused by an identical heterozygous CDMP-1 mutation in three unrelated families. Am J Med Genet A. 2003 Mar 1;117A(2):136–42. doi: 10.1002/ajmg.a.10924. [DOI] [PubMed] [Google Scholar]
- 13.Storm EE, Huynh TV, Copeland NG, et al. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994 Apr 14;368(6472):639–43. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 14.Grüneberg H, Lee AJ. The anatomy and development of brachypodism in the mouse. J Embryol Exp Morphol. 1973 Aug;30(1):119–41. [PubMed] [Google Scholar]
- 15.Chujo T, An HS, Akeda K, et al. Effects of growth differentiation factor-5 on the intervertebral disc--in vitro bovine study and in vivo rabbit disc degeneration model study. Spine (Phila Pa 1976) 2006 Dec 1;31(25):2909–17. doi: 10.1097/01.brs.0000248428.22823.86. [DOI] [PubMed] [Google Scholar]
- 16.Walsh AJ, Bradford DS, Lotz JC. In vivo growth factor treatment of degenerated intervertebral discs. Spine (Phila Pa 1976) 2004 Jan 15;29(2):156–63. doi: 10.1097/01.BRS.0000107231.67854.9F. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Kroeber M, Hanke M, et al. Release of active and depot GDF-5 after adenovirus-mediated overexpression stimulates rabbit and human intervertebral disc cells. J Mol Med. 2004 Feb;82(2):126–34. doi: 10.1007/s00109-003-0507-y. [DOI] [PubMed] [Google Scholar]
- 18.Cui M, Wan Y, Anderson DG, et al. Mouse growth and differentiation factor-5 protein and DNA therapy potentiates intervertebral disc cell aggregation and chondrogenic gene expression. Spine Journal. 2008;8:287–295. doi: 10.1016/j.spinee.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Settle SH, Jr, Rountree RB, Sinha A, et al. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003 Feb 1;254(1):116–30. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 20.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001 Oct;2(10):743–55. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 21.Harfe BD, Scherz PJ, Nissim S, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004 Aug 20;118(4):517–28. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999 Jan;21(1):70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 23.Nieto MA, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–35. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- 24.Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13(2):225–37. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chugh R, Tawbi H, Lucas DR, et al. Chordoma: the nonsarcoma primary bone tumor. Oncologist. 2007 Nov;12(11):1344–50. doi: 10.1634/theoncologist.12-11-1344. [DOI] [PubMed] [Google Scholar]
- 26.Vujovic S, Henderson S, Presneau N, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006 Jun;209(2):157–65. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- 27.Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999 May 1;209(1):11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- 28.Francis-West PH, Abdelfattah A, Chen P, et al. Mechanisms of GDF-5 action during skeletal development. Development. 1999 Mar;126(6):1305–15. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 29.Sohn P, Cox M, Chen D, et al. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev Biol. 2010 Mar 9;10:29. doi: 10.1186/1471-213X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Maitre CL, Freemont AJ, Hoyland JA. Expression of cartilage-derived morphogenetic protein in human intervertebral discs and its effect on matrix synthesis in degenerate human nucleus pulposus cells. Arthritis Res Ther. 2009;11(5):R137. doi: 10.1186/ar2808. [DOI] [PMC free article] [PubMed] [Google Scholar]