Abstract
Mechanisms of signal transduction regulation remain a fundamental question in a variety of biological processes and diseases. Previous evidence suggests the primary cilium can act as a signaling hub1, but its exact role in many of its described pathways has remained elusive. Here, we investigate the mechanism of cilia regulation of the canonical Wnt pathway through systematic knock-down and knock-out approaches. We found that the primary cilium dampens canonical Wnt signaling through a unique spatial mechanism involving compartmentalization of signaling components. The cilium, through regulated intraflagellar transport (IFT), diverts Jouberin (Jbn), a ciliopathy protein and context specific Wnt pathway regulator2, away from the nucleus and limits β-catenin nuclear entry. This repressive regulation does not silence the pathway, but instead maintains a discrete range of Wnt responsiveness; cells without cilia have potentiated Wnt responses whereas cells with more than one cilium display inhibited responses. Furthermore, we show that this regulation occurs during embryonic development and is disrupted in cancer cell proliferation. Together these data explain a unique spatial mechanism of regulation of Wnt signaling which may provide insight into ciliary regulation of other signaling pathways.
Keywords: Signaling, cilia, Wnt, β-catenin, Jouberin, Ahi1, Intraflagellar transport, Kif3A, Dnchc2, ciliopathy
The Wnt signaling pathway functions in a variety of cellular processes, and mutations in this pathway have been identified in a broad array of illnesses, from developmental disorders to cancer3, 4. Thus, understanding the regulation of this pathway is vital to the goal of developing treatments for these diseases. Primary cilia have been connected with a variety of disorders known as ciliopathies5, 6, and have been implicated in negative regulation of the canonical Wnt pathway7–9. However, there is also convincing data from other models of cilia disruption in which such regulation has not been apparent10, 11, suggesting potential context or evolutionary dependency.
To examine this apparent contradiction, as well as subcellular mechanisms, we have assayed canonical Wnt signaling activity using a variety of assays and cell types, and following several genetic manipulations to disrupt specific aspects of ciliary function. First, we examined Wnt activity using a luciferase reporter assay12 in two cellular models that disrupt cilia. We tested Wnt activity in mouse embryonic fibroblasts (MEFs) following serum stimulation in order to induce ciliary retraction13. These deciliated cells (quantified in Supplementary Fig.1a) showed exaggerated Wnt activity following Wnt stimulation (Fig. 1a), similar to previously described in nonciliated cells8, 9, suggesting cilia retraction may prime the cell for an increased Wnt response. We also tested this effect in MEFs from mice harboring an early nonsense mutation in the retrograde transport motor dynein cytoplasmic heavy chain 2 (Dnchc2) gene14. These independently derived cells lacked cilia (Supplementary Fig. 1b, c), in contrast to other Dnchc2 MEFs generated by another group from a mouse with a different mutation in the gene11. Using the Wnt reporter assay, we observed an increase in Wnt response to Wnt3a conditioned media in mutant cells (Fig. 1b), similar to previously described for other IFT mutant MEFs9. Together, these data support a role for cilia in repressing cellular Wnt responsiveness.
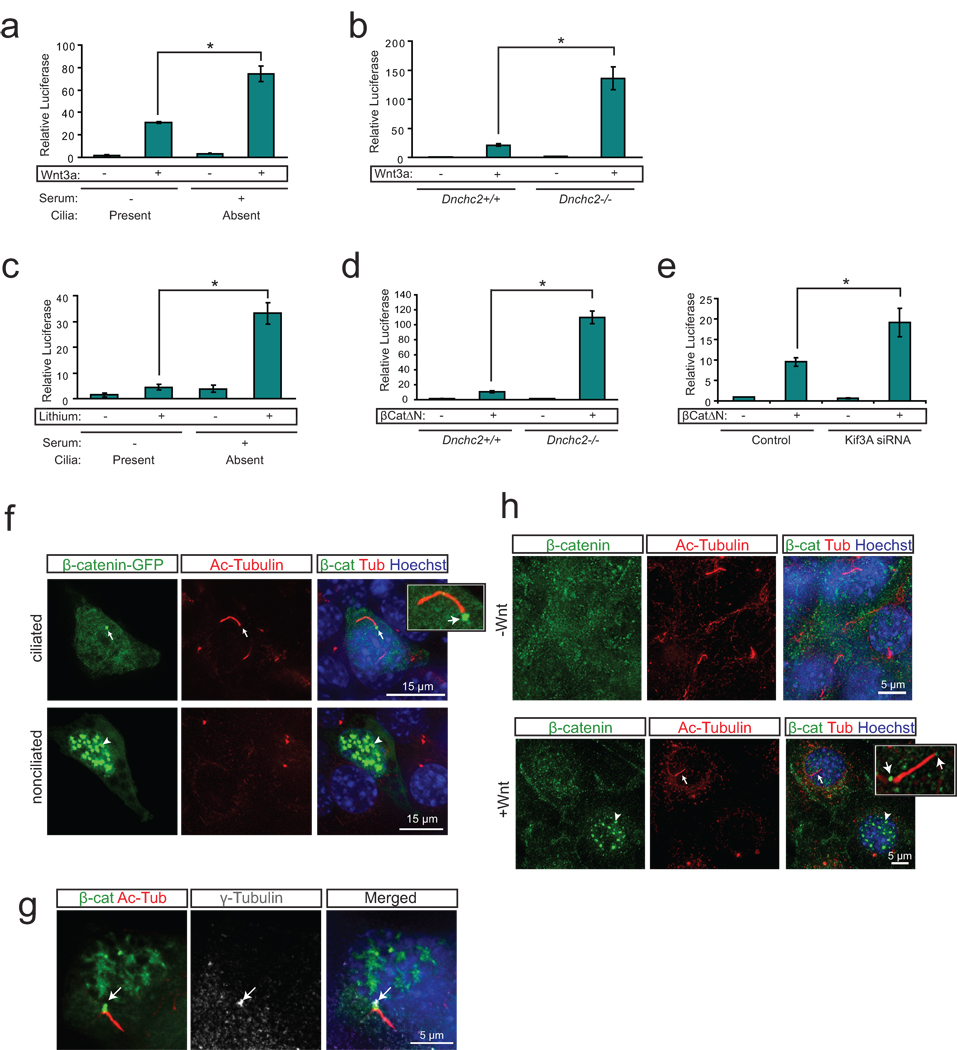
Figure 1. The primary cilium dampens Wnt activity by regulating β-catenin.
a. Luciferase response to Wnt3a conditioned media in a wild-type MEF cell line following serum starvation and re-addition. *P<0.05, n=3 independent experiments, Student’s t-test. b. Luciferase assay in Dnchc2 mutant MEFs and littermate control MEFs reveals increased Wnt response to Wnt3a conditioned media in Dnchc2−/− cilium mutant MEFs. *P<0.05, n=3 independent experiments, Student’s t-test. c. Luciferase response to LiCl in wild-type MEFs following serum starvation and re-addition. *P<0.05, n=3 independent experiments, Student’s t-test. d. Luciferase activity in Dnchc2 MEFs with activation of the pathway with transfected β-catΔN. *P<0.05, n=3 independent experiments, Student’s t-test. e. Luciferase activity in MEFs transfected with Kif3a siRNA and activation of the pathway with cotransfected β-catΔN. *P<0.05, n=3 independent experiments, Student’s t-test. Values for luciferase assays were normalized across measurements for each experiment and for transfection efficiency relative to β-gal. Error bars represent S.E.M. f. Localization of β-catenin-GFP (green) in wild-type MEFs treated with Wnt3a conditioned media and stained with acetylated tubulin (red) to mark cilia and Hoechst (blue) as a nuclear marker. Arrows point to basal body localization of β-catenin while arrowheads indicate nuclear localization in a nearby nonciliated cell. g. Colocalization of β-catenin-GFP (green) with the basal body marker γ-tubulin (grey, arrow) and cilia (acetylated tubulin, red). Hoechst (blue) labels the nucleus. h. Localization of endogenous β-catenin (green) in wild-type MEFs in the presence and absence of Wnt3a conditioned media. Cilia are labeled with acetylated-tubulin (red) and nuclei are labeled with Hoechst (blue). Arrow marks ciliary localization of β-catenin while the arrowhead indicates nuclear localization in a neighboring nonciliated cell when treated with Wnt3a.
We next questioned whether the primary cilium represses the Wnt pathway upstream or downstream of β-catenin stabilization by measuring Wnt activity following activation with the Gsk3β inhibitor, lithium15. Cells with retracted cilia and treated for 8 hrs with LiCl exhibited an increase in Wnt responsiveness compared with ciliated cells (Fig. 1c). Importantly, lithium can lead to modest cilia lengthening16, but we found no effect in these cells (Supplementary Fig. 1d). We further tested this effect using LiCl treatment (Supplementary Fig. 1e) or overexpression of a constitutively active β-catenin construct (β-CatΔN) in Dnchc2 mutant MEFs, which exhibited an increased response compared with control MEFs (Fig. 1d). A similar outcome was produced in wild-type MEFs transfected with Kif3a siRNA, which blocks formation of the primary cilium8 (Fig. 1e and Supplementary Fig. 2a, b). These findings suggest the primary cilium restrains the canonical Wnt pathway at least partially downstream of β-catenin cytosolic stabilization.
We next examined the subcellular localization of β-catenin in ciliated and nonciliated cells treated with Wnt3a conditioned media. We observed basal body localization of a GFP tagged β-catenin (Fig. 1f, g, Supplementary Fig. 2c) as well as endogenous β-catenin (Fig. 1h) in ciliated cells despite the presence of Wnt conditioned media, whereas nonciliated cells exhibited a more striking nuclear accumulation of β-catenin. Furthermore, an alternate ciliated cell type, mouse inner medullary collecting duct cells (IMCDs), treated with Wnt3a, exhibited stronger nuclear β-catenin staining in nonciliated cells compared with neighboring cells with cilia (Supplementary Fig. 3a, b). These results suggest the presence of the cilium represses nuclear accumulation of β-catenin.
We previously showed that Jbn, encoded by the gene AHI1 and mutated in the ciliopathy Joubert syndrome17–19, is a positive modulator of the canonical Wnt pathway through facilitation of β-catenin nuclear translocation2. Since Jbn is the only known modulator of β-catenin nuclear translocation that also has a role at the cilium, we hypothesized that the mechanism of ciliary inhibition of β-catenin nuclear accumulation may involve the Jbn protein. To test this hypothesis, we first examined Jbn and β-catenin colocalization in ciliated and nonciliated cells. We found that, whereas ciliated MEFs exhibited basal body localization of Jbn and β-catenin, Dnchc2 mutant MEFs instead exhibited a prominent increase in nuclear levels of both Jbn and β-catenin (Fig. 2a). These results were further supported by nuclear extraction, which revealed increased nuclear β-catenin and Jbn in Dnchc2 mutant MEFs upon activation of the Wnt pathway (Supplementary Fig. 3c), whereas total Jbn levels were not increased in mutant MEFs (Supplementary Fig. 3d). This suggests that Jbn’s role in the Wnt pathway may be inhibited by the presence of the primary cilium.
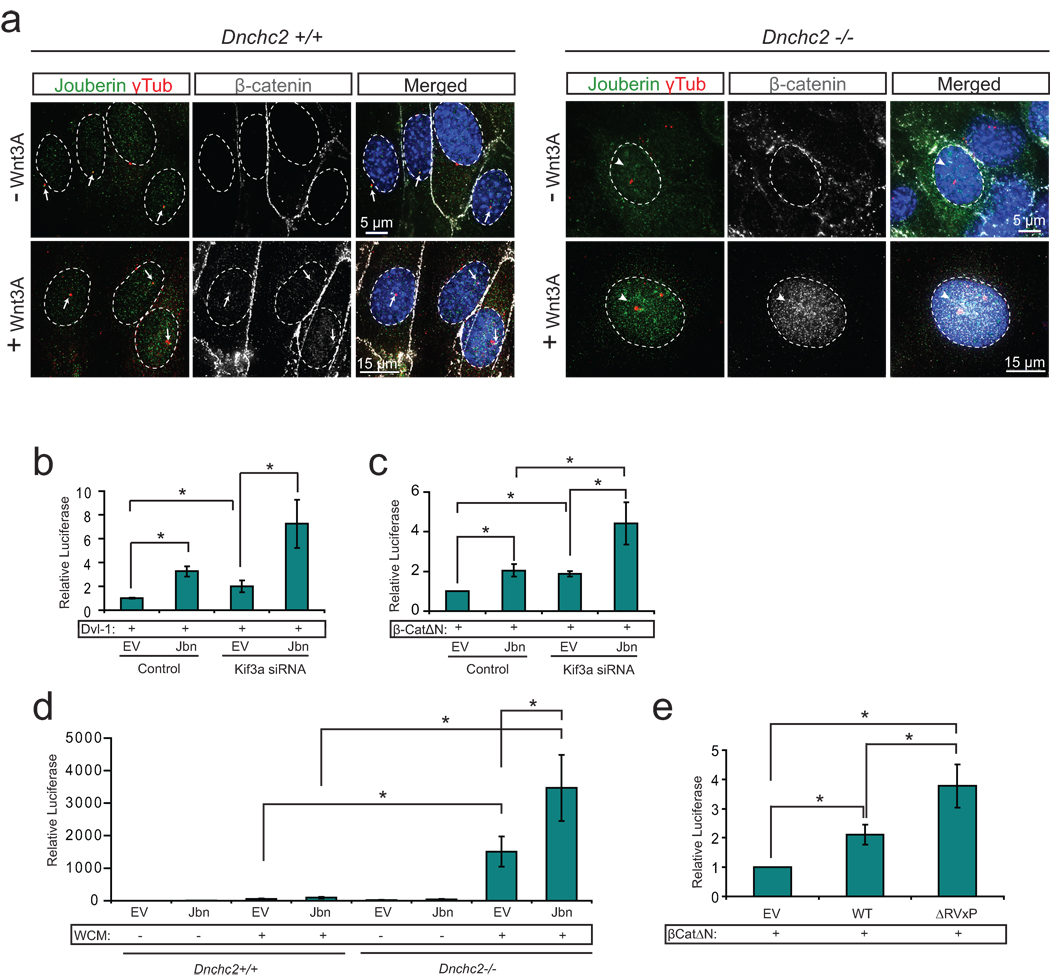
Figure 2. The primary cilium inhibits Jouberin mediated Wnt pathway regulation.
a. Localization of endogenous β-catenin (grey) and Jbn (green) in Dnchc2 MEFs. Control MEFs (top) exhibit basal body localization (γ-tubulin, red) of β-catenin and Jbn (arrows) when treated with Wnt3A conditioned media, whereas Dnchc2−/− MEFs exhibit increased nuclear (Hoechst, blue) staining (arrowheads) of both β-catenin and Jbn. b. Luciferase activity in 293T cells transfected with Kif3a siRNA and overexpressed Jbn or empty vector (EV) with activation of the pathway by cotransfection of Dvl-1. *P<0.05, n=3 independent experiments, Student’s t-test. c. Luciferase activity in 293Ts transfected with Kif3a siRNA and overexpressed Jbn or EV with activation of the pathway with cotransfected β-catΔN. *P<0.05, n=3 independent experiments, Student’s t-test. d. Luciferase activity in Dnchc2 MEFs transfected with Jbn or EV with activation of the pathway with Wnt3a conditioned media (WCM). *P<0.05, n=3 independent experiments, Student’s t-test. e. Luciferase activity in MEFs with overexpression of EV, wild-type Jbn or ΔRVxP mutant construct and cotransfected β-catΔN. *P<0.05, n=3 independent experiments, Student’s t-test. Values for luciferase assays were normalized across measurements for each experiment and for transfection efficiency relative to β-gal. Error bars represent S.E.M.
To test this hypothesis, we measured Wnt activity in ciliated and nonciliated cells transfected with a Jbn overexpression construct. We observed an increase in potentiation of the pathway by Jbn MEFs following serum-induced cilia retraction (Fig. 2a and Supplementary Fig. 4). This effect was also evident in ciliated 293T cells8 (Supplementary Fig. 4a, b) or MEFs transfected with Kif3a siRNA (Fig. 2b, c and Supplementary Fig. 4c), and was also evident in Dnchc2 mutant MEFs (Fig. 2d) with activation of the pathway upstream or at the level of β-catenin stabilization. Furthermore, we identified a region within the Jbn protein previously described to act as a ciliary targeting motif in other cilia proteins, which has the sequence RVxP20. We mutated all three key residues of this sequence within the Jbn protein and examined both ciliary localization and Wnt activity. We found that mutation of this ciliary targeting motif resulted in a failure to localize to cilia compared to wild-type (Supplementary Fig. 4d) yet elicited an increased Wnt response in ciliated MEFs (Fig. 2e). These results support the hypothesis that the presence of the primary cilium and subsequent ciliary localization of Jbn suppress Jbn’s role in the Wnt pathway.
Since the cilium appears to regulate subcellular localization of both Jbn and β-catenin, we hypothesized that the cilium may act to spatially sequester Jbn. Although Jbn does not exhibit constitutive ciliary axoneme staining2, we identified an association with microtubules (Supplementary Fig. 4e) consistent with the possibility that Jbn is transiently transported along microtubules and potentially within the cilium. We therefore performed an assay to transiently disrupt retrograde IFT, resulting in a redistribution of IFT transported components due to their inability to return to the basal body21–24. We performed siRNA knockdown of Dnchc2 in ciliated fibroblasts to inhibit retrograde transport but leave ciliary structure intact (Supplementary Fig. 4f). Using this approach, we observed a redistribution of Jbn along the length of the primary cilium (Fig. 3a). Importantly, although there was still some cytosolic Jbn present with Dnchc2 knockdown, levels of Jbn within the cilium were significantly increased compared with basal body localized Jbn seen with control siRNA (Figure 3a quantification). This suggests a sequestration of the basal body localized pool of Jbn along the ciliary axoneme with inhibition of Dnchc2.
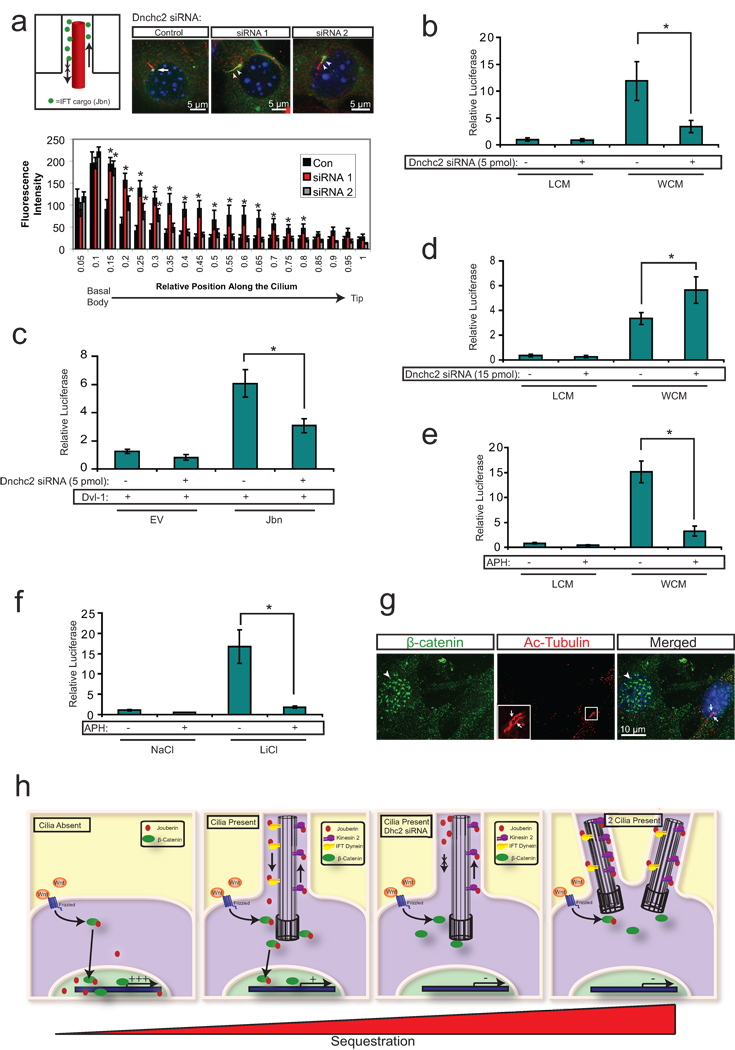
Figure 3. The primary cilium sequesters Jouberin and β-catenin away from the nucleus.
a. 3T3s transfected with two siRNAs to Dnchc2 exhibit accumulation of Jbn-GFP (green) along the length of the cilium (labeled with antibody to acetylated tubulin, red). The nucleus is labeled with Hoechst (blue). Schematic represents the experimental approach and interpretation of these results. Quantification of the green fluorescence along the cilium reveals a shifted localization along the ciliary axoneme in siRNA transfected cells with significantly increased levels of GFP beyond the basal body. Average fluorescence intensities of cells transfected with negative control siRNA (n=7), Dnchc2 siRNA #1 (n=10), or Dnchc2 siRNA #2 (n=11) were normalized and displayed in arbitrary grey value units. Position along the cilium was normalized for total cilia length and results were binned into 20 equal segments. b. Luciferase activity in MEFs transfected with 5 pmol of Dnchc2 siRNA and treated with Wnt3a conditioned media (WCM) or control L-cell conditioned media (LCM). *P<0.05, n=7 from 6 independent experiments, Student’s t-test. c. Wnt activity as reported by luciferase in 293T cells with overexpression of wild-type Jbn or EV and cotransfection of 5 pmol negative control siRNA or Dnchc2 siRNA. Cells were cotransfected with Dvl-1 to activate the Wnt pathway. *P<0.05, n=6 from three independent experiments, Student’s t-test. d. Luciferase activity in MEFs transfected with 3-fold increased Dnchc2 siRNA dose and treated with WCM or LCM. *P<0.05, n=7 from 6 independent experiments, Student’s t-test. e. Luciferase activity in MEFs treated with 1 µg/ml aphidicolin (APH) following serum starvation and treated with WCM or LCM. *P<0.05, n=6 from 5 independent experiments, Student’s t-test. f. Luciferase activity in MEFs treated with 1 µg/ml APH following serum starvation and treated with LiCl or NaCl. *P<0.05, n=3 independent experiments, Student’s t-test. Values for luciferase assays were normalized across measurements for each experiment and for transfection efficiency relative to β-gal. Error bars represent S.E.M. g. Localization of endogenous β-catenin (green) in MEFs treated with APH. Arrows point to two cilia (red, acetylated tubulin) within one cell while arrowheads point to nuclear β-catenin in a neighboring nonciliated cell. Hoechst (blue) labels nuclei. h. Model of the proposed mechanism of ciliary inhibition of canonical Wnt signaling. Jbn is proposed to interact with β-catenin upon Wnt activation and facilitate its translocation into the nucleus. In the absence of the cilium, this translocation is uninhibited compared to ciliated cells where nuclear translocation is highly regulated. This regulation is dependent upon intact retrograde IFT of Jbn down the cilium to the basal body where it can then interact with β-catenin and facilitate its nuclear accumulation. Furthermore, the presence of two cilia more strictly regulates the nuclear import of β-catenin, further dampening the pathway.
To test whether this sequestration affects Wnt pathway responsiveness, we performed luciferase experiments in wild-type ciliated MEFs transfected with Dnchc2 siRNA. We observed a decrease in Wnt response with knockdown of Dnchc2 suggesting intact retrograde IFT is required for the Wnt response in ciliated cells (Fig. 3b). This effect was also evident in ciliated 293T cells transfected with a Jbn overexpression construct, which failed to potentiate the pathway following Dnchc2 knockdown to the same degree as in control siRNA transfected cells (Fig. 3c).
We next examined the basis for the difference in effect between siRNA knockdown of Dnchc2 and null mutation of Dnchc2 in the MEFs. First, we examined whether the effect of Dnchc2 knockdown was cilia specific by testing Wnt response following knockdown of Dnchc2 in nonciliated cells. We tested MEFs that were not subjected to serum starvation, and therefore lacked cilia, and found that in these nonciliated MEFs, knockdown of Dnchc2 had no effect (Supplementary Fig. 5a). We next tested whether the degree of knockdown may be the basis for the difference between null MEFs and MEFs transfected with siRNA. We found that increasing the dosage of Dnchc2 siRNA by 3-fold resulted in a loss of cilia rather than the more subtle retrograde transport defects seen with lower siRNA concentrations (Supplementary Fig. 5b). Furthermore, this high dose siRNA transfection led to a switch in Wnt responsiveness such that cells transfected with increased siRNA exhibited an increase in Wnt responsiveness (Fig. 3d).
Our results suggest that, whereas retrograde IFT is required in ciliated cells for Wnt activity, complete loss of the cilium leads to increased Wnt responsiveness. This suggests the cilium regulates canonical Wnt signaling through dual roles in order to restrict the range of Wnt activity. This is reminiscent of the dual role cilia play in hedgehog signaling recently described25, 26. In order to test this hypothesis further, we examined cells with more than one cilium. We found that treatment of MEFs with the DNA polymerase inhibitor aphidicolin led to the appearance of many cells with two cilia, similar to the topoisomerase inhibitor described by Anderson and Stearns27 (Supplementary Fig. 5c, d). We therefore tested whether application of this drug affected Wnt responsiveness. We found that cells treated with aphidicolin exhibited a dramatic reduction in Wnt activity compared with cells treated with vehicle alone (Fig. 3e, f). Furthermore, we examined β-catenin localization in cells treated with aphidicolin and found strikingly decreased levels of nuclear β-catenin in biciliated cells (Fig. 3g).
These findings suggest a model in which the primary cilium maintains a restricted range of Wnt activity through spatial constraint of Jbn and β-catenin, thus limiting nuclear entry (Fig. 3h). This model would fit with the observation that ciliated cells exhibited muted Wnt activity but not a complete lack of response. In the absence of the cilium, Jbn and its partner β-catenin enter the nucleus more freely, resulting in an exaggerated downstream transcriptional response following stimulation. Specific inhibition of retrograde IFT instead leads to increased sequestration and therefore a decrease in Wnt response, suggesting a fine balance of Wnt responsiveness in ciliated cells through spatial regulation of Wnt components. Biciliation further restricts this range resulting in a further muted response.
We tested the physiological relevance of this regulation by examining Dnchc2 mutant (Dnchc2Q397Stop allele14); BATgal+28 mice for Wnt reporter activity during embryonic development. We selected this mutant because of the specific effect on retrograde IFT described previously14, which we hypothesized would lead to increased sequestration as we have seen in fibroblasts with knockdown of Dnchc2. First, we examined BATgal reporter activity by whole-mount X-gal staining of embryos at E9.5 and E11.5. We observed a decrease in Wnt activity in a subset of tissues at these time points, such as the limb buds, developing hair follicles and mammary glands, whereas other regions such as nasal folds and brain tissue exhibited increased Wnt activity (Fig. 4a and Supplementary Fig. 6a). In order to examine the cilia in these mutants, we stained whole embryo sections for both β-galactosidase and for the presence of cilia using the Arl13b antibody29 (Fig. 4b). β-galactosidase antibody staining in control embryos revealed specific staining in Wnt responsive tissues of BATgal+ embryos compared with BATgal- embryos (Supplementary Fig. 6b). Dnchc2; BATgal+ mutants instead exhibited several regions (including limb bud, metanephros, and developing lung) that displayed decreased Wnt activities (Fig. 4b and Supplementary Fig. 6c) with abundant but shortened cilia (Supplementary Fig. 6d). Furthermore, we performed immunofluorescence staining for β-catenin to examine nuclear localization, along with Arl13b staining on serial sections, and similarly observed a decrease in Wnt activity in developing kidney of Dnchc2 mutants (Supplementary Fig. 6e). These results suggest that loss of the retrograde motor leads to inhibition of Wnt signaling in specific ciliated tissues consistent with our model of ciliary regulation of the canonical Wnt pathway.
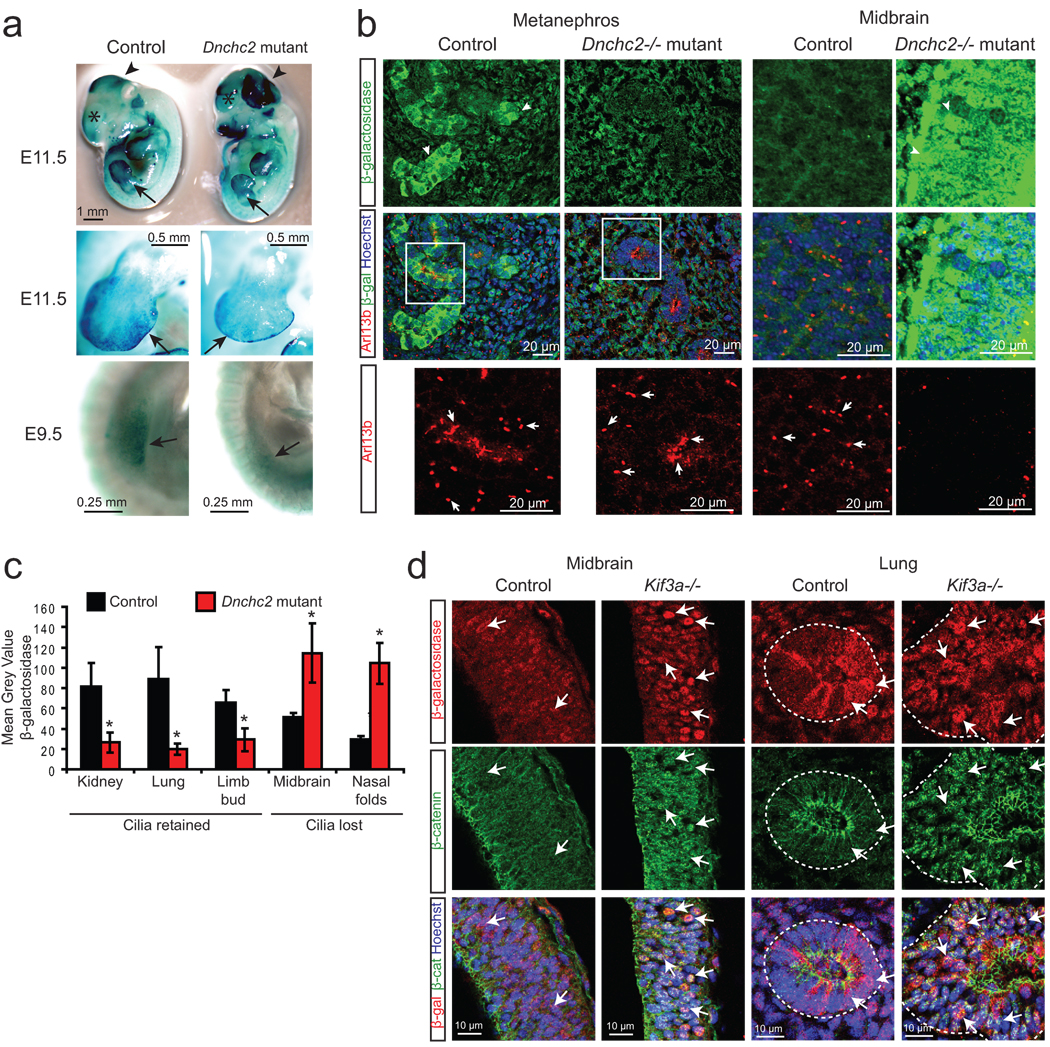
Figure 4. Wnt is regulated by the primary cilium during embryonic development.
a. X-gal staining (blue) of Dnchc2 mutants at E11.5 and E9.5. Arrows point to developing limb bud, arrowheads demarcate the mid-hindbrain region, and asterisks label the developing telencephalon. b. Representative E11.5 embryonic sections of the developing kidney (metanephros) and midbrain stained for cilia (Arl13b, red) and Wnt activity (β-galactosidase, green). Hoechst labels nuclei (blue). Arrows point to cilia in control and mutant tissue, which are shown as magnified insets for the metanephros. Images for control and mutant tissues were acquired and analyzed identically. c. Quantification of average staining intensity for β-galactosidase in representative tissues from three E11.5 littermate control and Dnchc2 mutant embryos. Tissues that are ciliated (kidney, lung, limb bud) and nonciliated (midbrain, nasal folds) in the mutant are shown for comparison of the effect of loss of cilia compared with specific retrograde IFT defect. *P<0.05, Student’s t-test. Error bars represent S.E.M. d. β-galactosidase staining (red) and β-catenin staining (green) in E9.5 midbrain and lung from Kif3a mutant and littermate control embryos. Arrows point to β-galactosidase reporter activity and β-catenin nuclear staining, both of which are increased in Kif3a mutant tissues.
Interestingly, some regions, such as brain tissue and nasal folds, exhibited a loss of cilia in these mutants (Fig. 3b and data not shown) similar to the Dnchc2 MEFS, suggesting cell type specific sensitivity to the absence of Dnchc2 and its effect on cilia generation or maintenance. Although the basis of this difference is still under investigation, we observed an increase in Wnt activity in nonciliated tissues (Fig. 4b), which is also consistent with the model that cilia generally repress Wnt responsiveness. Furthermore, we observed variability in the severity of phenotypes with more severe mutants exhibiting more extensive cilia loss with associated increased canonical Wnt activity (data not shown). However, we universally observed decreased Wnt activity in those regions that maintained cilia, while regions with a loss of cilia exhibited increased Wnt activity (quantified in Fig. 4c). Finally, to further examine this effect of loss of cilia in vivo, we examined Kif3a−/−; BATgal+ embryos which completely lack cilia30, and we observed a similar increase in Wnt activity in all regions examined (Fig. 4d). Together, these results suggest that, whereas a specific disruption of retrograde IFT leads to a decrease in Wnt signaling in vivo, complete loss of the cilium instead results in increased Wnt signaling.
In addition to its roles in embryonic development, the canonical Wnt pathway is also a key regulator of proliferation and cancer transformation. Since the cilium has also recently been implicated in proliferation in the context of cancer25, 26, and Jbn has previously been identified as an oncogene in certain types of leukemia and lymphoma in mice and humans31, we hypothesized that the cilium and Jbn may regulate canonical Wnt signaling in cancer cell proliferation as well. To test this hypothesis, we first examined proliferation of ciliated MEFs transfected with Jbn-GFP or the cilia localization deficient ΔRVxP mutant Jbn. We observed an increase in proliferation upon overexpression of this mutant compared with wild-type Jbn (Fig. 5a), suggesting ciliary regulation of Jbn and the Wnt pathway may represent an important regulator of cellular proliferation in fibroblasts.
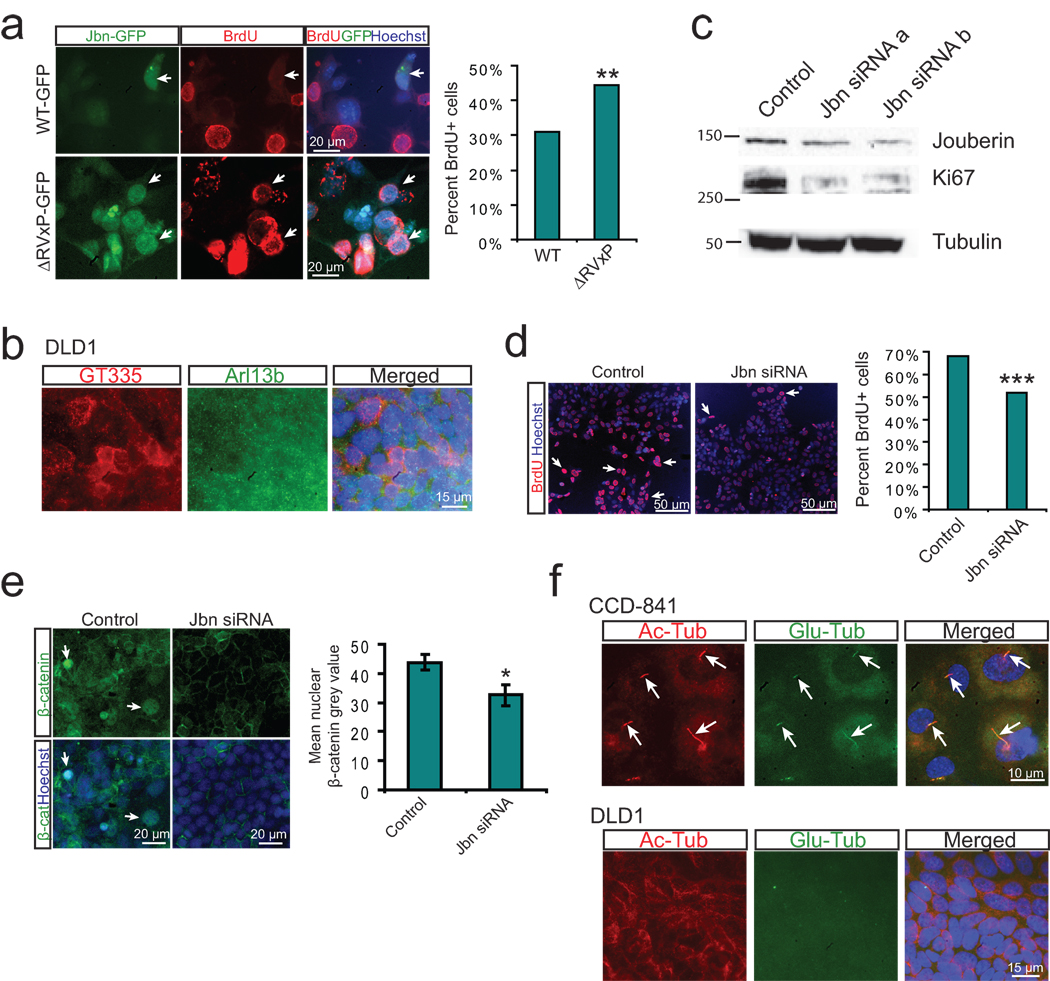
Figure 5. Regulation of cancer cell proliferation by cilia-localized Jouberin.
a. BrdU staining (red) in MEFs transfected with GFP tagged wild-type or ΔRVxP mutant construct (green). Hoechst labels nuclei (blue). Arrows point to GFP positive transfected cells. Quantification of the percent of 200 counted GFP positive cells which stained positive for BrdU is shown at right. **P<0.0005, Chi-square test. b. Staining for cilia in DLD1 cells reveals a lack of cilia. Cilia markers include GT335 (red), Arl13b (green), while Hoechst labels nuclei. c. Western blot analysis of Jbn and Ki67 expression from whole cell lysates of DLD1 cells transfected with Jbn siRNAs. Alpha-tubulin is shown as the loading control. d. BrdU staining (red) of DLD1 cells transfected with Jbn siRNA and treated for 16 hr with BrdU. Hoechst labels nuclei (blue). Quantification of 3000 cells from images acquired and analyzed identically is shown at the right. ***P<10−5, Chi-square test. e. β-catenin staining (green) in DLD1 cells transfected with Jbn siRNA. Hoechst labels nuclei (blue). Arrows point to nuclear β-catenin staining. Quantification of average nuclear staining intensity for β-catenin from over 100 cells in three regions of the well is shown at the bottom. *P<0.05, Student’s t-test. Error bars represent S.E.M. f. Cilia staining using two markers: acetylated tubulin (red) and glutamylated tubulin (green) in human colon CCD-841 cells which exhibited cilia in 75% of cells (over 200 cells examined) whereas DLD1 cells completely lacked cilia. Hoechst (blue) labels nuclei.
We next tested the potential role of Jbn in cancer cell proliferation by examining the well-described colon cancer DLD1 cell line which harbors a mutation in APC32 leading to overactivation of the Wnt pathway upstream of β-catenin, and which we found lacked cilia (Fig. 5b). We performed Jbn knockdown experiments in these cells and observed a decrease in proliferation based upon BrdU incorporation and Ki67 levels (Fig. 5c, d). Additionally, β-catenin nuclear levels were decreased in DLD1 cells treated with Jbn siRNA (Fig. 5e). These results suggest a role for Jbn in Wnt-mediated cancer cell proliferation and point to a potential novel therapeutic target downstream of most carcinogenic Wnt pathway mutations described, including the APC mutation in DLD1 cells. Since DLD1 cells lacked cilia, one intriguing hypothesis is that the loss of cilia may represent an important step in the transformation of cancer cells such as DLD1 cells, since it may be expected to exacerbate Wnt defects. Indeed, we found many cilia in healthy primary human colon cells, in contrast to DLD1 cells (Fig. 5f).
Our findings implicate a new role for the cilium in spatial compartmentalization of signaling components, and specifically a mechanism for inhibition of a transcriptional regulator through sequestration at a discrete subcellular location. Strikingly, we found that although the presence of the cilium dampens Wnt responses, retrograde IFT is necessary for this same response in ciliated cells. This dual positive and negative regulation sets up a discrete range of responsiveness in ciliated cells and may help explain why particular cells (ie. in the developing embryo) respond to Wnt ligands within a specific range of activity and why others (ie. cancer cells) exhibit out-of-control responses. Although we describe a novel point of ciliary regulation in the pathway, others have described regulation of Dvl9 and proteasomal degradation of β-catenin8 as possible mechanisms, suggesting multiple points of regulation for this organelle. Whether similar spatial control is involved at these other points in the pathway or whether this type of regulation is at play in other signaling pathways remains to be determined, but our results suggest a new spatial facet to the conventional view of the Wnt signaling cascade.
Additionally, our results point to a specific role for Jbn in cilia mediated Wnt signaling regulation. It is important to point out that although Ahi1 mutant mice exhibit Wnt defects, these occur only in specific tissues and defined stages of development (manuscript in press) and homeostasis2, whereas severe developmental Wnt phenotypes are spared. Furthermore, we have examined Ahi1−/−; BATgal+ mice which exhibit grossly normal embryonic Wnt activity in the majority of tissues (data not shown). Thus, it is clear that Jbn is not unique in its Wnt regulatory role, and other proteins have been suggested to regulate β-catenin nuclear translocation33, though potential roles at the cilium have not been examined. Moreover, there are now over 35 genes which cause the range of ciliopathy phenotypes6, including many that overlap with Wnt phenotypes, so there is likely a high degree of compensation between the ciliopathy proteins, including Jbn. Finally, these results along with our previous findings2 point to Jbn acting as a positive modulator, rather than as a broadly required or permissive factor. Overall, our findings suggest the primary cilium may act as a detour to the nucleus, thereby segregating cytosolic components to keep the Wnt pathway in check. This model may in part explain the predisposition of certain types of cancer cells to lose the primary cilium34 since this may allow for positive selection of cells with hyperactivation of the oncogenic Wnt pathway.
Methods
Plasmid constructs and materials
GFP tagged Jbn was used as described previously2. β-catΔN expression construct was obtained from M.G. Rosenfeld, Super Topflash construct was provided by R.T. Moon, and Dvl-1 and β-catenin constructs were obtained from K. Willert. Jbn siRNA, Dnchc2 siRNA, and Kif3a siRNA oligonucleotides were purchased from Invitrogen (Stealth RNAi). Dnchc2 siRNA #1: GGGCGGAUAUUUGUUGGGUUGGUAA. Dnchc2 siRNA #2: GAGCAGUGUUCUCACUGAUUGAUUA. Dnchc2 siRNA #3: ACUGCUGUGUCAGUUUGCAU GGUUU. Kif3a siRNA #1: UCAUGUGCCUUAUCGUAACUCUAAA. Kif3a siRNA #2: GACUCGUCUUCUUCAGGAUUCCUUA. Kif3a siRNA #3: GCGAUAAUGUGAAGGUUGUUGUUAG. Jbn siRNA-a corresponds to previously2 used oligo #2 and siRNA-b corresponds to previously used oligo #3. These siRNAs were used because of homology to the human sequence in DLD-1 cells. Low GC content siRNA control oligo was used as a negative control (Invitrogen). Dnchc2 siRNA #1 was used for luciferase in MEFs and 293Ts. Kif3a siRNA #1 was used for luciferase in 293T while Kif3a siRNA #3 was used for MEFs because of greater homology (100% in the seed region). ΔRVxP mutant construct was generated by QuikChange Site-Directed Mutagenesis Kit (Stratagene) to generate the following combination of mutations: R692G, V693A, and P695A. Wnt3A conditioned media (WCM) was obtained from stably transfected L cells with Wnt3A expression vector (provided by K. Willert) and used as described35. Control media was obtained from untransfected L cells. Aphidicolin (Sigma-Aldrich, A0781) was used at a concentration of 1 µg/ml from a stock solution of 1 mg/ml dissolved in DMSO. DMSO was used as vehicle control.
Mouse lines and tissue preparation
Dnchc2Q397Stop allele mice14 were kindly provided by A.S. Peterson, and Kif3a+/− mice were kindly provided by L.S.B. Goldstein. Both lines were then crossed with BATgal+ mice28 (a gift from S. Piccolo). We obtained embryos from timed matings which were fixed for 2 hours in 4% paraformaldehyde (PFA) followed by whole mount X-gal staining and/or embedding in 10%/7.5% gelatin/sucrose and cryosectioning at 20 µm for immunostaining. Mouse work was carried out in compliance with Institutional Animal Care and Use Committee approved protocols. X-gal staining was performed according to a previously published protocol36.
Immunofluorescence
Cilia were induced in fibroblasts by serum starvation for 24 hrs in OptiMEM (Invitrogen). MEFs and IMCDs were treated with WCM or control L-cell conditioned media (LCM) diluted 1:3 in OptiMEM for 4 hrs for localization studies. MEFs, HEK293T, NIH3T3, DLD-1, CCD-841 (CoN, untransformed) and IMCD cells (ATCC) or Dnchc2 mutant and control MEFs (obtained from A.S. Peterson) were fixed with 4% PFA for 15 min at room temperature, washed in PBS and blocked in 4% donkey serum/0.1% TritonX. BrdU labeling experiments were performed by incubation with BrdU (10 µM) in the medium (serum free for MEFs) for 16 hrs with WCM followed by fixation and treatment with 2N HCl for 15 min at 37°C before staining. Fixed cells and tissue sections were stained with primary antibodies: mouse anti-acetylated tubulin (Zymed 32–2700, 1:500 dilution), mouse anti-β-catenin (BD Transduction Labs, 610153, 1:200), rabbit anti-β-catenin (Cell Signaling, 95825, 1:200), goat anti-γ-tubulin (Santa Cruz Biotech., sc7396, 1:100), rabbit anti-Arl13b (gift from T. Caspary, 1:1500), rabbit anti-glutamylated tubulin (detyrosinated tubulin, Millipore, AB3201, 1:750), chicken anti-β-galactosidase (Abcam, ab9361, 1:250), rat anti-BrdU (Abcam, ab6326, 1:100) mouse anti-polyglutamylated tubulin (GT335 antibody, a gift from C. Janke, 1:1000) and rabbit anti-Jbn2 (Quality Controlled Biochemicals, 1:200). Samples were washed and incubated in secondary antibodies (AlexaFluor donkey anti-mouse 594, donkey anti-rat 594, donkey anti-chicken 488, donkey anti-rabbit 488, and donkey anti-goat 647, Molecular Probes, 1:500). Hoechst was used as a nuclear stain (Molecular Probes, H3570). Images were acquired using a DeltaVision Spectris deconvolution microscope or FV1000 Spectral Deconvolution Confocal microscope (UCSD Neuroscience Microscopy Core) and controls and Dnchc2 mutant tissues were imaged using identical settings and exposures. Quantification was performed with ImageJ software (NIH). For quantification of nuclear β-catenin levels, ImageJ was used to generate a binary mask from Hoechst staining which was then applied to the green channel to quantify only nuclear levels.
Cell culture and luciferase assays
293Ts, 3T3s, DLD-1 and IMCDs were grown in DMEM with 10% fetal bovine serum (FBS), and MEFs were grown in 15% FBS. For transient transfections of 293Ts, 3T3s or MEFs, Lipofectamine 2000 (Invitrogen) was used according to manufacturer’s protocol. For luciferase assays, 293T cells, Dnchc2 mutant and littermate control MEFs and an independent wild-type MEF cell line were grown in 12-well plates and transfected with Super Topflash and β-Galactosidase expression plasmid (β-gal) at a 5:1 ratio. Cells were co-transfected in certain experiments with Jbn expression plasmid or empty vector2 and/or with Dvl-1 expression plasmid or β-CatΔN at an amount equal to Topflash plasmid DNA. For experiments involving siRNA, 5 pmol siRNA (or 15 pmol for high dose experiments) was cotransfected with 400 ng total DNA per well according to the manufacturer. Twenty-four hrs following transfection, cells were serum starved for 12-36 hrs then treated with WCM or LCM diluted 1:3 in OptiMEM, or 20 mM Lithium (LiCl), or 20 mM NaCl as a negative control, to stimulate the Wnt pathway. Aphidicolin treatments (1 µg/ml) were performed with Wnt or LiCl treatments. For serum withdrawal and re-addition experiments, cells were serum starved for 24 hrs followed by addition of media with 15% serum. This was followed by WCM/LCM treatment or LiCl/NaCl addition for 8 hrs. The luciferase assay was performed according to a previously published protocol37 and β-gal activity was measured using the Tropix Galacto-light Plus kit (Applied Biosystems, T1007). β-gal was used to normalize data for transfection efficiency, and data were normalized for variability across experiments to control untreated or empty vector transfected cells. Statistical analyses included 2-tailed, unpaired Student’s t-test, and Chi-square test to calculate the p-value.
Western blot and microtubule association assay
For the microtubule association assay, five P5 mice were dissected and the cerebellum was removed and homogenized in MES buffer followed by centrifugation at 25,000 rpm for 15 min. Supernatant was subsequently centrifuged at 75,000 rpm for 90 minutes and samples were then treated with 100 mM GTP and 1 mM Taxol for 30 min at 37°C. Samples werecentrifuged through a 10% sucrose cushion and the pellet was resuspended in MES buffer and loaded on a 12% SDS-PAGE. Nuclear extraction was performed as previously described2. Western blotting was performed on lysates prepared in modified RIPA buffer using the following antibodies: rabbit anti-Jbn (Quality Controlled Biochemicals), goat anti-DCX (Santa Cruz Biotech., SC-8067), rabbit Cyclin D1 (Santa Cruz Biotech, SC-718), mouse anti-α-tubulin (Sigma, T-6074), rabbit anti-total ERK (Upstate, 06-182), rabbit anti-Dnchc1 and rabbit anti-Dnchc2 antibodies (both kindly provided by R.B. Vallee), mouse anti-Kif3a (Covance, MMS-198P), mouse mab414 (Abcam, 24609), rabbit anti-Ki67 (Novocastra, NCL-Ki67p). All antibodies were used at 1:1000 dilution in 4% milk.
Supplementary Material
a. Staining for cilia (acetylated tubulin, red, arrow) in serum starved MEFs with reapplication of serum. These results are quantified at the right for at least 200 cells. ***P<0.0005, Chi-square test. Nuclei are labeled with Hoechst (blue). b. Staining for the primary cilium (acetylated tubulin, red, arrow) in Dnchc2 MEFs reveals loss of cilia in Dnchc2−/− MEFs. These results are quantified in the panel on the right for at least 200 cells. ***P<0.0005, Chi-square test. c. Quantification of percent ciliated Dnchc2 mutant MEFs using an antibody for an alternative cilia marker Arl13b. ***P<0.0005, Chi-square test. d. Staining for cilia (acetylated tubulin, red, arrows) in MEFs serum starved and treated with NaCl or LiCl. Average cilia length was quantified from over 30 cells from three regions of the well for each treatment. e. Luciferase activity in Dnchc2 MEFs with activation of the pathway with lithium (LiCl). *P<0.05, n=3 independent experiments, Student’s t-test. Error bars represent S.E.M.
a. Western blot analysis of whole cell lysates from 293Ts transfected with Kif3A siRNA. Alpha-tubulin is shown as a loading control. b. Staining for cilia (acetylated tubulin, red) in MEFs transfected with Kif3a siRNA #3. Arrows point to cilia in control transfected cells. Hoechst (blue) labels nuclei. Results are quantified at the right from at least 200 cells for each. ***P<0.0005, Chi-square test. c. β-catenin-GFP (green) localization in untreated ciliated (acetylated tubulin, red) and nonciliated MEFs. Arrow points to basal body localized β-catenin in ciliated cell. Hoechst (blue) labels nuclei.
a. Staining for endogenous β-catenin (green) in ciliated (actylated tubulin, red) and nonciliated (arrow) IMCDs revealing increased nuclear (Hoechst, blue, outlined) localization of β-catenin in nonciliated cells compared with ciliated neighboring cells. b. Quantification of nuclear β-catenin staining in ciliated and nonciliated IMCDs. Mean grey values were calculated using ImageJ software (NIH) to create a binary mask from Hoechst staining as a nuclear boundary for measurement of β-catenin levels. *P<0.05, n=20 nonciliated and n=48 ciliated cells, Student’s t-test. Error bars represent S.E.M. c. Western blot (WB) for β-catenin and Jbn from nuclear preps of Dnchc2+/+ and −/− MEFs treated with Wnt3a conditioned media. The increase in nuclear β-catenin levels in Dnchc2−/− cells is more pronounced compared with +/+ cells. Additionally, Jbn exhibits increased nuclear levels in Dnchc2 −/− cells. Mab414 is shown as a nuclear loading control and Alpha-tubulin is a cytosolic control. d. Western blot for Jbn from total cell lysates from Dnchc2 mutant and control MEFs treated with Wnt3a conditioned or L cell conditioned media. Alpha-tubulin is shown as loading control.
a. 293T cells stained for cilia with antibody to acetylated tubulin (red) display clear cilia with Jbn-GFP (green) localized to the basal body similar to previously described in other ciliated cells2, 38. Hoechst was used to stain the nucleus (blue). b. Cilia staining of 293T cells with two independent markers: acetylated tubulin and Arl13b. Arl13b reveals a greater number of cilia (44% of cells) than acetylated tubulin staining (10% of cells). c. Luciferase activity in MEFs transfected with Kif3a siRNA and overexpressed Jbn or EV with activation of the pathway with cotransfected β-catΔN. *P<0.05, n=3 independent experiments, Student’s t-test. Error bars represent S.E.M. d. Localization of wild-type and ΔRVxP mutation (GFP tagged constructs, green) to primary cilia (labeled by acetylated tubulin, red) of MEFs. Hoechst (blue) labels nuclei. Arrows point to cilia. Arrowheads point to basal body localization of Jbn wild-type construct. e. Microtubule association assay in P5 mouse cerebellum lysates followed by western blotting reveals endogenous Jbn associated with repolymerized microtubules in the Pellet and Pellet + Taxol fractions. Blots were probed for Doublecortin (DCX) as a positive control, and total ERK as a negative control. α-tubulin is enriched in pellet fractions. f. Western blot analysis of whole cell lysates from 3T3s transfected with Dnchc2 siRNA constructs. Dnchc2 levels were specifically decreased in siRNA-1 and siRNA-2 transfected cells without an effect on dynein heavy chain 1 (Dnchc1) which is also shown as a loading control.
a. Luciferase activity in MEFs without serum starvation and transfected with 5 pmol Dnchc2 siRNA and treated with WCM or LCM. N.S.=not significant, n=3 independent experiments, Student’s t-test. Error bars represent S.E.M. b. Staining for cilia (acetylated tubulin, red, arrow) in MEFs transfected with low dose and high dose (3-fold higher) Dnchc2 siRNA. Hoechst (blue) labels nuclei. These results are quantified in the panel on the right for at least 200 cells. ***P<0.0005, Chi-square test. c. Staining for cilia (acetylated tubulin, red, arrows) in serum starved MEFs treated with 1 µg/ml aphidicolin (APH) or vehicle. d. Quantification of percent of cells with two cilia from at least 200 cells for each treatment. ***P<0.0005, Chi-square test.
a. Whole mount images of X-gal stained E9.5 control and Dnchc2 mutant embryos revealing increased Wnt activity in the mid-hindbrain region and nasal folds (arrows). b. β-galactosidase staining (green) of E9.5 BATgal+ and BATgal− control mesonephros (outlined). Hoechst (blue) labels nuclei. c. Wnt reporter staining (β-galactosidase, green) in representative E11.5 lung and limb bud of Dnchc2 mutant and control littermate. Arl13b (red) stains cilia in these tissues. Cilia (arrows) are retained in lung and limb bud. Hoechst labels nuclei (blue). d. High magnification image revealing cilia (red, Arl13b staining) in control and Dnchc2 mutant embryos. Hoechst labels nuclei (blue). Arrows point to cilia, while arrowhead points to apparently bulbous cilia of Dnchc2 mutant embryo. e. β-catenin staining (red) in E11.5 metanephros (outlined) of littermate control and Dnchc2 mutant embryos revealing decreased nuclear staining (arrows) in mutants compared with controls. Adjacent sections (bottom panels) were also stained for cilia (Arl13b, green). Hoechst staining (blue) labels nuclei.
Acknowledgements
We are grateful to members of the Gleeson lab for technical expertise and feedback and the UCSD Neuroscience Microscopy Core. We would like to thank C. Kintner for helpful feedback on the manuscript. We also thank K. Willert for reagents and technical expertise. We are grateful to S. Piccolo at the Departments of Histology, Microbiology and Medical Biotechnologies, University of Padua, Italy for the BATgal mice and to A.S. Peterson at the Ernest Gallo Clinic and Research Center, Emeryville, California for the Dnchc2 mutant mice and MEFs as well as L.S.B. Goldstein for Kif3a mutant mice. We also thank M.G. Rosenfeld at the School of Medicine, University of California, San Diego for the β-catΔN construct, and R.T. Moon at the Department of Pharmacology, University of Washington for the Super Topflash construct, as well as T. Caspary at the Department of Human Genetics, Emory University School of Medicine for the Arl13b antibody, C. Janke at Curie Institut, Paris, France for GT335 antibody, and R.B. Vallee at the Department of Pathology and Cell Biology, Columbia University for Dnchc1 and Dnchc2 antibodies. We also thank P. Mellon for β-galactosidase expression construct for luciferase assays. M.A.L. received support from the Bear Necessities Pediatric Cancer Foundation. This work was supported by the US National Institutes of Health, and the Burroughs Wellcome Fund in Translational Research (J.G.G.). J.G.G. is an investigator with Howard Hughes Medical Institute.
Footnotes
Author contributions. J.G.G. and M.A.L. conceived and designed the experimental approach, interpreted data, and wrote the manuscript. M.A.L. and J.S. carried out experiments. J.G.G. directed and supervised the project.
Competing financial interests statement. The authors declare no competing financial interests.
References
- 1.Berbari NF, O'Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lancaster MA, et al. Impaired Wnt-beta-catenin signaling disrupts adult renal homeostasis and leads to cystic kidney ciliopathy. Nat Med. 2009;15:1046–1054. doi: 10.1038/nm.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai SL, Chien AJ, Moon RT. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 2009;19:532–545. doi: 10.1038/cr.2009.41. [DOI] [PubMed] [Google Scholar]
- 5.Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- 6.Lancaster MA, Gleeson JG. The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev. 2009;19:220–229. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 9.Corbit KC, et al. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 10.Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–3098. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ocbin PJ, Tuson M, Anderson KV. Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS One. 2009;4:e6839. doi: 10.1371/journal.pone.0006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaykas A, et al. Mutant Frizzled 4 associated with vitreoretinopathy traps wild-type Frizzled in the endoplasmic reticulum by oligomerization. Nat Cell Biol. 2004;6:52–58. doi: 10.1038/ncb1081. [DOI] [PubMed] [Google Scholar]
- 13.Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May SR, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 15.Hedgepeth CM, et al. Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev Biol. 1997;185:82–91. doi: 10.1006/dbio.1997.8552. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi K, Kasahara K, Miyazaki I, Asanuma M. Lithium treatment elongates primary cilia in the mouse brain and in cultured cells. Biochem Biophys Res Commun. 2009;388:757–762. doi: 10.1016/j.bbrc.2009.08.099. [DOI] [PubMed] [Google Scholar]
- 17.Louie CM, Gleeson JG. Genetic basis of Joubert syndrome and related disorders of cerebellar development. Hum Mol Genet. 2005;14(Spec No. 2):R235–R242. doi: 10.1093/hmg/ddi264. [DOI] [PubMed] [Google Scholar]
- 18.Dixon-Salazar T, et al. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferland RJ, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 20.Geng L, et al. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci. 2006;119:1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- 21.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 22.Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Signor D, et al. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. The Journal of cell biology. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pazour GJ, Dickert BL, Witman GB. The DHC1B (DHC2) isoform of cytoplasmic dynein is necessary for flagellar maintenance as well as flagellar assembly. Mol Biol Cell. 1999;10S:396a. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong SY, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han YG, et al. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol. 2009;19:1498–1502. doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maretto S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci U S A. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennah E, et al. Identification of tyrosine kinase, HCK, and tumor suppressor, BIN1, as potential mediators of AHI-1 oncogene in primary and transformed CTCL cells. Blood. 2009;113:4646–4655. doi: 10.1182/blood-2008-08-174037. [DOI] [PubMed] [Google Scholar]
- 32.Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117:6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- 33.Townsley FM, Cliffe A, Bienz M. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat Cell Biol. 2004;6:626–633. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- 34.Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer research. 2008;68:2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 36.Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- 37.Nelson SB, Lawson MA, Kelley CG, Mellon PL. Neuron-specific expression of the rat gonadotropin-releasing hormone gene is conferred by interactions of a defined promoter element with the enhancer in GT1-7 cells. Mol Endocrinol. 2000;14:1509–1522. doi: 10.1210/mend.14.9.0521. [DOI] [PubMed] [Google Scholar]
- 38.Eley L, et al. Jouberin localizes to collecting ducts and interacts with nephrocystin-1. Kidney Int. 2008 doi: 10.1038/ki.2008.377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a. Staining for cilia (acetylated tubulin, red, arrow) in serum starved MEFs with reapplication of serum. These results are quantified at the right for at least 200 cells. ***P<0.0005, Chi-square test. Nuclei are labeled with Hoechst (blue). b. Staining for the primary cilium (acetylated tubulin, red, arrow) in Dnchc2 MEFs reveals loss of cilia in Dnchc2−/− MEFs. These results are quantified in the panel on the right for at least 200 cells. ***P<0.0005, Chi-square test. c. Quantification of percent ciliated Dnchc2 mutant MEFs using an antibody for an alternative cilia marker Arl13b. ***P<0.0005, Chi-square test. d. Staining for cilia (acetylated tubulin, red, arrows) in MEFs serum starved and treated with NaCl or LiCl. Average cilia length was quantified from over 30 cells from three regions of the well for each treatment. e. Luciferase activity in Dnchc2 MEFs with activation of the pathway with lithium (LiCl). *P<0.05, n=3 independent experiments, Student’s t-test. Error bars represent S.E.M.
a. Western blot analysis of whole cell lysates from 293Ts transfected with Kif3A siRNA. Alpha-tubulin is shown as a loading control. b. Staining for cilia (acetylated tubulin, red) in MEFs transfected with Kif3a siRNA #3. Arrows point to cilia in control transfected cells. Hoechst (blue) labels nuclei. Results are quantified at the right from at least 200 cells for each. ***P<0.0005, Chi-square test. c. β-catenin-GFP (green) localization in untreated ciliated (acetylated tubulin, red) and nonciliated MEFs. Arrow points to basal body localized β-catenin in ciliated cell. Hoechst (blue) labels nuclei.
a. Staining for endogenous β-catenin (green) in ciliated (actylated tubulin, red) and nonciliated (arrow) IMCDs revealing increased nuclear (Hoechst, blue, outlined) localization of β-catenin in nonciliated cells compared with ciliated neighboring cells. b. Quantification of nuclear β-catenin staining in ciliated and nonciliated IMCDs. Mean grey values were calculated using ImageJ software (NIH) to create a binary mask from Hoechst staining as a nuclear boundary for measurement of β-catenin levels. *P<0.05, n=20 nonciliated and n=48 ciliated cells, Student’s t-test. Error bars represent S.E.M. c. Western blot (WB) for β-catenin and Jbn from nuclear preps of Dnchc2+/+ and −/− MEFs treated with Wnt3a conditioned media. The increase in nuclear β-catenin levels in Dnchc2−/− cells is more pronounced compared with +/+ cells. Additionally, Jbn exhibits increased nuclear levels in Dnchc2 −/− cells. Mab414 is shown as a nuclear loading control and Alpha-tubulin is a cytosolic control. d. Western blot for Jbn from total cell lysates from Dnchc2 mutant and control MEFs treated with Wnt3a conditioned or L cell conditioned media. Alpha-tubulin is shown as loading control.
a. 293T cells stained for cilia with antibody to acetylated tubulin (red) display clear cilia with Jbn-GFP (green) localized to the basal body similar to previously described in other ciliated cells2, 38. Hoechst was used to stain the nucleus (blue). b. Cilia staining of 293T cells with two independent markers: acetylated tubulin and Arl13b. Arl13b reveals a greater number of cilia (44% of cells) than acetylated tubulin staining (10% of cells). c. Luciferase activity in MEFs transfected with Kif3a siRNA and overexpressed Jbn or EV with activation of the pathway with cotransfected β-catΔN. *P<0.05, n=3 independent experiments, Student’s t-test. Error bars represent S.E.M. d. Localization of wild-type and ΔRVxP mutation (GFP tagged constructs, green) to primary cilia (labeled by acetylated tubulin, red) of MEFs. Hoechst (blue) labels nuclei. Arrows point to cilia. Arrowheads point to basal body localization of Jbn wild-type construct. e. Microtubule association assay in P5 mouse cerebellum lysates followed by western blotting reveals endogenous Jbn associated with repolymerized microtubules in the Pellet and Pellet + Taxol fractions. Blots were probed for Doublecortin (DCX) as a positive control, and total ERK as a negative control. α-tubulin is enriched in pellet fractions. f. Western blot analysis of whole cell lysates from 3T3s transfected with Dnchc2 siRNA constructs. Dnchc2 levels were specifically decreased in siRNA-1 and siRNA-2 transfected cells without an effect on dynein heavy chain 1 (Dnchc1) which is also shown as a loading control.
a. Luciferase activity in MEFs without serum starvation and transfected with 5 pmol Dnchc2 siRNA and treated with WCM or LCM. N.S.=not significant, n=3 independent experiments, Student’s t-test. Error bars represent S.E.M. b. Staining for cilia (acetylated tubulin, red, arrow) in MEFs transfected with low dose and high dose (3-fold higher) Dnchc2 siRNA. Hoechst (blue) labels nuclei. These results are quantified in the panel on the right for at least 200 cells. ***P<0.0005, Chi-square test. c. Staining for cilia (acetylated tubulin, red, arrows) in serum starved MEFs treated with 1 µg/ml aphidicolin (APH) or vehicle. d. Quantification of percent of cells with two cilia from at least 200 cells for each treatment. ***P<0.0005, Chi-square test.
a. Whole mount images of X-gal stained E9.5 control and Dnchc2 mutant embryos revealing increased Wnt activity in the mid-hindbrain region and nasal folds (arrows). b. β-galactosidase staining (green) of E9.5 BATgal+ and BATgal− control mesonephros (outlined). Hoechst (blue) labels nuclei. c. Wnt reporter staining (β-galactosidase, green) in representative E11.5 lung and limb bud of Dnchc2 mutant and control littermate. Arl13b (red) stains cilia in these tissues. Cilia (arrows) are retained in lung and limb bud. Hoechst labels nuclei (blue). d. High magnification image revealing cilia (red, Arl13b staining) in control and Dnchc2 mutant embryos. Hoechst labels nuclei (blue). Arrows point to cilia, while arrowhead points to apparently bulbous cilia of Dnchc2 mutant embryo. e. β-catenin staining (red) in E11.5 metanephros (outlined) of littermate control and Dnchc2 mutant embryos revealing decreased nuclear staining (arrows) in mutants compared with controls. Adjacent sections (bottom panels) were also stained for cilia (Arl13b, green). Hoechst staining (blue) labels nuclei.