Abstract
B cell developmental pathways in teleost fishes are poorly understood. In the absence of serological reagents, an alternative approach to dissecting teleost B cell development is to use transcription factors that are differentially expressed during B cell development. This review discusses the structure and function of six transcription factors that play essential roles during teleost B cell development: Ikaros, E2A, EBF, Pax5, Blimp1, and XbpI. Research on alternative splicing of both the Ikaros and Pax5 genes in rainbow trout is presented, including their functional significance. An application is discussed that should aid in elucidating teleost B cell development and activation, by using transcription factors as developmental markers in flow cytometric analysis. Possible future studies in teleost B cell development are suggested in the context of gene regulation. Lastly, broader impacts and practical applications are discussed.
1. Introduction
With the introduction of the jaw, the capacity to efficiently respond to infections increased greatly in vertebrate animals, and this correlates with the emergence of adaptive immunity (Matsunaga and Rahman, 1998). Consequently, the lymphocytes of fishes, including teleost species, are highly diverse, similar to what is seen in mammals. However, teleosts lack bone marrow, the mammalian site for lymphocyte development or lymphopoeisis; instead, they use the anterior kidney. Because, presumably, teleost species have not changed the location for lymphopoiesis over the past several hundred million years, the anterior kidney must provide an excellent site. To date, in the absence of serological tools, the process of lymphopoeisis in the anterior kidney remains quite elusive. Exploring the expression of (lymphoid-specific) transcription factors in anterior kidney cells should shed light on the processes that drive teleost lymphoid development. In this regard, it is of interest that many of the transcription factors that drive lymphocyte development in jawed fish are already present in jawless fish and even invertebrates, well before the emergence of adaptive immunity. Hence it appears that the structure, but not necessarily the function, of transcription factors is highly conserved. This review will focus on teleost transcription factors expressed during B cell development, by comparing extensive knowledge of the mammalian system with the limited information available in the teleost.
2. B cell development
After birth, mammalian hematopoiesis takes place exclusively in the bone marrow, a complex tissue that has been studied extensively. The bone marrow environment is necessary and sufficient to generate all immune cells, including hematopoietic stem cells, lymphoid and myeloid progenitors, as well as their more differentiated immune cell descendents, with the exception of T cell development, which takes place in the thymus. Mature, surface-IgM expressing B cells are generated through the B lymphopoiesis pathway; earliest progenitors include the common lymphoid progenitor (CLP), followed by pro-B cell, pre-BI, large and small pre-BII, and immature B cell stages, as shown in Figure 1 (reviewed in Melchers and Kincade, 2004). Immature B cells leave the bone marrow, and travel through the blood to the spleen to complete maturation (Melchers and Kincade, 2004). Small populations of (long-lived) Ig-secreting cells are stored in the bone marrow (reviewed in Rajewski and Radbruch, 2004) where they provide long-term humoral protection.
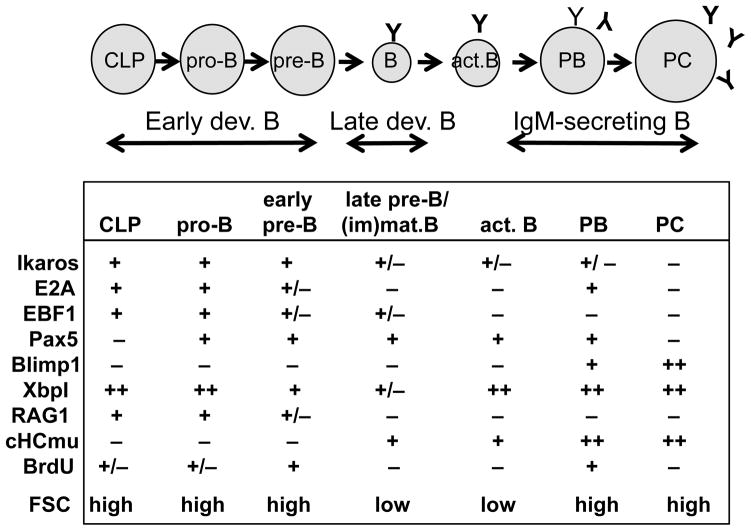
Figure 1.
Defining the major stages of B cell development in vertebrate species based on the combinatorial expression of (B cell-specific) transcription factors Ikaros, E2A, EBF1, Pax5, Blimp1, and XbpI, the recombinase RAG1, cytoplasmic immunoglobulin heavy chain mu (cHCmu), cell proliferation (BrdU), and cell size or forward scatter (FSC).
In the rainbow trout, the entire kidney contains immune cells: the anterior kidney has the highest concentration of developing B lymphoid cells, and also contains low levels of antibody-secreting cells (ASC) (Bromage et al., 2004; Zwollo et al., 2005, 2008, 2010), interdigitated with adrenal-like tissue, and has no renal function (it lacks nephrons) (Zapata and Cooper, 1990, Milano et al, 1997). In contrast, posterior kidney possesses both renal and immune tissue (Zapata and Cooper, 1990, Zwollo et al. 2005, 2008). The terms anterior and posterior kidney are often poorly defined in the scientific literature. In order to more accurately define teleost kidney function, we have proposed a nomenclature for rainbow trout, which can be applied to all Oncorhyncus species (Zwollo, unpublished observations) as follows: the kidney is (arbitrarily) divided into five segments of 7 vertebrate lengths each, named K1-K5, with K1 being the most anterior section, and K5 the most posterior (Zwollo et al., 2005). Hence, K1 is equivalent to head kidney or anterior kidney, but with a highly defined border.
The teleost spleen functions as a major secondary immune organ, as in mammalian species, with abundant IgM+ mature B cells; IgM-secreting cells are generated in LPS-activated cultures derived from splenic B cells (Zapata and Cooper, 1990, Kaattari and Irwin, 1985, Bromage et al, 2004, Zwollo et al., 2005, 2008). The blood of the rainbow trout also contains mature, LPS-sensitive IgM+ B cell populations (Bromage et al, 2004, Zwollo et al., 2005), however, ex vivo induction in PBLs is moderate compared to that of the spleen.
One approach to begin dissecting the molecular pathways of teleost B cell differentiation is to examine developmentally regulated expression of B cell-specific transcription factors in teleost immune tissues. This approach has proven fruitful in studies of mammalian B cell development (reviewed in Northrup and Allman, 2008). Importantly, the structure of transcription factors shows a remarkable level of conservation between vertebrate species. The most highly conserved region of transcription factors is typically the DNA binding domain, with inter-species sequence homologies often close to 100%. Hence, use of transcription factors as developmental markers in less well-studied animals such as the teleost provides an attractive comparative approach.
3. Transcription factors during vertebrate B cell development and activation
Transcription factors are proteins that interact with specific DNA sequences within the enhancers or promoters of target genes, and can modify the chromatin structure surrounding a target gene to change accessibility for transcription. Ultimately, transcription factors regulate the rate of transcription into RNA, either increasing it or decreasing it. The typical transcription factor consists of separate functional domains, including a DNA binding, activation/repressor, and, if it functions as a dimer, dimerization domain. Transcription factors are classified based on the type of DNA binding domain they possess, including the homeodomain, zinc-finger, leucine zipper, helix-loop helix, or winged-helix proteins.
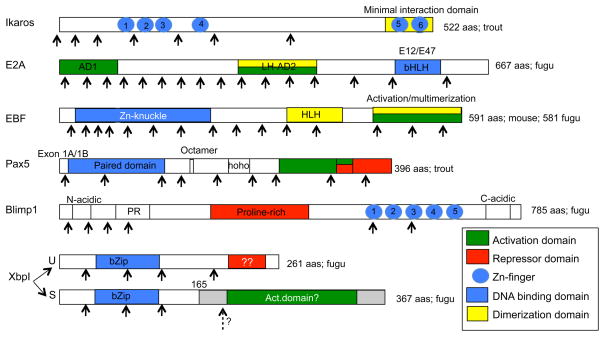
The major transcription factors that play roles in vertebrate B cell development, and which have been characterized at least to some extent in teleosts, include Ikaros, E2A, Early B cell Factor-1 (EBF1), Paired box-5 (Pax5), B lymphocyte-induced protein-1 (Blimp1), and X-box binding protein-1 (XbpI). This review will begin by briefly discussing each transcription factor separately below. The expression pattern for each transcription factor during B cell maturation is shown in Figure 1, while the organization of each transcription factor, including its functional domains, is shown in Figure 2.
Figure 2.
Organization of six transcription factors expressed during teleost B cell development. Coding regions for six transcription factors are shown. The species from which the sequence information was used, is indicated on the right. Arrows indicate approximate intron-exon boundaries. Colors indicte the functional domains. Dotted arrow: undetermined boundary. U: unspliced, S: spliced form of XbpI. Grey boxed: spliced form of XbpI (XbpI-S) has a frame-shift starting in aa position 165.
Ikaros
In mammals, Ikaros is first expressed during the earliest stage of immune cell development, in hematopoeietic stem cells (HSCs) and is necessary for T, B, and NK development (reviewed in Georgopoulos et al 1997). Within the B cell lineage, Ikaros is highly expressed in CLP, pro-B, and pre-B cells (Figure 1; Liberg et al. 2003). Other members of the Ikaros gene family include Aiolos, Helios and Eos. Ikaros is a zinc-finger type transcription factor and forms homo-and heterodimers with its family members, leading to highly diverse functions (Hahm et al., 1994, Georgepoulos et al. 1992). Ikaros commonly functions as a gene silencer, and regulates proliferation and maturation of lymphocytes through chromatin remodeling complexes and histone deacetylation interference (reviewed in Liberg et al., 2003; illustrated in Figure 3). Ikaros has a total of 6 zinc fingers, 4 N-terminal, and 2 C-terminal (Hahm et al.,1994, and Georgopolous et al, 1992), as shown in Figure 2. The N-terminal Zn-fingers are necessary for DNA binding, and the C-terminal fingers for dimerization (Georgeopolous et al. 1994). Zinc-finger containing Ikaros genes are present in all vertebrates (Haire et al., 2000). Ikaros is extensively alternatively spliced, as will be discussed in the next section.
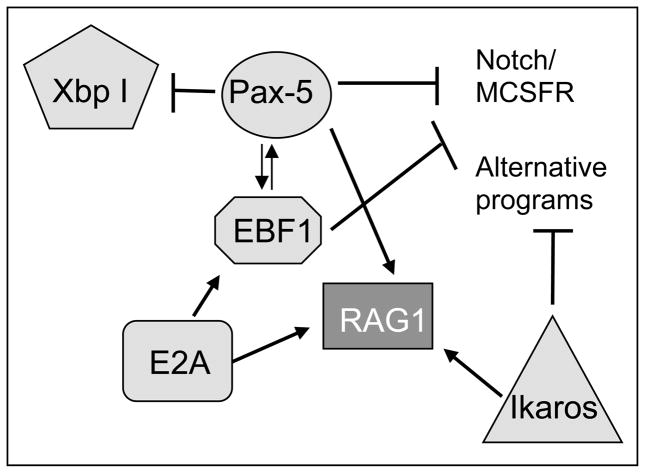
Figure 3.
Functional relationships between transcription factors during vertebrate B cell development. Transcription factors are shown in light grey boxes, the recombinase RAG1 in dark grey. Arrows indicate activation, T indicates repression.
The first teleost Ikaros homolog was isolated and characterized in the rainbow trout (Hansen et al., 1997). Comparison between mouse and trout Ikaros cDNAs revealed a strong conservation of the DNA binding domain (98% homology) as well as conservation of the relative location of the other functional domains, and the number of exons (Figure 2). Trout Ikaros has been detected in both primary immune tissues (thymus, anterior kidney) and secondary immune tissues (spleen) of juvenile trout (Hansen et al., 1997), and as such Ikaros expression is highly conserved between mouse and trout.
An Ikaros homolog has also been isolated in zebrafish. During embryonic development, Ikaros is expressed in the ventral side of the dorsal aorta (the hematopoiesis site in birds and mammals), and near the forming thymus (Willett et a., 2001). In adult zebrafish, transcripts have been detected in primary immune sites anterior kidney and thymus, while low and varying levels were detected in spleen (Willett et al., 2001). One study investigated the function of Ikaros using zebrafish that made a mutant form of the Ikaros protein, lacking the two C-terminal Zn-fingers and as a result, no longer able to dimerize with other Ikaros proteins (Schorpp, et al., 2006). Previously, it had been suggested that zebrafish and rainbow trout have two different B cell lineages, one expressing IgM, and the other IgT (Danilova et al, 2005, Hansen et al., 2005) and recently it was shown that these B cell lineages have functionally different properties (Zhang et al., 2010). Interestingly, the zebrafish expressing the mutant Ikaros protein lacked IgT expressing cells. Low levels of IgM+, oligoclonal B cells were detected in the anterior kidney, although these cells formed later during embryonic development and had a lower frequency of productive Ig HC rearrangements. Hence, at least in zebrafish, proper dimerization of Ikaros is required for IgT-rearrangement, and facilitates IgM-rearrangement.
E2A
This gene is one of three mammalian E protein family members, which also include HEB and E2-2. All 3 genes encode helix-loop-helix type transcription factors (reviewed in Engel and Murre, 2001). E-proteins can form homo- and heterodimers with each other and with other members of the HLH family (Engel and Murre, 2001). Transcriptional activity of E proteins, specifically the ability to bind target DNA, is inhibited through heterodimerization with Id proteins (Engel and Murre, 2001). The E2A gene encodes two proteins, E12 and E47, through alternative splicing of the HLH domain (Figure 2; Murre et al., 1989). In developing B cells, the main functional E-protein is the E47 homodimer (Murre et al., 1991).
E2A drives early B cell development and regulates expression of Lamda5, EBF1, TdT and RAG1 genes (Kee and Murre, 1998; Figure 3), but is also essential for Ig rearrangement and expression during terminal differentiation, and plays a role during class switching (Murre et al., 2001, Quong et al., 1999). Not surprisingly then, high expression of E2A is seen during early B cell development and then again in proliferating germinal center B cells (Murre et al., 2001, Quong et al., 1999).
Of the many important scientific contributions by Greg Warr, sorting out the structure and function of teleost E-proteins was just one. His work, in collaboration with others, showed that while mammals only have three E-protein genes, puffer fish have five, including HEB, E2-2, E2A1, E2A2, and EX (Hikima et al., 2005a). While puffer fish possess two different E2A homologs, E2A1 (the E12/E47 homolog shown in Figure 2), and E2A2 (which lacks alternative splicing), only one E2A gene sequence has been isolated for catfish (Hikima et al., 2005a, 2005b).
Interestingly, while in mammals E2A is the major E-protein in B cells, in catfish it is CFEB1, the catfish homolog of HEB (Hikima et al., 2005b). Also, a teleost-specific difference is found in the coding region of teleost E2A, which has a 17 aa insertion in its inhibitory domain (which functions in preventing homodimerization), shared with zebrafish and Xenopus but not mouse, human or chicken (Hikima 2005b). Differences in this domain likely result in important changes in target gene expression. As Greg Warr emphasized throughout his work on teleost E-proteins, the latter underscores the importance of comparative molecular studies in elucidating B cell developmental pathways.
Early B cell factor 1 (EBF-1)
This transcription factor, like E2A, also drives B lineage commitment and B cell determination, and is considered an epigenetic pioneer factor, directing chromatin modifications for transcriptional activation (reviewed in Lukin et al., 2008 and Northrup and Allman, 2008). EBF1 is a member of the Collier/Olf-1/EBF family (COE) family, which includes EBF1-4, Collier/knot, and Unc-3 (Lukin et al. 2008). The EBF proteins contain a highly conserved DNA binding domain at the N-terminal region of the protein (Figure 2), which contains an atypical Zn-binding motif. EBF binds to DNA as a homodimer through HLH domains, and has a transactivation/multimerization domain at its C terminus (Figure 2).
Mammalian EBF1 expression is high during early B cell development, particularly in CLPs, pro-B cells, and (im)mature B cells (Figure 1). EBF1 is also expressed in non-lymphoid tissues such as adipocytes, forebrain neurons, plasmacytoid dendritic cells, BM stromal cells, and osteoblasts (Lukin et al., 2008). EBF1 and another B cell-specific transcription factor, Pax5, regulate each other’s expression during B cell development (Roessler et al., 2007; Figure 3); in cooperation with E2A, EBF1 also regulates expression of components of the (pre)BCR (CD79α and Vpreβ1), while in cooperation with E47 it regulates VDJ recombination (Sigvardsson et al., 1997, Lukin et al. 2008, Northrup and Allman 2008). Lastly, EBF1 cooperates with both E2A and Pax5 on several other target genes (Lukin et al., 2008). EBF has also been shown to reprogram myeloid progenitors, and as such, enforces B cell specification, blocking alternative differentiation pathways (Pongubala et al. 2008). This complex transcriptional circuitry underlying B cell fate is illustrated in Figure 3 (Lukin et al. 2008, Northrup and Allman, 2008).
EBF1 gene orthologs have been identified in zebrafish (GenBank XM680898) and a cartilaginous species, clearnose skate (Anderson et al, 2004). The DNA binding domain of EBF1 is highly conserved, with close to 100% homology between zebrafish, skate and mouse (Anderson et al., 2004). In the skate, EBF1 is expressed in spleen, Leydig and epigonal tissues, thymus, and adipose tissue (Anderson et al., 2004). In the rainbow trout, EBF1 expression has been detected using a mouse EBF1-specific antibody: EBF1 is expressed both in kidney tissue and PBLs. In K1, EBF1 is expressed in HCmu–but not HCmu+ lymphoid cells, often co-expressed with RAG-1 (Zwollo et al., 2010).
The Pax5 gene encodes a homeodomain type transcription factor, the B cell-specific activator protein (BSAP), and has been extensively studied by several groups including our own (reviewed in Hagman and Lukin, 2007, and Cobaleda et al., 2007). Pax5 has been identified in both mammalian and non-mammalian species and contains a highly conserved, N-terminal paired domain (Figure 2). Pax5 is expressed in the vertebrate B cell lineage as well as in adult testis, while during embryonic development, it is expressed in the brain (reviewed in Cobaleda et al., 2007). In lower vertebrates including zebrafish and Xenopus, Pax5 is faintly expressed in the developing inner ear, but mammals lack expression at this site (Heller and Brandli,1999). Within the mammalian B cell lineage, Pax5 is expressed from the pro-B cell through mature and activated B cell stages, is downregulated during terminal differentiation and absent at the plasma cell stage (Figure 1; Cobaleda et al, 2007). Recently, Pax5 was also shown to regulate the adhesion and migration properties of murine B cells (Schebesta et al. 2007).
Pax5 directs B cell fate: it represses both macrophage-colony-stimulating factor receptor (M-CSFR) expression, thereby blocking myeloid cell fate, and Notch1 expression, thereby constraining T cell development (reviewed in Northrup and Allman, 2008). Additionally, downregulation of Pax5 correlates with the (reversible) trans-differentiation of a B lymphoma cell into a macrophage upon in vitro culturing with M-CSF (Hodawadekar et al., 2007). This is quite fascinating since teleost B cells reportedly have phagocytic properties (Li et al., 2006), although it is unclear whether such cells simultaneously share both myeloid and B lymphoid characteristics or rather fluctuate between the two states.
Pax5 binds to DNA as a monomer. A large number of potential target genes have been identified for Pax5, with Pax5 acting either as activator or repressor. For example, Pax5 is an activator of EBF1, and acts as a repressor for XbpI, Blimp1, and IgHC (shown in Figures 3 and 4, and reviewed in Cobaleda et al., 2007). Pax5 induction in pro-B cells is dependent on expression of E2A and EBF1, while Pax5 itself can further activate EBF1 expression (Northrup and Allman, 2008; Figure 3). Both the activation and repressor domains of Pax5 are present at the C-terminus of the protein (Figure 2) and are common targets for alternative splicing, as discussed below.
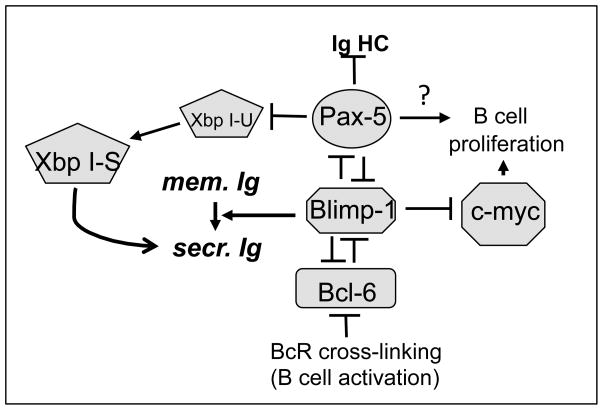
Figure 4.
Functional relationships between transcription factors during vertebrate B cell activation. Transcription factors are shown in grey boxes. Pax5 inhibits full expression of the immunoglobulin heavy chain (Ig HC) in mature B cells. Membrane immunglobulin, mem Ig; secreted immunoglobulin, sec. Ig. XbpI-U: unspliced, XbpI-S, spliced form of XbpI.
During B cell activation, suppression of Pax5 is necessary to complete terminal differentiation into a plasma cell, and this is done through the transcriptional repressor Blimp1 (Kallies and Nutt, 2007), as shown in Figure 4. In addition, in resting, mature B cells Pax5 directly represses the transcription factor Xbp-1, thereby blocking terminal differentiation until the B cell is induced into terminal differentiation through engagement between its BcR and antigen (Reimold et al. 1996).
Pax5 cDNA has been isolated from the B cells of three teleost species, including rainbow trout, puffer fish, and zebrafish (Zwollo et al., 2008, Ohtani et al. 2006, Pfeffer et al. 1998). The locations of functional domains of the Pax5 gene are highly conserved between species. Additionally, teleost Pax5 paired domain shares almost one hundred percent homology with that of mouse, hence, as for the other transcription factors reviewed here, the DNA binding domain is highly conserved. Two interesting differences were discovered between teleost Pax5 (rainbow trout, puffer fish, and zebrafish) and non-teleost Pax5 (mouse, human, chicken, and Xenopus): all teleosts share a 25 aa (in-frame) deletion of their partial homeodomain, resulting in a truncated version of the domain (Zwollo et al., 2008). Two regulatory proteins, TATA-binding protein and retinoblastoma (Rb), have been shown to interact with the partial homeodomain (Eberhard and Busslinger, 1999) and hence, a shortened version of it likely correlates with unique functions in teleost Pax5 activity. A second difference concerns a 30 aa in-frame insertion at the C-terminus of all 3 teleost Pax5 proteins, which is lacking in non-teleost Pax5. (Zwollo et al., 2008). This insertion expands the repressor domain of teleost Pax5, and likely affects expression of target genes.
Blimp1
The Zn-finger type transcription factor Blimp1 is a master regulator of cell differentiation, including both terminal B cell and macrophage differentiation (reviewed in John and Garrett-Sinha, 2009). The five Zn-finger-containing, C-terminal DNA binding domain of Blimp1 (see Figure 2) is highly conserved among vertebrates, with more than 93% homology between zebrafish, Xenopus, pufferfish, and mouse (Ohtani et al., 2006). Blimp1 functions as a transcriptional repressor and associates with Groucho and histone deacetylases (John and Garrett-Sinha, 2009). Together with XbpI, Blimp1 is sufficient to trigger terminal differentiation of B cells. Blimp1 expression is induced in B cells through downregulation of BCL-6 (Figure 4); Blimp1 not only represses both Pax5 and c-myc directly but also regulates the shift from membrane to secreted forms of Ig RNA in activated B cells (Figure 4). Of interest, Blimp1 expression is directly repressed by Pax5, (Kallies and Nutt, 2007), possibly providing the cell with a mechanism to regulate its rate of terminal B cell differentiation
Maturation from transient plasmablasts to long-lived ASC in the bone marrow has been shown to correlate with quantitative increases in Blimp1 expression in individual cells, as studied using heterozygous Blimp1gfp mice (Kallies et al., 2004). In these mutant mice, Ig-secreting, Pax5− plasma cells contained the highest levels of Blimp1; plasmablasts had intermediate Blimp1 levels, while PBLs had the lowest Blimp1 expression per cell. These data are especially promising for future studies in teleosts, because they show that nuclear Blimp1 levels can be used as tools to determine terminal differentiation state of B cells.
Blimp1 cDNA has been isolated from two teleosts, zebrafish and pufferfish and a highly conserved location of Blimp1 functional domains was noted (Ohtani et al., 2006). Partial Blimp1 cDNA sequence (covering exons 6–7) has also been isolated from rainbow trout spleen tissue (Zwollo, unpublished data). Comparison between Blimp1 DNA binding domains from mouse, zebrafish, pufferfish and rainbow trout showed a highly conserved sequence, with >92% homology between species.
XbpI
This basic leucine zipper type transcription factor is, together with Blimp1, necessary for terminal B cell differentiation (Figure 4) (Reymold et al., 1996). Because XbpI is directly repressed by Pax5, XbpI expression levels are opposite to those of Pax5 (Reymold et al., 1996; Figures 1 and 3). Hence the ratio of Pax5 to XpbI provides a indicator for B cell differentiation state.
XbpI is a key regulator of the unfolded protein response (UPR) (Yoshida et al., 2001). In response to ER stress, 26 ribonucleotides are removed from the XbpI (U) transcript, which results in a frame-shift, translating into a more potent transcription factor (XbpI (S) with a different C-terminus (Yoshida et al., 2006, Hu et al., 2007; Figure 2). XbpI-S translocates to the nucleus where it activates certain UPR genes and plays essential roles in cell survival. This is particularly important for the plasma cell, which is in a state of “acute stress” (Lenci and Sitia, 2007), synthesizing tremendous amounts of antibodies as part of the humoral immune response. XbpI-S levels increase during differentation from plasmablasts into plasma cells, and this induces a (apparent paradoxical) decrease in proteosome capacity, which in turn induces apoptosis in plasma cells (Lenci and Sitia,. 2007). Hence, XbpI-S plays important roles in regulating the half-life of plasma cells (Lenci and Sitia, 2007).
Teleost XbpI cDNA sequences have been determined for pufferfish, Atlantic salmon, and zebrafish (Ohtani et al., 2006, Hu et al, 2007, Leong et al., 2010), but not much else is known about the factor in teleosts. Again, the DNA binding domains are highly conserved among the vertebrate species, and share almost 100% homology at the protein level
4. Alternative splicing of transcription factors
Alternative RNA splicing is a commonly used, but poorly understood mechanism that adds an additional level of regulation of gene expression (De la Grange et al. 2007). Not surprisingly, the great majority of genes encoding transcription factors use alternative splicing to regulate their activity. Alternative RNA splicing leads to mRNAs that lack one or more exons. Hence, in alternatively spliced transcription factors, one gene can generate multiple, partially identical, proteins with different, and often opposite, functions. Alternative splicing can reduce DNA binding strength of the resulting protein (through a modified or absent DNA binding domain), reduce interaction with co-repressors or -activators, or change interaction with basal transcription factors. In other words, alternative splicing of a transcription factor directly affects the expression patterns of its target genes. As such, alternative splicing likely plays important roles during B cell differentiation.
For most of the six transcription factors discussed here, examples of alternative splicing can be found in vertebrate species. One transcription factor that relies heavily on alternative splicing is Ikaros. In mammals, eight Ikaros splice forms have been reported, which differ in the number of Zn-fingers they possess (Hahm et al.,1994, Georgopolous et al, 1992, Sun et al. 1996). Splice variants that do not include at least three of the four internal zinc fingers do not bind single Ikaros binding sites and functionally interfere with the activity of the longer isoforms (Hahm et al.,1994). Differential expression of Ikaros isoforms has been observed during mammalian lymphoid development; for example, expression of the smallest isoform, IK6 (which lacks exons 4–7 and is unable to bind to DNA), is associated with the onset of lymphoid commitment (Klug et all 1998).
The Ikaros gene in rainbow trout has at least 8 isoforms, six of which are shared with the mouse homolog, and two that are unique to trout: both isoforms lack exon 7, while one also lacks the the most N-terminal Zn-finger; both isoforms retain ability to bind DNA (Hansen et al. 1997). Zebrafish have several Ikaros isoforms in common with the mouse, but also express one of the trout-specific isoforms (lacking both exon 7and Zn-finger 1) (Willett et al., 2001).
Like Ikaros, the Pax5 gene is also extensively alternatively spliced. Mouse splenic B cells, in addition to expressing the full-length (Pax5a) isoform, express at least three alternatively spliced forms, including one that lacks exon 2 and is unable to bind DNA, one that lacks both the partial homeodomain and transactivation/repressor domains, and one that lacks all three domains (Zwollo et al., 1997). Two of those isoforms (5d and 5e) have been shown to have opposing roles in B cell growth, supporting their functional significance (Lowen et al et al., 2001). In human PBLs, five alternatively spliced Pax5 isoforms have been reported. All have deletions around the transactivation/repressor domains, resulting in changes in transactivating potential (Robichaud et al., 2004). Studies on alternative splicing for Pax5 in the rainbow trout have also revealed multiple isoforms, with deletions in the DNA binding domain and/or the activation/repressor domains (Zwollo, unpublished data). Xenopus Pax5 has at least four isoforms that lack either DNA binding or activation domains (Heller et al., 1999).
Although beyond the scope of this review, several of the genes described here have two active promoters in B cells resulting from alternative usage of exon 1, and hence, their expression maybe regulated by two (overlapping) sets of transcription factors.
From the limited number of studies on alternative splicing in teleost B cells, it appears that the mechanism is conserved among vertebrate species. Interestingly, while some isoforms for a particular transcription factor are shared between species, others appear unique to teleost. However, such differences may turn out to be quantitative rather than qualitative, reflecting tissue or stage-specific differences in isoform abundance. Whether or not teleost-specific isoforms exist, only future studies will tell.
So far, all reported studies on alternative splicing of transcription factors have used RNA or protein from pooled cells as their source for analysis. Hence, the frequency of cells containing a particular isoform, or the level of expression for a given isoform in individual cells, cannot be determined using such an approach. It is possible that during vertebrate B cell development, individual cells within each developmental stage express distinct sets of isoforms, and that such isoforms are essential for proper progress through the B cell developmental program. For now, this hypothesis remains untested.
5. Using expression patterns of transcription factors to dissect teleost B cell maturation; B cell signatures
To be able to determine the frequency of distinct B cell subsets during teleost immune development, our laboratory has developed an approach to analyze single cells using flow cytometric analysis. The approach is based on the presence or absence of two differentially expressed transcription factors in individual B cells, with the goal to identify the developmental stage of each cell, and hence, the frequency of cells with specific “transcription factor-phenotypes” within an immune tissue. By using carefully selected combinations of markers based on the developmental state of the B cell (Figure 1) one can distinguish between early B cell progenitors, late developing B cells, resting B cells, activated B cells, plasmablasts, and plasma cells. Based on the highly conserved nature of transcription factors, as reviewed herein, this approach can be applied to any teleost species, with the advantage that antibodies against mammalian transcription factors are readily available. We recently proposed use of the term “B cell signatures” to refer to the specific patterns of transcription factor expression in a given immune organ or tissue (Zwollo et al., 2010).
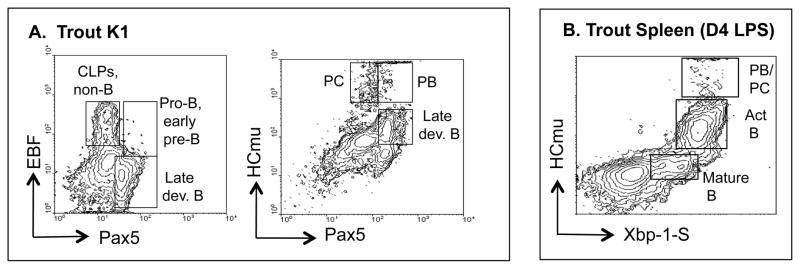
This approach is illustrated in Figure 5. The figure shows contour graphs derived from flow cytometric analysis of fixed and permeabilized trout immune cells. It shows the distribution of cells in a given tissue based on the expression of two different transcription factors (or other B cell markers) in individual cells. For example, the hematopoietic tissue of rainbow trout, K1, was analyzed for patterns of both developing and IgM-secreting B cell subsets. Two sets of antibodies were used: to transcription factors Pax5 and EBF1 (Figure 5A, left panel), or to cytoplasmic IgM (cHCmu) and RAG-1 (Figure 5A, right panel). The contours show the frequency of developing B cells (including CLPs, pro-B cells/early pre-B cells, and late developing B cells), as well as IgM-secreting plasmablasts and plasma cells, all of which can be quantified (Zwollo et al., 2010). Each contour represents one B cell signature for a given tissue, in a given fish. Fish have different B cell signatures if they differ in the relative abundance of their CLP, early progenitor, late progenitor, PB, or PC cell population.
Figure 5.
Contour graphs from flow cytometric analysis of freshly isolated, fixed and permeabilized primary and secondary trout immune tissues for the establishment of B cell signatures. CLP, Common Lymphoid progenitor; PB, Plasmablast; PC, plasma cell, act. B, activated B cell, late dev. B, late developing B cell. A. Contours of trout K1 cell suspension, using either EBF and Pax5 (left panel) or HCmu and Pax5 (right panel) antibodies. B. Contour graph of trout spleen cell suspension 4 days after LPS-induction (Day 4 LPS), using XbpI-S and HC-mu antibodies.
A second example shows the validity of this approach as a way to measure the level and rate of trout B cell activation in a secondary immune tissue. Splenic B cells can be induced with LPS ex vivo. In this case, the level of expression of both XbpI-S and cytoplasmic IgM in individual splenic B cells identifies the state of terminal B cell differentiation (Figure 1). This can be measured at specific timepoints after LPS-induction, for example, on day 4 (Figure 5B). A different rate for XbpI-S and HCmu induction translates into a different B cell signature for that fish (Zwollo and Barr, unpublished data).
6. Future directions
To increase our understanding of the teleost antibody response, future studies should include the identification of all relevant transcription factors expressed during the B cell developmental program for each teleost species of interest, including expression of alternative spliced isoforms. Additionally, it will be important to focus on identification and function of cis-acting elements for each transcription factor, including comparative studies in other teleost and non-teleost species.
In this regard, Greg Warr has been a true inspiration in the field, using similar approaches to dissect the complex regulation of the teleost IgH locus by E-proteins, particularly the functional interaction of catfish and pufferfish E-binding proteins with the Emu enhancer (Hikima et al., 2005a, b, 2008). By first identifying novel E-protein members for catfish, and then using this information to revisit the mammalian model of IgH regulation, his work showed clear evidence of the diversity, similarities, and differences in gene regulation between mammalian and teleost species.
This review emphasizes the conserved nature of transcription factors throughout vertebrate evolution, especially the high similarity of the DNA binding domain. However, it will be equally important to focus on species-specific differences in cis-acting regulatory sequences outside of the coding regions, as such elements drive species-specific developmental programs. In the end, as Greg understood so well, it is the differences in number, relative position, and/or orientation of cis-acting elements that lead to the right amount of gene product, at the right time, and in the right place. Ultimately, this is what makes a rainbow trout a rainbow trout, and a catfish a catfish.
From the data already available, it seems that studies on transcription factor function in the teleost B cell would benefit from information on alternative splicing. Alternative splicing for Pax5 and Ikaros is particularly extensive and commonly used in the teleost, but few studies have focused on it so far. If it can be determined when during B cell development certain isoforms are expressed, then that information can be integrated into the teleost B cell signature. For example, if specific combinations of Pax5 isoforms correlate with specific B cell stages, then we could use the “Pax5 signature” of a fish to define its immune state, and similarly, we could use “Ikaros signatures” for the same purpose. The advantage of this approach would be that only one gene (Pax5 or Ikaros) would have to be identified for each teleost species, providing a powerful tool to explore humoral immunity in less well-known fish species.
In spite of our very limited knowledge in the area of transcriptional regulation of the teleost B cell, much progress has been made over the past 10 years. Furthermore, we now have the ability to determine frequencies of B cell subsets in teleost, allowing for some important practical applications. These could include measuring effects of environmental stress (global warming, pollution) on teleost B cell maturation and antibody responses, or monitoring vaccination efficiencies in farmed fishes. No doubt future studies will lead not only to a greater appreciation of the complexity of gene regulatory circuits during fish B cell development, but also to an increased understanding of the vertebrate B cell response.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bromage ES, Kaattari IM, Zwollo P, Kaattari SK. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol. 2004;173:7317–7323. doi: 10.4049/jimmunol.173.12.7317. [DOI] [PubMed] [Google Scholar]
- Carrotta S, Nutt SL. Losing B cell identity. BioEssays. 2008;30:203–207. doi: 10.1002/bies.20725. [DOI] [PubMed] [Google Scholar]
- Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Letters. 2007;581:3652–3657. doi: 10.1016/j.febslet.2007.04.031. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Danilova N, Bussmann K, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish; identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- Davidson GA, Lin S-H, Secombes CJ, Ellis AE. Detection of specific and “constitutive” antibody secreting cells in the gills, head kidney, and peripheral blood leucocytes of dab (Limanda limanda) Vet Immunol Immunopath. 1997;58:363–374. doi: 10.1016/s0165-2427(97)00017-2. [DOI] [PubMed] [Google Scholar]
- De la Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinformatics. 2007;4(8):180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Busslinger M. Transcription factor Pax5 is an interaction motif for retinoblastoma and TATA-binding protein. Cancer Res. 1999;59:1716–17. [PubMed] [Google Scholar]
- Engel I, Murre C. The function of E-proteins and Id proteins in lymphocyte development. Nat Rev Immunol. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- Georgepoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- Georgopolous K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–146. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Ann Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- Hagman J, Lukin K. “Hands-on” regulation of B cell development by the transcription factor Pax5. Immunity. 2007;27:8–10. doi: 10.1016/j.immuni.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale S. The lymphoid transcription factor Lyf-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:8292. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire RN, Miracle AL, Rast JP, Litman GW. Members of the Ikaros gene family are present in early representative vertebrates. J Immunol. 2000;165:306–312. doi: 10.4049/jimmunol.165.1.306. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Strassburger P, Du Pasquier L. Conservation of a master hematopoietic switch gene during vertebrate evolution: isolation and characterization of Ikaros from teleost and amphibian species. Eur J Immunol. 1997;27:3049–58. doi: 10.1002/eji.1830271143. [DOI] [PubMed] [Google Scholar]
- Hansen J, Landis ED, Phillips RB. Discovery of a unique Ig heavy chain isotype (IgT) in rainbow trout; implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci USA. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikima J, Lennard M, Wilson M, Miller NW, Clem LW, Warr GW. Evolution of vertebrate E protein transcription factors: comparative analysis of the E-protein gene family in Takifugu rubripes and humans. Physiol Genomics. 2005a;21:144–151. doi: 10.1152/physiolgenomics.00312.2004. [DOI] [PubMed] [Google Scholar]
- Hikima J, Middleton D, Wislon MR, Miller N, Clem LW, Warr GW. Regulation of immunoglobulin gene transcription in teleost fish: identification, expression, and functional properties of E2A in channel catfish. 2005. Immunogenetics. 2005b;57:273–282. doi: 10.1007/s00251-005-0793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikima J, Richard ML, Wilson MR, Miller NW, Warr GW. Interaction between E-protein and Oct transcription factors in the function of the catfish IGH enhancer. Dev Comp Immunol. 2008;32:1105–1110. doi: 10.1016/j.dci.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodawadekar S, Yu D, Cozma D, Freedman B, Sunyer O, Atchison M, Thomas-Tikhonenko A. B-lymphoma cells with epigenetic silencing of Pax5 trans-differentiate into macrophages, but not other hematopoietic lineages. Exp Cell Res. 2007;313:331–340. doi: 10.1016/j.yexcr.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Gong HY, Lin GH, Hu SY, Chen M, Huang SJ, Liao CF, Wu JL. XbpI, a key regulator of unfolded protein response, activates transcription of IGF1 and Akt phosphorylation in zebrafish embryonic cell line. Bioch Bioph Res Com. 2007;359:778–783. doi: 10.1016/j.bbrc.2007.05.183. [DOI] [PubMed] [Google Scholar]
- Irwin MJ, Kaattari SL. Salmoinid B lymphocytes demonstrate organ- dependent functional heterogeneity. Vet Immunol Immunopath. 1986;12:39–45. doi: 10.1016/0165-2427(86)90108-x. [DOI] [PubMed] [Google Scholar]
- John SA, Garrett-Sinha LA. Blimp1: a conserved transcriptional repressor crirical for differentiation of many tissues. Exp Cell Res. 2009;315:1077–1085. doi: 10.1016/j.yexcr.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Kaattari SL, Irwin MJ. Salmonid spleen and kidney harbor populations of lymphocytes with different B cell repertoires. Dev Comp Immunol. 1985;9:433–444. doi: 10.1016/0145-305x(85)90006-0. [DOI] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton M, et al. Plasma cell ontogeny defined by quantitative changes in Blimp-1 expression. J Exp Med. 200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Nutt S. Terminal differentiation of lymphocytes depends on Blimp-1. Curr Opin Immunol. 2007;19:156–162. doi: 10.1016/j.coi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF1) and multiple B lineage genes by the basic-helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JS, Jantzen SG, von Schalburg KR, et al. Salmo salar and esox lucius full-length cDNA sequences reveal changes in evolutionary pressures on a post-tetraploidization. BMC genomics. 2010;11:279. doi: 10.1186/1471-2164-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Barreda D, Zhang YA, Boshra H, Gelman AE, LaPatra S, Tort L, Sunyer JO. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol. 2006;7:1116–1124. doi: 10.1038/ni1389. [DOI] [PubMed] [Google Scholar]
- Liberg D, Smale ST, Merkenschlager M. Upstream of Ikaros. Trends Immunol. 2003;24:567–570. doi: 10.1016/j.it.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Lowen M, Scott G, Zwollo P. Functional analyses of two alternative isoforms of the transcription factor Pax-5. J Biol Chem. 2001;276:42565–42574. doi: 10.1074/jbc.M106536200. [DOI] [PubMed] [Google Scholar]
- Lukin K, Fields S, Hartley J, Hagman J. Early B cell factor: regulator of B lineage specification and commitment. Semin Immunol. 2008;20:221–227. doi: 10.1016/j.smim.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Rahman A. What brought the adaptive immune system to vertebrates? The jaw hypothesis and the seahorse. Immunol Rev. 1998;166:177–186. doi: 10.1111/j.1600-065x.1998.tb01262.x. [DOI] [PubMed] [Google Scholar]
- Melchers F, Kincade P. In: Molecular Biology of B cells. Honjo Tasuku, Alt Frederick W, Neuberger Micheal., editors. Chpt 7. Elsevier Academic Press; 2004. pp. 101–117. [Google Scholar]
- Milano GE, Basari F, Chimenti C. Adrenocortical and adrenomedullary homologs in eight species of adult and developing teleosts: morphology, histology, and immunohistochemistry. Gen Comp Endocrinol. 1997;108:483–96. doi: 10.1006/gcen.1997.7005. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhaver binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C, Voronova A, Baltimore D. B-cell and myocyte-specific E2-box binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup D, Allman D. Transcriptional regulation of early B cell development. Immunol Res. 2008;42:106–117. doi: 10.1007/s12026-008-8043-z. [DOI] [PubMed] [Google Scholar]
- Ohtani M, Miyadai T, Hiroishi S. Identification of genes encoding critical factors regulating B-cell terminal differentiation in torafugu (Takifugu rubripes) Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:109–114. doi: 10.1016/j.cbd.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nature Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewski K, Radbruch A. In: Molecular Biology of B cells. Honjo Tasuku, Alt Frederick W, Neuberger Micheal., editors. Chpt 16. Elsevier Academic Press; 2004. pp. 247–254. [Google Scholar]
- Reimold A, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, Glimcher LH. Transcription factor factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KI, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–36. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud GA, Nardini M, Laflamme M, Cuperlovic-Culf M, Ouellette RJ. Human Pax-5 C-terminal isoforms possess distinct transactivation properties and are differentially modulated in normal and malignant B cells. J Biol Chem. 2004;279:49956–49963. doi: 10.1074/jbc.M407171200. [DOI] [PubMed] [Google Scholar]
- Roessler S, Gyory I, Imhof S, et al. Distinct promoters mediate regulation of Ebf1 expression. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger G, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Schorpp M, Bialecki M, Diekhoff D, Walderich B, Odenthal J, Maischein HM, Zapata AG, Boehm T Tubingen 2000 Screen Consortium, Freiburg Screening group. Conserved functions of Ikaros in vertebrate lymphocyte development: genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J Immunol. 2006;177:2463–2476. doi: 10.4049/jimmunol.177.4.2463. [DOI] [PubMed] [Google Scholar]
- Sigvardsson M, O’Riordan M, grosscgedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- Sun L, Liu A, Georgopolous K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin C, Shapiro MA, Calame KL. Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter. Nucl Acids Res. 2000;28:4846–4855. doi: 10.1093/nar/28.24.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett CE, Kawasaki H, Amemiya CT, Lin S, Steiner LA. Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Devel Dynamics. 2001;222:694–698. doi: 10.1002/dvdy.1223. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XbpI mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–889. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zapata AG, Cooper EL. Comparative Histopathology. Chichester, UK: John Wiley and Sons; 1990. The Immune system. [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, Lapatra SE, Batholmew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nature Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P, Arrietta H, Ede K, Molinder K, Desiderio S, Pollock R. The Pax-5 gene is alternatively spliced during B-cell development. J Biol Chem. 1997;272:10160–10168. doi: 10.1074/jbc.272.15.10160. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Cole S, Bromage E, Kaattari SL. B cell heterogeneity in the teleost kidney: evidence for a maturation gradient from anterior to posterior kidney. J Immunol. 2005;174:6608–16. doi: 10.4049/jimmunol.174.11.6608. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Haines A, Rosato P, Juliann Gumulak-Smith J. Molecular and cellular analysis of B cell populations in the rainbow trout using Pax5 and immunoglobulin markers. Dev Comp Immunol. 2008;32:1482–1496. doi: 10.1016/j.dci.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P, Mott K, Barr M. Comparative analyses of B cell populations in trout kidney and mouse bone marrow; establishing “B cell signatures”. Dev Comp Immunol. 2010;34:1291–1299. doi: 10.1016/j.dci.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]