Abstract
In contrast to other animal cell lines, the chicken pre-B cell lymphoma line, DT40, exhibits a high level of homologous recombination, which can be exploited to generate site-specific alterations in defined target genes or regions. In addition, the ability to generate human/chicken monochromosomal hybrids in the DT40 cell line opens a way for specific targeting of human genes. Here we describe a new strategy for direct isolation of a human chromosomal region that is based on targeting of the chromosome with a vector containing a yeast selectable marker, centromere, and an ARS element. This procedure allows rescue of the targeted region by transfection of total genomic DNA into yeast spheroplasts. Selection for the yeast marker results in isolation of chromosome sequences in the form of large circular yeast artificial chromosomes (YACs) up to 170 kb in size containing the targeted region. These YACs are generated by homologous recombination in yeast between common repeated sequences in the targeted chromosomal fragment. Alternatively, the targeted region can be rescued as a linear YACs when a YAC fragmentation vector is included in the yeast transformation mixture. Because the entire isolation procedure of the chromosomal region, once a target insertion is obtained, can be accomplished in ∼1 week, the new method greatly expands the utility of the homologous recombinationproficient DT40 chicken cell system.
Human/rodent hybrid cell lines containing single copies of human chromosomes are widely used for chromosome mapping and identification of specific genes. The use of these lines for modification and disruption of human genes is limited because of the low efficiency of gene targeting by homologous recombination in mammalian cells. Recently, a set of human monochromosomal hybrids has been developed in DT40 chicken pre-B cell lymphoma (Koi et al. 1997). The DT40 cells are highly proficient for homologous recombination, which allows the specific targeting of genes similar to that in yeast cells (Bezzubova et al. 1997). Because human chromosomes propagated in chicken cells can also be specifically targeted by homologous recombination (Dicken et al. 1996; Koi et al. 1997), such monochromosomal cell lines provide an opportunity to modify human genes in this background for further analysis.
In many cases, a modified chromosomal region needs to be isolated from genomic DNA. A general method of isolation of a targeted region is based on rescue of the targeting vector along with flanking genomic sequences. Because a vector usually contains a bacterial selectable marker and an origin of replication, rescue is accomplished by subsequent steps of endonuclease digestion and ligation of genomic DNA, followed by transformation into Escherichia coli cells. With this technique, fragments of genomic DNA up to 20 kb can be isolated (Smithies et al. 1985; Zakour et al. 1986; Grant et al. 1990; Kurdi-Haidar and Friedmann 1996). Isolation of bigger chromosome fragments requires yeast artificial chromosome (YAC) or bacterial artificial chromosome (BAC) cloning techniques that include time-consuming steps of library construction and subsequent identification of the region of interest among random clones (Burke et al. 1987; Shizuya et al. 1992; Ioannou et al. 1994).
In this report, we describe a new strategy for direct isolation of targeted chromosomal regions in the form of large YACs. Development of the new method was stimulated by two recent observations made during selective isolation of human DNA from human/rodent hybrids by transformation-associated recombination (TAR) cloning (Larionov et al. 1996a,b). First, we observed that large fragments of mammalian chromosomes can undergo circularization in yeast cells, presumably as a result of recombinational interaction between common, repeat sequences (such as SINEs and LINEs) near the broken ends. Second, YACs containing human or mouse DNA are capable of generating new telomeres when they lose one of their telomeric sequences (Larionov et al. 1996a). A new telomere is presumably generated from frequent (TG)n repeats present in genomic DNAs that are similar to yeast telomeric sequences. These two features form the basis of two related methods for rescue of targeted human sequences as described in this work.
RESULTS
A Strategy for Rescue of a Targeted Human Chromosome Region as YACs
A prerequisite for rescue of a targeted chromosomal region by recombination in yeast is the presence at the site of interest in the human chromosome a targeting vector with a yeast cassette containing a yeast selectable marker (TRP1), a centromere (CEN6), and an ARS element. This was achieved by site-directed homologous recombination of a yeast vector into a human chromosome in a chicken cell line with a high efficiency for homologous recombination.
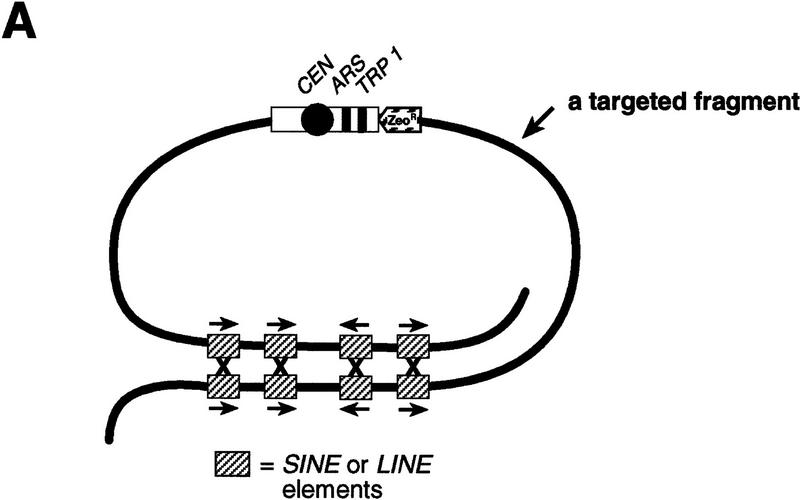
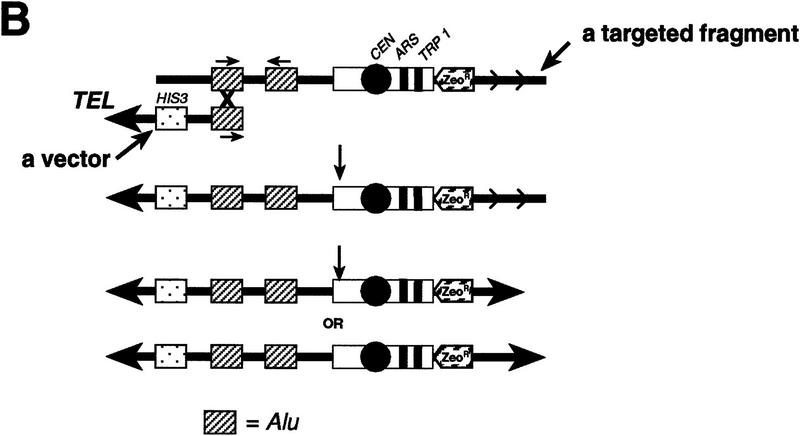
On the basis of our previous observations that (1) mammalian chromosome fragments can undergo circularization in yeast, and (2) linear YACs containing human or mouse DNA are capable of repairing broken ends (Larionov et al. 1996a,b), two different scenarios for rescue of a targeted chromosomal region were considered. According to the first scenario (Fig. 1A), total genomic DNA is directly transformed into yeast spheroplasts. Because a targeted chromosomal fragment contains the minimum requirement for its propagation in yeast cells (CEN, ARS, and TRP1), circularization leads to the generation of mitotically stable, circular YACs. The presence of multiple, repeated sequences in the targeted chromosomal fragment (such as SINEs and LINEs) provides multiple opportunities for circularization by homologous recombination. As a result, different size YACs are generated. Alternatively, genomic DNA can be transformed into yeast spheroplasts along with a YAC fragmentation vector containing a yeast telomere at one end and a common human Alu repeat at the other (Fig. 1B). Recombination between Alu repeats in the vector and the targeted chromosome fragment leads to establishment of a linear YAC containing one telomere. Healing of the other end of the YAC occurs by yeast-like telomeric repeats such as (TG)n present in the targeted fragment.
Figure 1.
Isolation of a targeted human chromosome region as circular or linear YACs by yeast transformation. (A) A scheme to rescue the targeted chromosome region as a series of circular YACs. Competent yeast spheroplasts are transformed with genomic DNA isolated from cells containing a targeted chromosome. The targeting vector (open box) incorporated into a specific region of a human chromosome contains a yeast centromere, CEN, a yeast origin of replication, ARSH4, a yeast selectable marker, TRP1, and a mammalian selectable marker, ZeoR. The position and orientation of several repeats (SINE or LINE) located near the end of the targeted chromosomal fragment are shown. The presence of multiple repeated sequences in the targeted chromosomal fragment provides many opportunities for homologous recombination. Recombination between inverted repeats leads to the establishment of series of different size circular YACs. Arrows indicate the orientation of the repeats in the targeted fragment. (B) A scheme to isolate the targeted chromosome region by recombination with a YAC fragmentation vector. Yeast spheroplasts are transformed with genomic DNA along with a YAC fragmentation vector marked by the HIS3 gene. This vector contains a yeast telomere (TEL) on one end and an Alu sequence at the other end. Recombination between the Alu sequence in the vector and an Alu sequence in the targeted chromosome fragment leads to the establishment of a linear semi-YAC (TEL–HIS3–CEN–ARS–TRP1) with one telomere. Because there are many yeast-like telomere sequences in the fragment (shown by small arrows) a series of different size linear YACs are generated from the semi-YAC by healing of the broken end. The position and orientation of several Alu sequences within the targeted fragment are shown.
Generation of a Hybrid Chicken Cell Line with a Specifically Targeted Human Chromosome 11
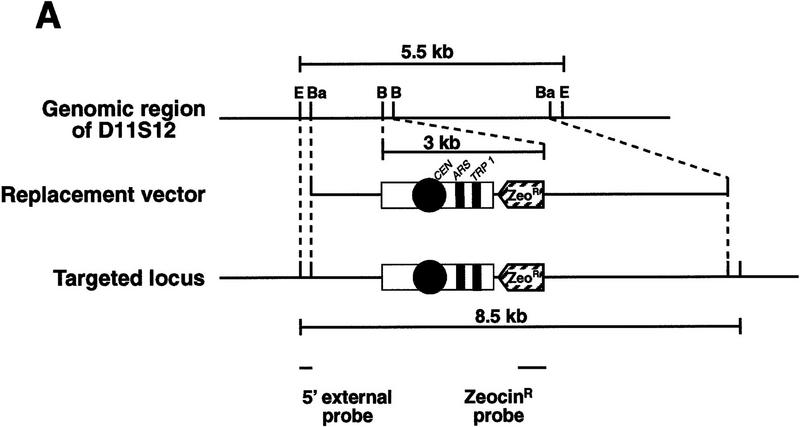
To test the rescue schemes proposed above, a gene targeting vector, pTV-3, was constructed for targeting of the D11S12 region of human chromosome 11. D11S12 was chosen for targeting because a novel tumor suppressor gene was mapped nearby (Koi et al. 1993; Bepler and Garcia-Blanco 1994). One end of the vector contains a 3.0 kb of the 5′ D11S12 sequence and the other contains a 2.0 kb of the 3′ D11S12 sequence. The targeting sequences were separated by a cassette containing mammalian and yeast selectable markers (ZeoR and TRP1), a yeast centromere (CEN6), and an ARS element (Figs. 1 and 2A). The linearized form of pTV-3 was electroporated into the DT40 chicken/human hybrid cell line containing two copies of human chromosome 11.
Figure 2.
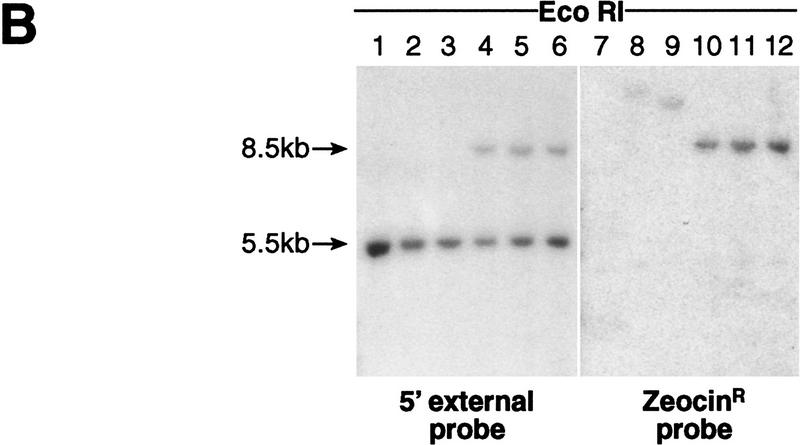
Targeting of the D11S12 locus. (A; Top) The genomic 5.5-kb EcoRI fragment includes a 5.0-kb BamHI-targeted region. (Middle) Scheme of the pVT-3 targeting vector linearized with EcoRI and NotI. Yeast sequences, CEN6, ARSH4, and TRP1, and the mammalian ZeoR gene are shown. The vector can replace the D11S12 region by double crossover. (Bottom) Structure of the targeted locus. As a result of the targeting replacement, the size of the EcoRI fragment becomes 8.5 kb. (B) Southern blots of EcoRI-digested DNA from the parental DT40 11-3 line (lane 1) and five drug-resistant clones (lanes 2–6). DNA was probed with a 5′ external probe (a 5′ external 0.5-kb fragment of the D11S12 locus), showing a 5.5-kb parental fragment and the expected 8.5-kb targeted fragment in three of the five clones (lanes 4–6). The same filter was probed with the Zeo probe (a 1.3-kb NaeI-ZeoR gene-containing fragment), showing an 8.5-kb ZeoR-containing fragment in the same three clones (lanes 10–12). (E) EcoRI; (Ba) BamHI; (B) BclI.
As shown in Figure 2A, a double crossover event would lead to replacement of the D11S12 targeted sequence by the ZeoR/CEN6/ARSH4/TRP1 cassette. To examine the targeting events, the EcoRI-digested DNA of 10 zeocin-resistant clones was hybridized with a probe external to the targeted region and the Zeo probe derived from the zeocin-resistance gene (Fig. 2B). If the correct targeting event had occurred, both probes would detect a 8.5-kb EcoRI fragment in the recombinants. If the illegitimate event had occurred, the D11S12 external probe would detect only the parental 5.5-kb EcoRI fragment, whereas the Zeo probe would detect a new chromosomal fragment in which the targeting vector was integrated. Among 10 clones analyzed, three showed the 8.5-kb band as expected for the specific targeting. In addition, a 5.5-kb band was observed corresponding to the second copy of chromosome 11 in which no integration had taken place (Fig. 2B).
Rescue of a Targeted Chromosomal Region as Circular YACs by Intramolecular Recombination in Yeast
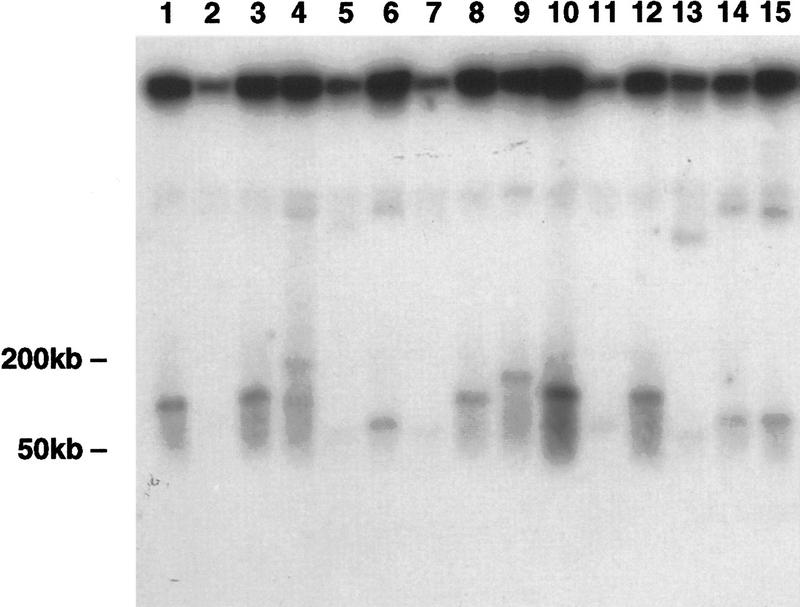
Total genomic DNA was isolated from the hybrid cell line DT40 11-3 containing the targeted human chromosome 11 and presented to yeast spheroplasts as described previously for a single gene isolation by TAR cloning (Larionov et al. 1997). Transformants were selected on synthetic medium lacking tryptophan. Fourteen Trp+ transformants were obtained in twenty independent transformations when 5 μg of genomic DNA was used with 2 × 109 yeast spheroplasts in each experiment. To demonstrate the presence of the targeted chromosomal region in the transformants, chromosomal size DNA from the Trp+ clones was separated by transverse alternating field electrophoresis (TAFE) gel electrophoresis, blotted, and hybridized with the D11S12 probe (see Methods). All the clones contained predicted sequences outside of the targeting vector. As expected for circular molecules (Larionov et al. 1996b), the D11S12-hybridizing material was retained in the starting wells of the TAFE gel. A low dose of ionizing radiation (5 kilorads) was used to induce, on average, less than one random break per circular molecule. The largest linear molecules, corresponding to a single break, varied from 70 to 170 kb in size for different transformants (Fig. 3).
Figure 3.
Characterization of circular YACs rescued in yeast. Chromosomal size DNA was isolated from 14 yeast transformants, exposed to a low dose of γ-rays, separated by TAFE, blotted, and hybridized with the D11S12 probe. The strong signals at the positions of the starting wells correspond to large circular molecules. The lagging bands correspond to circular molecules linearized by radiation. The weak hybridizing bands migrating between the 200-kb marker and the wells correspond to covalently closed forms of YAC DNAs. Each lane except lane 2 corresponds to an independent Trp+ isolate. (Lanes 2,15) The same isolate. The size of the linearized YACs was determined by use of λ oligomers.
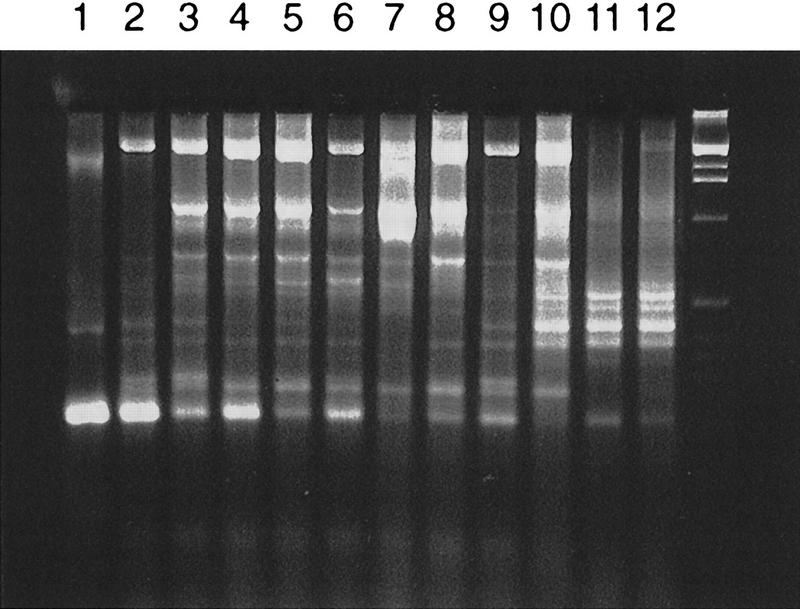
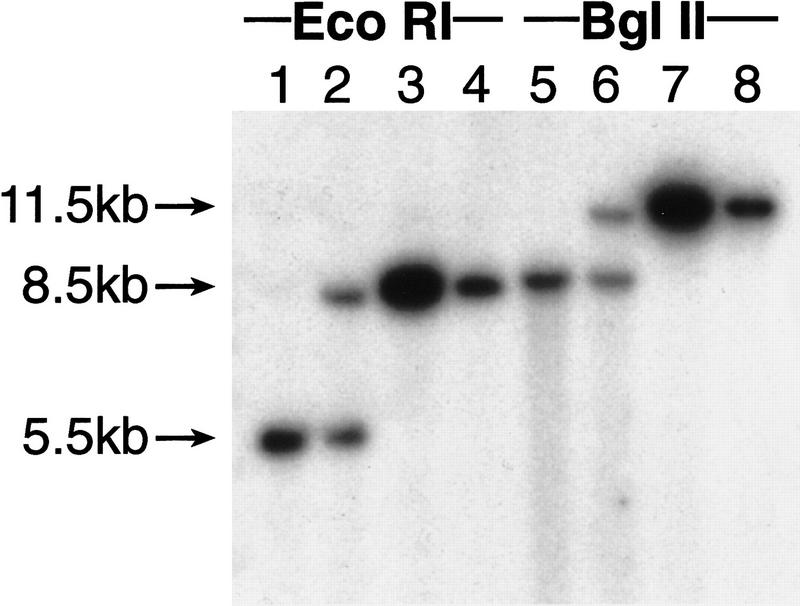
All Trp+ transformants contained the ZeoR gene as determined by PCR analysis and exhibited related Alu profiles (data not shown). Inter-Alu PCR analysis of the YACs revealed several common bands (Fig. 4) confirming that these YAC clones derived from the same chromosomal region. The presence of common genomic sequences flanking a targeting vector was also shown by hybridization of the EcoRI- or BglII-digested DNA isolated from the transformants with a probe 5′ upstream of the D11S12 sequence used for construction of the targeting vector (see Fig. 2). The probe identifies the predicted genomic fragments (Fig. 5). We concluded from these results that all the Trp+ transformants contained circular YACs carrying the targeting vector plus the human genomic DNA sequences flanking the targeted site.
Figure 4.
Alu PCR analysis of YACs containing the targeted chromosome region. Nine isolates of circular Trp+ YACs and three isolates of Trp+ His+ linear YACs were characterized. Total yeast DNA was purified from each isolate and PCR amplified by Alu-specific primers. (Lanes 1–9) Trp+ YACs; (lanes 10–12) Trp+ His+ YACs. The last lane corresponds to a 1-kb ladder.
Figure 5.
Comparison of the YACs with genomic DNA. DNA was isolated from Trp+ yeast transformants and chicken cells containing the targeted human chromosome 11, digested by EcoRI or BglII, and hybridized to the 5′ external probe shown in Fig. 2. (Lanes 1,5) Parental DT40 11-3 line. (Lanes 2,6) DT40 11-3 cells carrying the targeted human chromosome 11. (Lanes 3,4,7,8) Two different YAC isolates. In prediction that the YACs contain genomic sequences flanking the targeting vector, digestion identifies identical bands in YACs and genomic DNA.
Rescue of the Targeted Chromosomal Region as Linear YACs by Intermolecular Recombination in Yeast
To investigate rescue of the targeted chromosomal region by intermolecular recombination in yeast, spheroplasts were transformed with genomic DNA isolated from the DT40 11-3 cells along with the YAC fragmentation vector pBP465 (Alu–HIS3–Telomere) (Pavan et al. 1991). Transformants were selected on synthetic medium lacking histidine. Approximately 3000 His+ transformants were obtained in 10 standard transformation experiments (with 1 μg of the vector, 5 μg of genomic DNA, and 2 × 109 yeast spheroplasts in each experiment). The transformants were checked for the presence of the unselected TRP1 marker. Three Trp+ clones were identified. In all these clones, TRP1 was linked to HIS3. TAFE gel electrophoresis showed that these transformants contained linear YACs that hybridized with the D11S12 probe. Sizes of the YACs were 70, 100, and 150 kb (data not shown). Inter-Alu PCR products obtained with the Trp+His+ YACs were similar to those obtained for circular Trp+ YACs (Fig. 4). Thus, we conclude that these YACs were also derived from the D11S12 targeted region.
DISCUSSION
In this report, we have demonstrated that a human chromosomal region targeted by a vector containing a yeast selectable marker, centromere, and an ARS element can be selectively isolated as circular or linear YACs.
Direct transformation of genomic DNA into yeast spheroplasts and selection of the transformants for the yeast marker leads to isolation of the targeted region as a circular DNA molecule up to 170 kb in size. These YACs are likely the result of intrachromosomal recombination between free DNA ends. Because there are many potential sites for recombination within a given genomic fragment (on the basis of the frequency of repeated elements in human DNA), circular molecules of different sizes are generated from the same region. These circular YACs propagate stably in yeast cells and can be isolated by the alkaline lysis procedure (Devenish and Newlon 1982) for further analysis.
An alternative method for rescue of the targeted chromosomal fragment is transformation of genomic DNA into yeast spheroplasts along with an Alu-containing YAC fragmentation vector. Recombination between the vector and the targeted chromosomal fragment results in generation of an unstable, linear semi-YAC containing only one telomere, which is stabilized through new telomere formation at endogenous yeast telomere-like sequences. Although the nature of these stabilizing sequences is not yet established, such sequences are frequent in the human genome (Larionov et al. 1996a; N. Kouprina and V. Larionov, unpubl.). Use of the Alu-containing YAC fragmentation vector allows isolation of chromosomal fragments up to 150 kb flanking the targeting vector.
Given that the average distance between Alus is 4 kb, it is surprising that the YACs obtained by both methods were >100 kb. This result can be explained if recombination at homologous sequences occurs preferentially near broken ends.
On the basis of our previous results, no or few rearranged/chimeric clones are generated during TAR cloning (Larionov et al. 1996b, 1997; Kouprina et al. 1998; N. Kouprina and V. Larionov, unpubl.). In accordance with that observation, none of the YACs hybridized with chicken DNA (data not shown). Occurrence of human–human chimeras seems unlikely because human DNA represents a small percentage of the total DNA in chicken–human hybrid cells. However, even if some clones were rearranged, a genomic copy could be identified easily among multiple isolates.
The new method for isolation of targeted chromosomal regions greatly expands the utility of the homologous recombination-proficient DT40 chicken cell system. Human genes modified by targeting in chicken cells, can be rescued by yeast transformation and then transferred back into mammalian host cells for further functional studies.
The approach described here has many other potential uses. Because large fragments can be isolated rapidly, it provides an opportunity to identify new genes linked to a targeted site. This method can also be employed for the isolation of chromosomal regions containing cis-active elements controlling mitotic chromosome transmission (Brown et al. 1994). Furthermore, because a functional copy of a mammalian LINE retransposon was isolated recently (Moran et al. 1996; Sassaman et al. 1997), our method has more general application. It can be used for isolation of chromosome regions targeted by retransposon. Thus, the method can greatly facilitate a large-scale analysis of human and mouse genes after transposon-induced mutagenesis. Such an approach has been very powerful for analysis of the yeast Saccharomyces cerevisiae genome (Burns et al. 1994; Ross-Macdonald et al. 1997).
METHODS
Cell Lines
Recombination-proficient chicken DT40 11-3 cells containing two copies of human chromosome 11 were described previously (Koi et al. 1997). The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (500 ml, GIBCO BRL) supplemented with tryptose phosphate (30 ml, 2.95%, GIBCO BRL), new born calf serum (60 ml, GIBCO BRL), chicken serum (24 ml, GIBCO BRL), 1 m HEPES (6 ml, pH 7.3, GIBCO BRL), Nystatin (0.5 ml, GIBCO BRL), and 1 mg/ml of G418 (GIBCO BRL). The cells were grown in a 10% CO2/90% air incubator at 37°C.
Construction of the Targeting Vector
The targeting vector pTV-3 for replacement of the D11S12 locus on human chromosome 11p15.5 (Feder et al. 1985) was constructed with a 5.0-kb BamHI fragment of the D11S12 locus from plasmid pADJ762 (Barker et al. 1984). This fragment was cloned into a unique BamHI site of pBluescript II SK to generate plasmid pK11. A 1.7-kb cassette containing a yeast centromere (CEN6), a yeast origin of replication (ARSH4), and a yeast selectable marker (TRP1) was PCR amplified from plasmid pRS314 (Sikorski and Hieter 1989) with two primers: 1–49 (5′- GCCGGATCCCCCCGAAAAGTGCCACCTGGGTCCTTTTCATCACGTGCTA-3′) and 3–35 (5′-GCCGGATCCCGCATCTGTGCGGTATTTCACACCGC-3′). 1–49 and 3–35 correspond to positions 4181–4222 and 1216–1192 of pRS314, respectively, (GenBank accession no. U03440). The PCR product generated by the 1–49 and 3–35 primers was treated with BamHI and cloned into the BamHI-linearized pZeoSV plasmid (Invitrogen, San Diego) carrying a mammalian selection marker, the ZeoR gene, for selection in animal cells. The resulting plasmid pYZT containing CEN6, ARSH4, TRP1, and the ZeoR gene was cleaved with BglII. A 3.0-kb BglII fragment containing the ZeoR gene and the yeast cassette was gel purified and cloned into pK11 linearized at the BclI site. The resulting 10.5-kb targeting vector pTV-3 was digested by EcoRI and NotI before electroporation into the DT40 11-3 cells. Double digestion leads to release of the ZeoR/CEN6/ARSH4/TRP1 cassette flanked by D11S12-specific sequences (2.0 and 3.0 kb in size on the basis of the position of the BclI site). The Alu-containing YAC fragmentation vector pBP465 (Alu–HIS3–Telomere) (Pavan et al. 1991) was used for rescue of the targeted chromosome region via intermolecular recombination in yeast. The vector was cut with SalI to yield a molecule bounded by Alu and telomere sequences on opposite ends. Both vectors were purified by CsCl/ethidium bromide centrifugation for these experiments.
Gene Targeting and Southern Analysis
Twenty-five micrograms of the pTV-3 vector DNA treated with EcoRI plus NotI was added to 107 DT40 11-3 cells resuspended in 1.0 ml of serum-free RPMI1640 medium. The cells were transferred into an electroporation cuvette and incubated for 10 min at room temperature before electroporation. The cells were electroporated by use of a Gene Pulse apparatus (Bio-Rad) at 25 μF with 550 V. After electroporation, the cells were kept on ice for 10 min and then plated into 24 well dishes. Two days later, the cells were selected with zeocin (200 μg/ml) (Invitrogen) with medium changes every two days. After 3–4 weeks of selection, drug-resistant clones were isolated, expanded, and analyzed. High molecular weight DNA was isolated from the transformants and analyzed by Southern blot hybridization with 32P-labeled probes. The 5′ external probe for D11S12 was a 0.5-kb BamHI–EcoRI fragment isolated from pADJ762 (Barker et al. 1984). The ZeoR gene probe was a 1.3-kb NaeI fragment from pZeoSV plasmid. The probes were labeled by the random-priming method (Feinberg and Vogelstein 1994).
Host Yeast Strain and Spheroplasts Transformation
For transformation, the highly transformable S. cerevisiae strain VL6-48 (MATa, his3-Δ200, trp1-Δ1, ura3-52, lys2, ade2-101, met14) (Larionov et al. 1997), which has the TRP1 and HIS3 genes deleted was used. Spheroplasts that enable efficient transformation were generated by use of a previously described protocol (Larionov et al. 1996a,b). Agarose plugs (100 μl) containing ∼5 μg of high molecular weight genomic DNA were prepared from the chicken DT40 11-3 hybrid cells containing the targeted human D11S12 locus and were used for yeast transformation. Yeast transformants were selected on synthetic complete medium plates lacking either tryptophan or histidine.
Characterization of YAC Clones
Chromosomal size DNAs from yeast transformants were separated by TAFE, blotted, and hybridized with the specific probes as described previously (Larionov et al. 1996a,b). To estimate the size of circular YACs, agarose DNA plugs were exposed to a low dose of γ-rays (5 kilorads) before TAFE analysis. To identify fragments containing Alu sequences (Alu profiles), 1 μg of total yeast DNA was digested to completion with TaqI. Samples were run by gel electrophoresis, transferred to a nylon membrane, and hybridized with an Alu probe (Larionov et al. 1996a,b). Inter-Alu PCR was carried out in a 50-μl final volume containing 50 ng of total yeast DNA isolated from yeast transformants as described previously (Larionov et al. 1996b). The Alu PCR primer sequences (A1 and B1), were 5′-GGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGA-3′ and 5′-GGAGGCTGAGGCAGGAGAATCGCTTGAACCCGGGAGGCGG-3′, respectively. Two primers Zeo217A (5′-AGTGCCGTTCCGGTGCTCAC-3′) and Zeo217B (5′-ACTCGGCGTACAGCTCGTCC-3′) were used for identification of the ZeoR gene in transformants. These primers amplify a 217-bp sequence of the ZeoR-coding region.
Acknowledgments
We gratefully acknowledge critical discussions with M.A. Resnick during this work. We thank R. Schaaper and R. Wiseman for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL larionov@niehs.nih.gov; FAX (919) 541-7593.
E-MAIL koi@niehs.nih.gov; FAX (919) 541-7666.
REFERENCES
- Barker D, Holm T, White RA. A locus on chromosome 11p with multiple restriction site polymorphisms. Am J Hum Genet. 1984;36:1159–1171. [PMC free article] [PubMed] [Google Scholar]
- Bepler G, Garcia-Blanco MA. Three tumor suppressor regions on chromosome 11p identified by high-resolution deletion mapping in human non-small-cell lung cancer. Proc Natl Acad Sci. 1994;91:5513–5517. doi: 10.1073/pnas.91.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde JM. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, Brown WRA. Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum Mol Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- Burke DT, Carle GF, Olson MV. Cloning of large segments of DNA into yeast by means of artificial chromosome vectors. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes & Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Devenish RJ, Newlon CS. Isolation and characterization of yeast ring chromosome III by a method applicable to other circular DNAs. Gene. 1982;18:277–288. doi: 10.1016/0378-1119(82)90166-4. [DOI] [PubMed] [Google Scholar]
- Dieken ES, Epner EM, Fiering S, Fournier RE, Groudine M. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nature Genet. 1996;12:174–182. doi: 10.1038/ng0296-174. [DOI] [PubMed] [Google Scholar]
- Feder J, Yen L, Wijsman E, Wang L, Wilkins L, Schroder J, Spurr N, Cann H, Blumenberg M, Cavalli-Sforza LL. A systematic approach for detecting high-frequency restriction fragment length polymorphisms using large genomic probes. Am J Hum Genet. 1985;37:635–649. [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. (Addendum) Anal Biochem. 1994;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nature Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- Koi M, Johnson LA, Kalikin LM, Little PFR, Nakamura Y, Feinberg AP. Tumor cell growth arrest caused by subchromosomal transferable DNA fragments from chromosome 11. Science. 1993;260:361–364. doi: 10.1126/science.8469989. [DOI] [PubMed] [Google Scholar]
- Koi M, Lamb PW, Filatov L, Feinberg AP, Barrett JC. Construction of chicken x human microcell hybrids for human gene targeting. Cytogenet Cell Genet. 1997;76:72–76. doi: 10.1159/000134519. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Annab L, Graves J, Afshari C, Barrett JC, Resnick MA, Larionov V. Functional copies of a human gene can be directly isolated by TAR cloning with a small 3′ end target sequence. Proc Natl Acad Sci. 1998;95:4469–4474. doi: 10.1073/pnas.95.8.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi-Haidar B, Friedmann T. Simplified plasmid rescue of host sequences adjacent to integrated proviruses. Gene. 1996;168:199–203. doi: 10.1016/0378-1119(95)00753-9. [DOI] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci. 1996a;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Resnick MA. Highly selective isolation of human DNAs from rodent-human hybrid cells as circular yeast artificial chromosomes by transformation-associated recombination cloning. Proc Natl Acad Sci. 1996b;93:13925–13930. doi: 10.1073/pnas.93.24.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Solomon G, Barrett JC, Resnick MA. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc Natl Acad Sci. 1997;94:7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian Jr HH. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Pavan WJ, Hieter P, Sears D, Burkhoff A, Reeves RH. High-efficiency yeast artificial chromosome fragmentation vectors. Gene. 1991;106:125–127. doi: 10.1016/0378-1119(91)90576-w. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Sheehan A, Roeder GS, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:190–195. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian Jr HH. Many human L1 elements are capable of retrotransposition. Nature Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Zakour RA, Schaaper RM, Glickman BW. Introduction, rescue and expression of plasmid genes in mammalian cells and Escherichia coli. Mutat Res. 1986;163:3–13. doi: 10.1016/0027-5107(86)90052-7. [DOI] [PubMed] [Google Scholar]