Abstract
Type 1 diabetes (IDDM) is a complex disorder with multifactorial and polygenic etiology. A genome-wide screen performed in a BC1 cohort of a cross between the nonobese diabetic (NOD) mouse with the diabetes-resistant feral strain PWK detected a major locus contributing to diabetes development on the distal part of chromosome 6. Unlike the majority of other Idd loci identified in intraspecific crosses, susceptibility is associated with the presence of the PWK allele. Genetic linkage analysis of congenic lines segregating PWK chromosome 6 segments in a NOD background confirmed the presence of the Idd locus within this region. The genetic interval defined by analysis of congenic animals showed a peak of significant linkage (P = 0.0005) centered on an ∼9-cM region lying between D6Mit11 and D6Mit25 genetic markers within distal mouse chromosome 6.
[Genetic markers polymorphic between the NOD and PWK strains are available as a supplement at http://www.genome.org]
Insulin-dependent diabetes mellitus (IDDM) in human is a clinically heterogeneous disorder that is characterized by severe insulin deficiency due to autoimmune destruction of the insulin-producing β cells of the endocrine pancreas, by both humoral and cell-mediated mechanisms (Bottazzo 1986). Therapy with exogenous insulin is required to avoid severe hyperglycemia and ketoacidosis, which can result in diabetic coma and death. Its onset is most common in the young with prevalences of up to 1% recorded in some geographic areas, especially in Northern Europe (Mimura 1982).
Both genetic and nongenetic factors contribute to disease development, and as such it represents a multifactorial disorder. Although the importance of genetic factors in causing diabetes is well established, both the number and identity of diabetogenic genes involved is uncertain.
In human, loci containing genes that confer susceptibility to the IDDM have been identified in the HLA region of chromosome 6 (Davies et al. 1994; Hashimoto et al. 1994) and near the insulin gene on chromosome 11p15 (Bell et al. 1984; Julier et al. 1991). Other regions, also have been reported as containing potential susceptibility loci (Hashimoto et al. 1994; for review, see Julier et al. 1996). Recently, evidence for the presence of an additional diabetogenic locus mapping to human chromosome 6, but outside the HLA region, has been identified (Luo et al. 1995; Delepine et al. 1997). At present, the locus best characterized at the molecular level, other than the HLA locus, is the 4.1-kb segment located near the insulin gene (Lucassen et al. 1993; Julier et al. 1994; Bennett et al. 1995; Homo-Delarche and Boitard 1996).
The task of understanding the genetics of complex human traits can be facilitated by the use of experimental animal models. For IDDM, two animal models have been described and widely used, the nonobese diabetic (NOD) mouse (Castano and Eisenbarth 1990) and the BB (BioBreeding) rat (Jacob et al. 1992).
The NOD mouse develops insulin-dependent diabetes spontaneously with remarkable similarities to the human disease. As in human, the onset of clinical diabetes in the NOD mouse is preceded by progressive infiltration of the pancreatic islets of Langerhans by mononuclear cells, predominantly T lymphocytes and macrophages. The importance of both non-major histocompatibility complex (non-MHC) and MHC-linked susceptibility genes in disease development is another shared characteristic (Makino et al. 1980). However, NOD mouse diabetes shows distinctive features such as higher prevalence in female than in male animals.
The first Idd locus identified to be linked to and correlate with susceptibility to disease was the MHC locus on mouse chromosome 17 (Acha-Orbea and McDevitt 1987; Lucassen and Bell 1995), designated Idd1 (Todd et al. 1987). At least 16 additional Idd loci have since been identified in crosses between the NOD-susceptible strain and various diabetes-resistant inbred laboratory strains, although none of these non-MHC genes has yet been isolated (Wicher et al. 1987; Garchon et al. 1991; Chesnut et al. 1993; De Gouyon et al. 1993; Ghosh et al. 1993; Morahan et al. 1994; Serreze et al. 1994; McAleer et al. 1995; for review, see Vyse and Todd 1996). Loci such as the insulin locus, which appears to play an important role in human type 1 diabetes, have not been shown, to date, to play a major role in controlling susceptibility to diabetes in the mouse.
Attempts to define murine Idd loci more closely have been hampered by the reduced penetrance characteristic of multifactorial traits. Fine mapping, using standard backcrosses or intercrosses, has proven to be rather unproductive (Vyse and Todd 1996). An alternative approach for refining the genetic interval containing the disease gene is the construction of congenic strains. Such strains differ in principal from the parental strain solely by the genetic region that has been selected as specific for the chromosome segment containing the gene responsible for a given phenotype.
Previous studies on the genetics of interspecies crosses with the NOD mouse have proven to be of interest (De Gouyon et al. 1993; Hattori et al. 1993). Using species or subspecies different from the Mus laboratorus, to which the NOD strain belongs for genetic studies, allows the introduction of additional genetic variation. This should allow the detection of additional Idd loci, or segregate, as major disease modifiers, loci that were not identified to be of major importance in other crosses.
In this work we have studied the genetics of type 1 diabetes in a backcross between the NOD strain and the PWK feral inbred subspecies. The PWK strain belongs to the Mus musculus musculus subspecies, which separated from Mus musculus domesticus some 1 million years ago (Bonhomme and Guénet 1989). The experimental method chosen for phenotypic evaluation in our study consists of the induction of type 1 diabetes by cyclophosphamide (CY). CY is an alkylating agent used to suppress immune reactivity but may also increase immune responses, perhaps by selectively depleting regulatory mechanisms, allowing effector cells to become dominant (Haroda and Makino 1984). The diabetes induced by CY is characterized by all of the clinical futures seen in the spontaneous model. We assumed that CY induction of type 1 diabetes might reduce nongenetic variance due to environmental factors and increase penetrance of the diabetogenic genes. In addition, faster evaluation of the phenotypes can be accomplished as animals are used at an early age (6–8 weeks old), whereas >8 months of age is required for spontaneous phenotypes to develop.
Data presented in this report obtained from the analysis of (NOD × PWK)F1 × NOD backcross suggest that a major non-MHC locus controlling resistance/susceptibility to disease development is localized on distal chromosome 6.
To confirm and localize more precisely the chromosome 6 locus we have adopted a strategy based on the construction of congenic lines by introgressing PWK donor segments of distal chromosome 6 into the NOD receiver strain. Phenotypic and genetic linkage analyses of these congenic strains, using both intercrosses and intercongenic strain comparison, is simplified by the absence of segregating PWK alleles other than those present in the selected region of distal chromosome 6. This approach has both confirmed and refined the initial localization of this Idd locus to an ∼9-cM region centered around the D6Mit25 genetic marker.
RESULTS
Phenotypic Analysis of the BC1 Progeny Derived from a (NOD × PWK) × NOD Intersubspecific Cross
BC1 crosses allow easy detection of recessive susceptibility modifiers and are commonly used for genetic analysis studies of type 1 diabetes in the NOD mouse (Risch et al. 1993). We have produced first backcross (BC1) progenies by breeding diabetes-free (NOD × PWK)F1 females to the NOD parental strain, and diabetes phenotypes have been assessed after CY treatment. CY-treated PWK control animals remain normal with histology scores never being >1, whereas 77% of treated NOD control animals tested developed overt diabetes (histology score 6) in our colony.
Currently, spontaneous diabetes (histology score 6) affects 33% and 14.8% of female and male animals, respectively, in our NOD colony. Insulitis (histology scores 3–5) is observed in 66% of females and 45.1% of males, whereas 40% of males and only 1% of females remain unaffected (histology 1 and 2) at 1 year of age. This frequency reflects the nonspecific pathogen-free housing conditions in the Pasteur Institute animal facility.
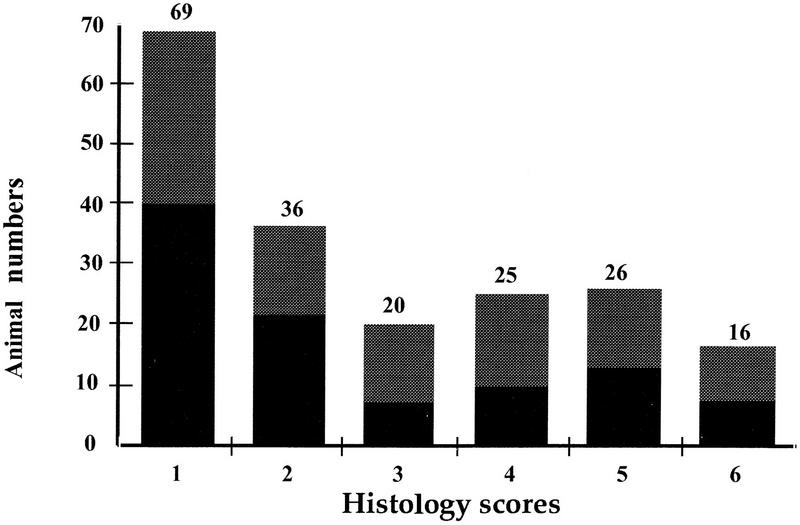
In the BC1 generation, 8% of the animals (seven males and nine females) developed diabetes after CY treatment (Fig. 1). Histological examination revealed that 35% of the animals (31 males and 36 females), developed insulitis (histology grades of ⩾4) (Fig. 1).
Figure 1.
Distribution of phenotypes in the BC1 progenies after CY treatment. Animal numbers are reported on the y-axis. Histology scores are 1–6 on the x-axis. (Shaded bars) Females; (solid bars) males.
Despite the use of CY the distribution of phenotypes observed in this (NOD × PWK)/NOD outcross closely ressembles those observed in crosses of the NOD mouse with other standard laboratory strains. When the laboratory mouse has been used as the resistant partner in crosses to NOD, 25%–50% of female animals have shown histology grades more than 4 for spontaneous insulitis (Ikegami and Makino 1993).
Genetic Analysis of the (NOD × PWK)NOD BC1 Progeny
DNA from the NOD and PWK parental strains was analyzed with 140 microsatellite primer pairs spread throughout the genome. A set of 107 (76%) polymorphic microsatellite markers were defined, of which 98 were used for mapping the BC1 progeny. These covered the mouse genome with a mean intermarker distance of 10.5 and 23.9 cM.
A multistep screening approach was adopted. A near complete genomic screen was made using a minimum of three evenly spaced informative markers per chromosome, followed by the use of additional markers lying close to known Idd loci. Where initial evidence of linkage was found, additional neighboring markers were also typed. Chromosome 6 was thus typed for 17 markers, and a precise anchored map covering the majority of the chromosome established (Davies et al. 1995). The order of loci on chromosome 6 was in agreement with that reported on the composite map of the Mouse Chromosome 6 Committee Report (Elliot and Moore 1997), suggesting that map positions of loci and genetic distances are unlikely to vary greatly in the NOD × PWK cross from those reported for other crosses.
Linkage analysis showed evidence of linkage to the histology scores with several markers covering 25 cM of the distal part of chromosome 6 with P values ranging from 0.05 for the most proximal marker D6Mit21 to 0.0013 for the most distal marker D6Mit14 (Table 1).
Table 1.
Polychotomous Linkage Analysis and Association of Distal Chromosome 6 Markers with Type 1 Diabetes Phenotype in BC1 (NOD/PWK × NOD) Progenies
| Locus | Normal (1 + 2) | Insulitis (3 + 4) | Severe insulitis (5) | Diabetes (6) | χ2 | P value |
|---|---|---|---|---|---|---|
| N/N : N/P | N/N : N/P | N/N : N/P | N/N : N/P | |||
| Sex combined | ||||||
| D6Mit21 | 54 : 50 | 23 : 22 | 11 : 15 | 5 : 11 | 3.98 | 0.05 |
| D6Mit53 | 57 : 48 | 21 : 24 | 10 : 16 | 5 : 11 | 7.96 | 0.01 |
| D6Mit11 | 59 : 46 | 21 : 24 | 9 : 17 | 5 : 11 | 12.15 | 0.00049 |
| Ly4 | 58 : 47 | 21 : 24 | 8 : 18 | 5 : 11 | 12.29 | 0.00045 |
| D6Mit25 | 60 : 45 | 20 : 25 | 9 : 17 | 5 : 11 | 12.11 | 0.0005 |
| D6Mit14 | 60 : 45 | 19 : 26 | 9 : 17 | 6 : 10 | 10.31 | 0.0013 |
| Males | ||||||
| D6Mit21 | 35 : 25 | 9 : 8 | 4 : 10 | 3 : 4 | 6.60 | 0.01 |
| D6Mit53 | 37 : 24 | 7 : 10 | 3 : 11 | 3 : 4 | 14.18 | 0.00016 |
| D6Mit11 | 35 : 26 | 8 : 9 | 3 : 11 | 3 : 4 | 11.32 | 0.00076 |
| Ly4 | 35 : 26 | 8 : 9 | 2 : 12 | 3 : 4 | 13.29 | 0.00026 |
| D6Mit25 | 37 : 24 | 8 : 9 | 2 : 12 | 3 : 4 | 13.08 | 0.00029 |
| D6Mit14 | 37 : 28 | 7 : 10 | 2 : 12 | 3 : 4 | 14.50 | 0.00014 |
| Females | ||||||
| D6Mit21 | 19 : 25 | 14 : 14 | 7 : 5 | 2 : 7 | 0.01 | 0.95 (N.S.) |
| D6Mit53 | 20 : 24 | 14 : 14 | 7 : 5 | 2 : 7 | 0.03 | 0.95 (N.S.) |
| D6Mit11 | 24 : 20 | 13 : 15 | 6 : 6 | 2 : 7 | 2.49 | 0.20 (N.S.) |
| Ly4 | 23 : 21 | 13 : 15 | 6 : 6 | 2 : 7 | 1.76 | 0.20 (N.S.) |
| D6Mit25 | 23 : 21 | 12 : 16 | 7 : 5 | 2 : 7 | 1.48 | 0.30 (N.S.) |
| D6Mit14 | 23 : 21 | 12 : 16 | 7 : 5 | 3 : 6 | 0.50 | 0.50 (N.S.) |
Higher P values (data not shown) were obtained by contingency table analysis based on the comparison of frequencies in the two extreme phenotypes: normal (histology scores 1+2) compared to severely diseased animals (histology scores 5+6). In our whole genome scan no markers other than those on chromosome 6 showed evidence of linkage at P ≤ 0.001. Usually in murine autoimmune disease models P ≤ 0.001 has been taken as sufficient evidence to name a locus in a genome scan and undertake additional experiments to confirm it (Lander and Kruglyak 1995; Vyse and Todd 1996).
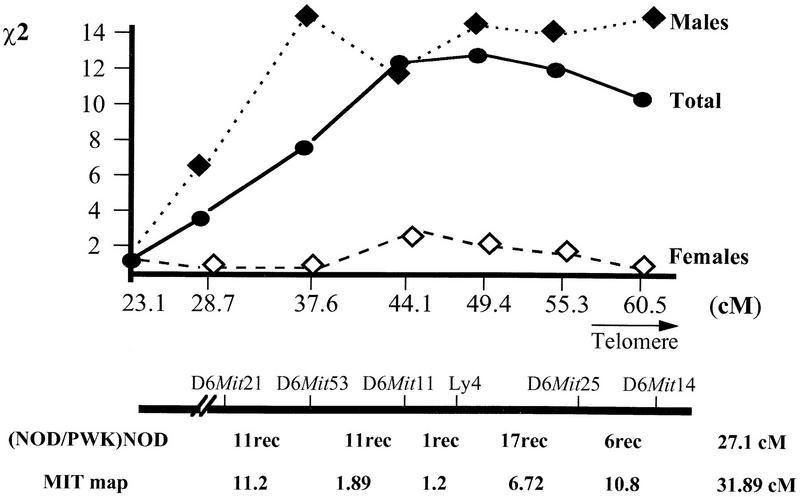
The linkage identified on distal chromosome 6 showed some interesting features: When cohorts were split according to sex, male animals showed clearly higher χ2 than females (Table 1). The majority of the markers located on distal chromosome 6 showed significant linkage (P < 0.001) to the disease phenotypes in the male group of animals but not in females. The differences between the linkage to histology scores in the two sexes were statistically significant for two markers: D6Mit53 and D6Mit14, for which the tests for sex differences gave probabilities P = 0.011 and P = 0.020, respectively (Fig. 2).
Figure 2.
Statistics for the contribution of distal chromosome 6 genetic interval to type 1 diabetes in the BC1 progenies. χ2 values are calculated by a polychotomous genetic analysis regression test (see Methods). Genetic distances between markers are calculated as reported previously (Davies et al. 1995). Numbers of recombinants are given for each interval between the genetic markers as calculated in the BC1 progenies. MIT map distances are from public data banks (Dietrich et al. 1994).
Inspection of the haplotypes corresponding to this part of the chromosome showed that 11 of the 16 animals (69%) with diabetes were NOD/PWK heterozygous for distal chromosome 6 alleles (Table 1, marker Ly4). An increase of NOD/PWK genotypes was also seen in severe insulitis (score 5) with 18 of 26 animals being NOD/PWK. When milder forms of the disease (insulitis scores 3+4), or normal animals (insulitis scores 1+2) were examined, the NOD/PWK genotype was found in <50% of the animals (Table 1). These data suggest that a NOD-resistance allele lies within the region.
The χ2 values in Table 1 indicate that a disease locus is located between D6Mit11 (χ2 = 12.15, at 43 cM from the centromere) and D6Mit14 (χ2 = 10.31, ∼60 cM from the centromere). No clear peak value was obtained when the χ2 values were plotted against genetic distances corresponding to this part of the chromosome (Fig. 2). The absence of an evident peak and the rather flat probability distribution within this interval suggested that additional markers corresponding to this region were unlikely to provide more refined evidence for linkage or allow the precise definition of the genetic interval corresponding to the disease-associated locus using the present set of animals. Consequently, other approaches were adopted to confirm and refine this localization.
Construction and Analysis of NOD.PWK6.I–IV Congenic Strains
To confirm and localize more precisely the diabetes-associated gene(s) lying in the distal part of chromosome 6, we have adopted a strategy based on the construction of congenic lines by introgressing PWK donor genetic material for this part of the chromosome into the NOD mouse genome. Segregation of PWK-derived alleles for distal chromosome 6 in the congenic lines allowed us to assess the implication of these alleles in the diabetes phenotypes in the absence of other PWK-derived genetic material. Repetitive backcrosses to NOD have been performed with breeder selection based on the presence of a PWK-derived allele for chromosome 6. Animals used as breeders were selected for heterozygosity at four markers, D6Mit21, D6Mit11, D6Mit25, and D6Mit26, located at positions 30, 43, 52, and 57 cM.
At the seventh backcross generation (N8), male and female heterozygous animals have been selected and used for brother/sister mating to produce the N8F1 generations. Recombinant animals are thus generated carrying the NOD/NOD, NOD/PWK, and PWK/PWK genotypes for distal chromosome 6 alleles. To produce additional NOD/NOD animals for distal chromosome 6 for use as an internal control, BC7 generation progenitors NOD/NOD, homozygous for this part of the chromosome, were selected in parallel. A total of 419 N8F1 congenic animals were CY-treated and assessed for phenotypes prior to genetic analysis. Both intercross analysis of congenic strains and intercongenic strain comparisons have been undertaken.
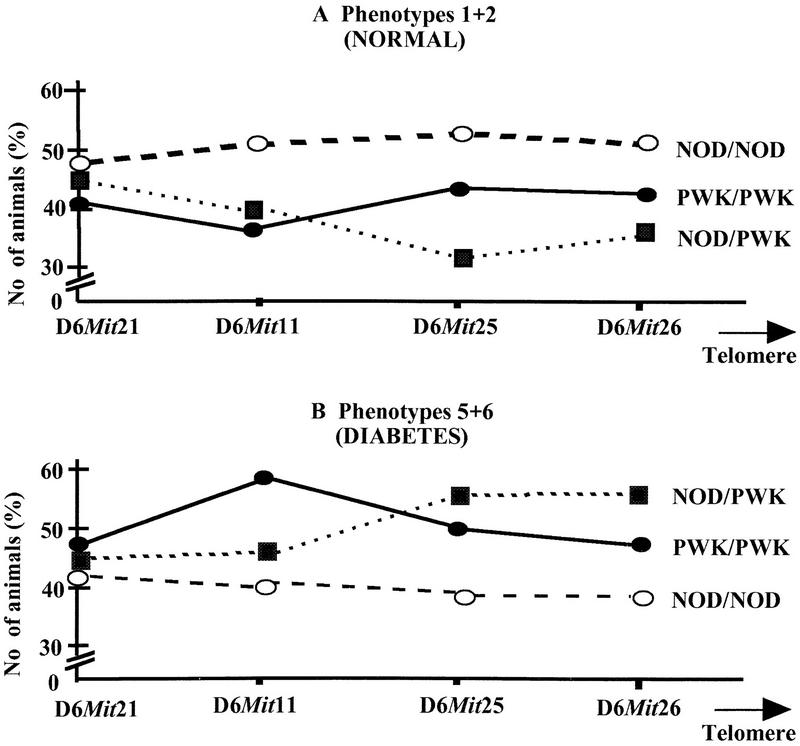
When the phenotypes of NOD/PWK heterozygotes were examined (Fig. 3B), an excess of animals (55% compared to 32%) developed the final stages of the disease (5+6) in comparison with those remaining unaffected (scores 1+2, Fig. 3A) (P = 0.002). The presence of a single PWK allele at loci between the D6Mit25 marker and the telomere is associated with an ∼20% increase in diabetes (Fig. 3A,B). In contrast, the presence of two NOD alleles within the distal part of the locus did not appear to increase diabetes or insulitis frequency. Considerably more animals (52%) with this genotype remained unaffected (1+2) after CY treatment than developed (40%) high histology scores (5+6) (see also Table 2). This is consistent with the presence of a NOD recessive resistance allele within this interval, affecting mainly the onset of severe insulitis and diabetes with only a small or no effect on the milder forms of insulitis (3+4) (Table 2). No such effect was associated with the most proximal part of the region examined around the D6Mit21 marker, as approximately equal numbers of NOD/PWK animals have been observed in the 1+2 (47%) and 5+6 (44%) phenotypic classes (Table 2; Fig. 3A,B).
Figure 3.
Genetic contribution of distal chromosome 6 in type 1 diabetes phenotypes as studied in congenic lines. (A) Normal (1+2) phenotypes; (B) severe disease phenotypes (5+6) of NOD.PWKchr6 congenics lines. The number of animals is reported as the percentage of the number observed for a given phenotype for each genotype compared to the total number of animals of this genotype (see also Table 2).
Table 2.
Genotype Distribution and Statistics for Distal Chromosome 6 in NOD.PWKchr6 Congenic Animals (N8F1 Generations)
| Locus | Normal (1 + 2) | Insulitis (3 + 4) | Severe phenotype (5 + 6) | P value |
|---|---|---|---|---|
| N/N : N/P : P/P | N/N : N/P : P/P | N/N : N/P : P/P | ||
| Combined | ||||
| D6Mit21 | 91 : 68 : 28 | 15 : 13 : 9 | 83 : 64 : 31 | N.S. |
| D6Mit11 | 119 : 47 : 19 | 18 : 17 : 1 | 97 : 53 : 29 | N.S. |
| D6Mit25 | 135 : 35 : 20 | 20 : 14 : 3 | 105 : 59 : 23 | 0.005 |
| D6Mit26 | 136 : 32 : 24 | 24 : 8 : 5 | 107 : 49 : 27 | 0.025 |
Comparison of PWK/PWK homozygotes with NOD/PWK heterozygotes for the most distal region seems to indicate that the presence of a second PWK allele decreases the effect of this region (D6Mit25–D6Mit26) on the disease. Fewer PWK/PWK animals (48%) showed severe phenotypes when compared to the NOD/PWK animals, whereas 55% developped the disease (Fig. 3B). This could be interpreted by the presence of a second gene acting as a diabetes NOD susceptibility allele in the most distal part of chromosome 6. Additional experimental evidence from the analysis of second-generation congenic lines constructed from these strains is required, however, to verify this hypothesis.
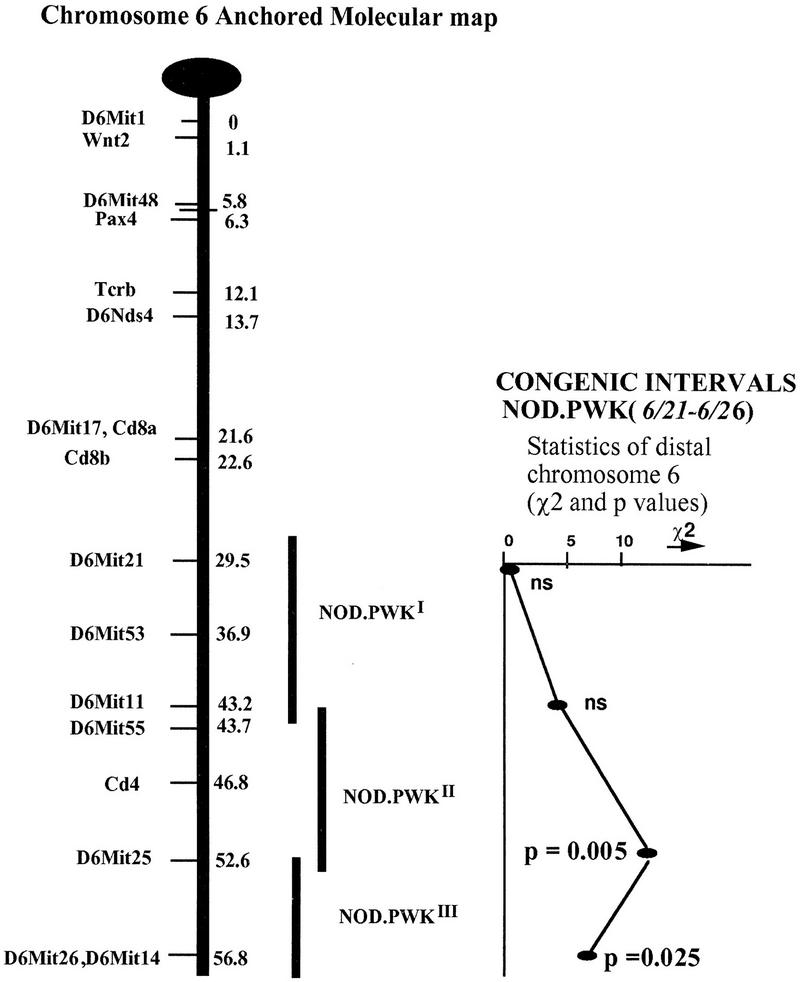
When the entire cohort was analyzed, significant linkage to diabetes was established for the D6Mit25 marker, with the presence of a PWK allele at this locus being associated with an increase in disease onset (P = 0.005) (Table 2). When female and male animals were analyzed separately, the significantly higher contribution of males to the disease, observed at the BC1 generation, was not observed in the congenic lines. Female animals showed significant linkage of the D6Mit25 marker to diabetes (P = 0.001) (data not shown). The sex difference observed in the congenic animals was not statistically significant. This descrepancy between the congenic and the BC1 data could be a reflection of the increased presence of the NOD genome in these lines, with the reappearance of a higher prevalence of the disease in female animals reflecting the known characteristic to the NOD strain. Three different methods of statistical analysis (see Methods) of the congenic data have given similar results (data not shown). The statistics corresponding to this Idd locus on the distal part of chromosome 6 are represented schematically on Figure 4. A clear peak for linkage is associated with the D6Mit25 marker locus (P = 0.005).
Figure 4.
Localization and statistics for markers on distal chromosome 6. NOD.PWKI,II or NOD.PWKIII corresponds to the subset of congenic animals used for evaluating the contribution of each interval (I, II, III) containing diabetes resistance and/or susceptibility allele(s) on the distal part of chromosome 6. P values correspond to contingency table analysis of the combined sex data (see also Table 2). The anchor map for mouse chromosome 6 is adapted from previously published data (Davies et al. 1995).
These data therefore confirm the presence of a diabetes-resistance gene within this region initially detected by the analysis of BC1 progeny. With the aim to better define the interval containing the diabetes gene(s) we have performed linkage analysis using pooled data corresponding to three different genetic intervals of distal chromosome 6. These intervals have been defined as follows: In interval I, animals were pooled according to NOD homozygosity, PWK homozygosity, or NOD/PWK heterozygosity, defined by the genotypes corresponding to the two most proximal markers D6Mit21 and D6Mit11. Interval II was defined similarly, but taking into consideration genotypes corresponding to the D6Mit11 and D6Mit25 genetic markers. Interval III corresponds to the genotypes within the D6Mit25 and D6Mit26 span for the most distal markers examined (Fig. 4).
Phenotypic analysis of animals corresponding to each interval has shown that NOD/PWK heterozygous and PWK/PWK homozygous animals for interval II have the highest incidence of severe phenotypes (51% and 62%, respectively), when compared with NOD/NOD homozygous (37%) for the same region (Table 3). Polychotomous linkage analysis comparing the NOD/NOD versus NOD/PWK + PWK/PWK genotypes showed that interval II, defined by the markers D6Mit11–D6Mit25, carries the highest significance for linkage (P = 0.0005; Table 4). It is evident that the diabetes incidence observed upon replacement of one or both NOD alleles suggests the presence of a NOD-resistance disease modifier within this interval. As the progenitors of the N8F1 congenic animals used in this analysis were chosen based on retained heterozygosity for a large segment (25 cM) of distal chromosome 6, neither the presence of more than one gene affecting phenotypes nor the presence of epistasis between different effector genes located within this region can be excluded. The polychotomous linkage analysis confirmed the identification of a single peak within the D6Mit11–D6Mit25 interval. The diabetes-associated gene(s) therefore must be localized to an interval of ∼9 cM within the distal part of chromosome 6. Additional congenic lines fixed for the presence of NOD or PWK alleles at loci within the genetic interval defined in our study will be necessary to establish with precision the exact location of the gene(s). Preliminary data from the analysis of second generation NOD.PWK congenic lines have shown a large increase in diabetes incidence when compared with congenic lines having NOD alleles fixed for the same region (E. Melanitou, unpubl.). Further analysis of these congenic lines will eventually allow confirmation of the presence of more than one gene(s) and enable the study of possible gene interactions within this region (our ongoing experiments).
Table 3.
Severe Phenotypes of Chromosome 6 Congenic Intervals (Histology Scores 5 + 6)
| Interval | Locus | N/N vs. total N/N (%) | N/P vs. total N/P (%) | P/P vs. total P/P (%) |
|---|---|---|---|---|
| I | D6Mit21–11 | 63/149 (42) | 28/63 (44) | 17/29 (58.6) |
| II | D6Mit11–25 | 77/206 (37) | 36/71 (51) | 18/29 (62) |
| III | D6Mit25–26 | 99/247 (40) | 40/73 (55) | 15/35 (43) |
Numbers of animals are reported for the corresponding genotype for each genetic interval. Percentages are calculated vs. the total number of animals observed for each genotype.
Table 4.
Genotype Distribution for Each Phenotype and Statistics for the Three Gene Intervals of Distal Chromosome 6
| Interval | Locus | Normal (1 + 2) | Insulitis (3 + 4) | Severe phenotype (5 + 6) | χ2 | Pvalue |
|---|---|---|---|---|---|---|
| N/N : N/P : P/P | N/N : N/P : P/P | N/N : N/P : P/P | ||||
| Sex combined | ||||||
| I | D6Mit21–11 | 75 : 26 : 11 | 11 : 9 : 1 | 63 : 28 : 17 | 1.55 | N.S. |
| II | D6Mit11–25 | 113 : 24 : 11 | 16 : 11 : 0 | 77 : 36 : 18 | 11.39 | 0.0005 |
| III | D6Mit25–26 | 128 : 25 : 18 | 20 : 8 : 2 | 99 : 40 : 15 | 2.82 | N.S. |
Modification of the polychotomous linkage analysis allowed the NOD/NOD genotypes to be assessed against the pooled NOD/PWK + PWK/PWK genotypes for each histology class. This is a 1 degree of freedom test. Congenic interval analysis for distal chromosome 6.
(NS) Not significant.
DISCUSSION
Our whole genome scan of the (NOD × PWK)F1 × NOD backcross cohort has provided interesting insights into the genetic control of autoimmune diabetes in the NOD mouse within an intersubspecific genetic background. Five loci showed weak linkage to diabetes phenotypes, with the most convincing evidence being obtained for a locus on distal chromosome 6. The greater genetic diversity introduced by the PWK strain and the possible segregation of additional resistance genes contributed by the PWK genome could, by disguising the effects of NOD-positive disease modifiers, account for the relatively weak linkages we have observed in the BC1 progeny. In crosses between NOD and more closely related standard laboratory strains, the number of resistance alleles shared between the parental strains may be greater. Nonsegregation of these loci may increase the resolving power of genetic analysis for those actually segregating and showing linkage to the disease.
Two of the initial loci identified in the BC1 progeny were on chromosome 3 (D3Mit7 and D3Nds1), one was on proximal chromosome 14 (D14Mit5), and one was on chromosome 17 (D17Mit9) close, but distal, to the MHC locus (data not shown). As high levels of significance were not attained for the chromosome 3, 14 and 17 loci, these findings will require independent confirmation. The large number of different Idd loci identified in crosses of NOD with different strains of the laboratory mouse suggests that extensive genetic heterogeneity characterizes autoimmune diabetes in the mouse. Major, that is, common to all crosses, loci affecting diabetes onset, with the exception of the MHC-linked gene(s), appear nonexistent or difficult to identify. It is worth noting, however, that three of the localizations detected in our cross lie near or within regions containing Idd loci identified in other cross designs (Todd et al. 1991; De Gouyon et al. 1993; Ghosh et al. 1993; Ikegami et al. 1995). It remains to be confirmed whether or not the same gene(s) within the identified regions are responsible for the disease in the different crosses.
We have also assessed the implication of the MHC in the diabetes phenotypes of the (NOD × PWK)/NOD outcross. We first showed that class II molecules expressed by the PWK strain are clearly distinct from NOD class II molecules. The PWK strain not only expresses I–E unlike NOD (Acha-Orbea and McDevitt 1987) but also carries an I–A molecule distinct from the one carried by NOD (Hattori et al. 1986; data not shown). In our cross configuration, 25% and 66% of the diabetic and prediabetic animals, respectively, inherited a single NOD allele at the MHC locus (data not shown). This rather weak association of the MHC with type 1 diabetes may reflect the segregation of other loci interfering with the classical expectation of a clear MHC association. Yet similar data have been obtained previously in a cross between the NOD and SEG strains (De Gouyon et al. 1993).
Genetic analysis of the BC1 progeny has shown linkage to five markers lying within 25 cM on distal chromosome 6. The data are consistent with the presence of a diabetes-resistance allele at a locus defined by the D6Mit25 marker (Table 1; P = 0.0005). The significance of linkage to the disease phenotypes, shown by the combined sex data, is contributed mainly by the male subgroup of BC1 progenies (Table 1; and P value for sex difference, P<0.01). Sex differences in linkage significance have not been reported previously for any of the 16 known Idd loci. However, the experimental designs of other investigators have been either centered on female progeny or have ignored the sex of the animals under analysis. It is unlikely that this sex linked effect is due to the use of CY for diabetes induction, as NOD animals treated with CY in our animal facility develop overt diabetes in similar numbers in both sexes. The sex difference observed concerning the chromosome 6 allele contribution to the disease onset in the BC1 generations seems to be rather specific to the presence of PWK genome in this cross. One hypothesis could be that environmental factors as well as the hormonal status of the animals (i.e., the presence or absence of male hormones) may influence the expression of the chromosome 6 diabetes-associated gene(s) in this genetic background.
The use of congenic lines has proven to be a very powerful approach in the dissection of complex characters, as it allows confirmation or rejection of a disease locus previously identified by genetic studies applied to the entire genome (Wicker et al. 1992, 1994; Lord et al. 1995; Podolin et al. 1997). In this study double emphasis has been given to the use of congenic strains both to provide a simplified intercross environment for linkage analysis as well as to allow intercongenic strain comparison.
The progenitors of our congenic animals were initially selected for heterozygosity over a region spanning 25 cM of distal chromosome 6. The study of NOD.PWKchr6 congenics has allowed us to restrain the candidate region found from the analysis of the BC1 cohort to an interval of 9 cM flanked by the D6Mit11 and D6Mit25 genetic markers. Analysis of additional congenic lines carrying more restricted regions of chromosome 6 derived by recombination will allow confirmation of the eventual presence of more than one locus and enable the study of possible gene interactions within distal chromosome 6 to be undertaken (our ongoing experiments).
Implicit in this analysis of the congenic lines is that the NOD diabetogenic genes were fixed by the N8 generation and that any differences observed in the phenotypes at this point can be attributed to the effects of gene(s) present in the region of distal chromosome 6. By generation N5 the prevalence of diabetes after CY treatment in animals that are NOD/PWK heterozygous for distal chromosome 6 exceeded the frequencies observed for spontaneous diabetes of the NOD parental strain in our colony (data not shown). Comparisons between Idd loci identified in different crosses are rendered complex both by the lack of precision in whole genome scan localizations and the eventual large number of Idd loci involved. Recent data concerning the Idd3 and Idd10 loci on mouse chromosome 3, using congenic lines, suggest interaction between at least three closely linked genetic determinants (Podolin et al. 1997).
Two previous studies have found evidence for linkage with markers located within the distal part of chromosome 6 (Ghosh et al. 1993). None of these loci seems to correspond to the locus studied here. In the case of the cross of NOD with the C57Bl/6 strain, which detected weak linkage to the disease with the D6Mit14 marker located on the most distal part of chromosome 6, a NOD susceptibility allele was identified (Ghosh et al. 1993). This gene was not detected in our cross. In another study, a nonobese nondiabetic (NON) susceptibility allele was detected within a similar region in an F2 cross of NOD with the NON strain (McAleer et al. 1995). Because this locus could not be identified in the BC1 progeny it seems again unlikely to correspond to the Idd PWK locus that we have identified (provisionally named Idd19).
Distal chromosome 6 contains a number of genes, which may represent excellent candidates for the disease, including loci associated with processes involved in the etiology of both autoimmunity and type 1 diabetes. Candidate genes include the locus controlling dexamethasone-induced apoptosis (Penha-Gonçalves et al. 1995) and the structural or regulatory gene implicated in the expression of an antigen recognized by an islet-specific and diabetogenic T-cell clone (Dallas-Pedretti et al. 1995).
Other candidate genes also lie within this region, including the gene coding for the tumor necrosis factor receptor (TNFR2) (Goodwin et al. 1991), the natural killer (NK) gene complex (Clark et al. 1981; McCoy et al. 1984), and the gene coding for hematopoietic cell phosphatase (HCP) (Plutzky et al. 1992). A locus controlling Bordetella pertussis-induced histamine sensitization also maps to the distal part of chromosome 6 (Sudweeks et al. 1993). B. pertussis toxin is used for induction of experimental allergic encephalomyelitis (EAE) in mice, which represents the primary animal model for multiple sclerosis, a human disorder with immunopathological etiology (Martin et al. 1992). Experimental approaches based on the genetic analysis of NOD.PWKchr6 congenic lines carrying restricted fragments of the donor strain combined with a candidate gene approach are being pursued to define at the molecular level the gene(s) responsible for the disease onset. Further refinement of the genetic interval containing the diabetes gene in this region of chromosome 6 should allow tightly targeted linkage studies to be undertaken in human. IDDM loci have yet to be identified within the region of human chromosome 12p12–13 homologous to distal mouse chromosome 6 (Baens et al. 1995).
METHODS
Animals
NOD/Lt mice at generation F48, were obtained from E. Leiter (The Jackson Laboratory) and maintained in our research colony by brother/sister mating. Female NOD animals were mated with males of the PWK feral-derived inbred strain, which is maintained in our colony by brother/sister mating. F1 females were subsequently backcrossed to NOD/Lt males to produce BC1 generations. A total of 192 BC1 animals were analyzed for their diabetes phenotypes (see below).
For the construction of the chromosome 6 congenic lines, first backcross males selected for NOD/PWK heterozygosity at the distal part of chromosome 6 were crossed with NOD females to produce BC2 progenies. In total, seven successive backcrosses to the NOD parent were performed by selecting progenitors for heterozygosity using four microsatellite markers (as indicated in Results) located within a 25-cM region of the distal part of chromosome 6. The progenitors used for the establishment of our congenic lines were also selected for NOD MHC homozygosity, using the D17Mit24 microsatellite marker, which maps to the MHC region on mouse chromosome 17 and detects allelic variation between NOD and PWK mice (Dietrich et al. 1994, 1996; Copeland et al 1993).
Congenic progeny was first tested at the BC4 generation for diabetes phenotypes after CY treatment. At BC7 (N8), both male and female animals selected for NOD/PWK heterozygosity were intercrossed to create the N8F1 generations. BC7 animals homozygous for NOD alleles at distal chromosome 6 loci were also selected for separate intercross breedings to generate large numbers of NOD.PWKchr6I–IIIn/n animals as internal controls for our experiments (see below). A total of 419 N8F1 congenic animals were derived, treated with CY, and subjected to phenotype assessment.
All animals used in our study were housed under the standard conditions pertaining at the Pasteur Institute.
CY Treatment
CY (Endoxan-Asta Laboratories) was prepared immediately before injection at a final concentration of 10 mg/ml in PBS. Eight-week-old BC1, BC4, and N8F1 animals received two intraperitoneal injections of 200 mg/kg at 14-day intervals. Animals were monitored for glucose detection in urine (Glucotest; Boehringer Mannheim) and assessed for glycemia 14, 21, and 28 days after the first injection by using test strips and a colorimetric assay (Haemogluctest and Reflolux F; Boehringer Mannheim). Diabetes was diagnosed when permanent-fasting glycemia of >3 grams/liter was observed.
Histopathology
Pancreata removed 14 days after the second CY injection or from untreated animals at least 40 weeks old were fixed in Bouin’s solution. Four-millimeter sections from at least three noncontiguous tissue levels were stained with hematoxylin and eosin, and insulitis was scored on a minimum of 15 islets per mouse. Histology grades were assigned on a scale of 1–6 based on the percentage of islets showing a lymphoid infiltrate within and around the islets. The following classification system was used: (1) normal islets; (2) peri-islet infiltration with no reduction of islet mass, no insulitis; (3) insulitis in 20% of the islets; (4) insulitis in 20%–80% of the islets; (5) extensive insulitis with significant reduction in islet mass (>80%); and (6) extensive insulitis with only few residual islets remaining and overt diabetes. In some experiments (see Results) the histology scores were grouped for simplicity as follows: scores 1+2 for normal phenotypes, 3+4 for insulitis, and 5+6 for severe forms of the disease, corresponding to animals with extensive insulitis, significant islet mass reduction, and the presence of hyperglycemia.
Histology scores were obtained from 192 BC1 and 419 N8F1 congenic animals after CY treatment. Twenty PWK and an equivalent number of NOD control animals were also treated with CY and analyzed histologically.
DNA Extraction
DNA was prepared from splenocytes as described previously (De Gouyon et al. 1993) and from mouse tails using standard methods with minor modifications : 1.5 cm of tail was cut and put into a tube containing 700 μl of lysis buffer (50 mm Tris at pH 8, 100 mm EDTA, 100 mm NaCl, 1% SDS). After addition of 35 μl of 10 mg/ml solution of proteinase K (Merck 24568, EM Biochemicals) dissolved in H2O, and incubation at 55°C for 8–18 hr, 20 μl of RNase A (Worthington no. 5679, 13 μg/ml) was added. DNAs were extracted once with phenol/chloroform (1:1), and the aqueous phases precipitated with isopropanol. Pellets were washed twice with ethanol (70% and 100%) and dried. DNAs were taken up in 500 μl of H2O.
Genotyping
DNA from NOD and PWK parental strains was analyzed with 140 microsatellite primer pairs. Sequences for the polymorphic PCR-based microsatellite loci primers, as described (Copeland et al. 1993; Dietrich et al. 1994, 1996), were synthesized by Genset (Paris, France). All reactions were carried out in 96-well U-bottom flexible plates with 50 ng of DNA in a total volume of 10 μl using PCR mix containing, 200 nm of each primer, 0.2 mm each dNTPs, 1.5 mm MgCl2, 1 unit of Taq polymerase (GIBCO BRL), and 0.03 μl of [32P]dCTP (3000 Ci/mmole). Amplifications were carried out in a Techne PH-3 DNA thermal cycler. An initial denaturation step of 3 min at 94°C, was followed by 30 cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 2 min, and elongation at 72°C for 2 min. This was followed by a final elongation step of 7 min. Two or more primer pairs were usually coamplified in the same reaction as described previously (Melanitou et al. 1996). Amplified samples were analyzed on 6% (wt/vol) acrylamide sequencing gels. The gels were directly exposed to X-ray autoradiographic film (XAR, Kodak, Rochester, NY). Exposure time was usually for 12 hr at −80°C.
Genetic Analysis
Histology scores were considered as ordered categories of severity, and linkage was evaluated by a log–linear polychotomous regression model applied to the multimonial data as described previously (Ghosh et al. 1993). This provides a likelihood ratio test for the trend of a genotype frequency to increase or decrease with increasing histology scores. Two genotypes are possible in the backcross cohort, and the log–linear polychotomous regression model has 1 degree of freedom (df) for the test of linkage. In the case of the N8F1 congenic cohort, three genotypes may be present and the log–linear polychotomous regression model has 2 df in the test of linkage.
Three different approaches were used for testing linkage to markers lying within the distal part of chromosome 6 in the congenic lines. Contingency table analysis was based on the comparison of frequencies of the three genotypes in the two extreme classes defined by histology scores 1+2 and 5+6. Polychotomous linkage analysis compared the frequency of NOD/NOD versus the NOD/PWK genotypes in all classes. Finally, a modification of the polychotomous analysis allowed the NOD/NOD genotypes to be assessed against the pooled NOD/PWK + PWK/PWK genotypes for each histology class, corresponding to the data reported in Tables 2 and 4.
Acknowledgments
We thank Dr. Philip Davies for participation in this work during his postdoctoral fellowship in the Mouse Molecular Genetics Unit and Marie-Françoise Richard for excellent technical assistance. Special thanks are also due to Jean-Marc Sebaoun for expert computer advice. This work was supported by grants from the Juvenile Diabetes Foundation International to P.A. and C.B., and the G.R.E.G (Groupement de Recherches et d’Études sur le Génome) and the French Ministry of Research and Technology (MRE) to P.A. A list of microsatellites (MIT markers) polymorphic between the NOD and PWK strains is available upon request from E.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eviemel@pasteur.fr; FAX 33 1 45 68 86 56.
REFERENCES
- Archa-Orbea H, McDevitt HO. The first external domain of the non-obese diabetic mouse class II I-Ab chain is unique. Proc Natl Acad Sci. 1987;84:2435–2439. doi: 10.1073/pnas.84.8.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baens M, Aerssens J, van Zand K, Van der Berghe H, Marynen P. Isolation and regional assignment of human chromosome 12p cDNAs. Genomics. 1995;29:44–52. doi: 10.1006/geno.1995.1213. [DOI] [PubMed] [Google Scholar]
- Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependant diabetes mellitus. Diabetes. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- Benett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, Kawasuchi Y, Dronsfield MJ, Pociot MJ, et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nature Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- Bonhomme J F, Guénet J-L. The wild house mouse and its relatives. In: Lyon MF, Searle AG, editors. Genetic variants and strains of the laboratory mouse. Oxford, UK: Oxford University Press; 1989. pp. 649–662. [Google Scholar]
- Bottazzo GF. Death of a beta cell: Homicide or suicide. Diabetic Med. 1986;3:119–130. doi: 10.1111/j.1464-5491.1986.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Castano L, Eisenbart GS. Type-1 diabetes: A chronic autoimmune disease of human, mouse and rat. Annu Rev Immunol. 1990;8:647–679. doi: 10.1146/annurev.iy.08.040190.003243. [DOI] [PubMed] [Google Scholar]
- Chesnut K, She J-X, Cheng I, Muralidharan K, Wakeland EK. Characterization of candidate genes from the diabetes-prone NOD mouse strain. Mamm Genome. 1993;4:549–554. doi: 10.1007/BF00361383. [DOI] [PubMed] [Google Scholar]
- Clark EA, Shultz LD, Pollack SB. Mutations in mice that influence natural killer cell (NK) activity. Immunogenetics. 1981;12:601–613. doi: 10.1007/BF01561700. [DOI] [PubMed] [Google Scholar]
- Copeland N G, Gilbert DJ, Jenkins NA, Nadeau JH, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Steen RG, Lincoln SE, et al. Genome Maps IV. Science. 1993;262:67–82. doi: 10.1126/science.8211131. [DOI] [PubMed] [Google Scholar]
- Dallas-Pedretti A, McDuffie M, Haskins K. A diabetes-associated T-cell autoantigen maps to a telomeric locus on chromosome 6. Proc Natl Acad Sci. 1995;92:1386–1390. doi: 10.1073/pnas.92.5.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SCL, Jenkins SC, Palmer SM, et al. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- Davies PO, Melanitou E, Asano M, Avner PR, Montaguitelli X. An anchored molecular map of mouse chromosome 6 with an analysis of interference. Mamm Genome. 1995;6:738–740. doi: 10.1007/BF00354297. [DOI] [PubMed] [Google Scholar]
- De Gouyon B, Melanitou E, Richard MF, Requarth M, Hahn IH, Guénet J-L, Demenais F, Julier C, Lathrop MG, Boitard C, Avner P. Genetic analysis of diabetes and insulitis in an interspecific cross of the nonobese diabetic mouse with Mus spretus. Proc Natl Acad Sci. 1993;90:1877–1881. doi: 10.1073/pnas.90.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepine M, Pociot F, Habita C, Hashimoto L, Froguel P, Rotter J, Cambon-Thomsen A, Deschamps I, Djoulah S, Weissenbach J, et al. Evidence of a non-MHC susceptibility Locus in Type 1 diabetes linked to HLA on chromosome 6. Am J Hum Genet. 1997;60:174–187. [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant M, Damron D, Nahf R, Gross A, Joyce D, Wessel M, Dredge R, et al. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nature Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O’Conner TJ, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Elliot RW, Moore KJ. Mouse Chromosome 6. Mamm Genome. 1997;7:S102–S120. doi: 10.1007/s003359900318. [DOI] [PubMed] [Google Scholar]
- Garchon HJ, Bedossa P, Eloy L, Bach JF. Identification and mapping to chromosome 1 of a susceptibility locus for periinsulitis in non-obese diabetic mice. Nature. 1991;353:260–262. doi: 10.1038/353260a0. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Palmer SM, Rodrigues NR, Cordell HJ, Hearne CM, Cornall RJ, Prins J-B, McShane P, Lathrop GM, Peterson LB, et al. Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nature Genet. 1993;4:404–409. doi: 10.1038/ng0893-404. [DOI] [PubMed] [Google Scholar]
- Goodwin RG, Anderson D, Jerzy R, Davis T, Brannan C, Copeland N, Jenkins NA, Smith CA. Molecular cloning and expression of the Type 1 and Type 2 murine receptors for tumor necrosis factor. Mol Cell Biol. 1991;11:3020–3026. doi: 10.1128/mcb.11.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Makino S. Promotion of spontaneous diabetes in NOD-prone mice by cyclophospharamide. Diabeteologia. 1984;27:604–611. doi: 10.1007/BF00276978. [DOI] [PubMed] [Google Scholar]
- Hashimoto L, Habita C, Beressi JP, Delepine M, Besse C, Campon-Thomsen A, Deschamps I, Rotter JI, Djouiah S, James MR, et al. Genetic mapping of a susceptibility locus for insulin-dependent diabetes mellitus on chromosome 11q. Nature. 1994;371:161–164. doi: 10.1038/371161a0. [DOI] [PubMed] [Google Scholar]
- Hattori M, Buse JB, Jackson RA, Glimcher L, Dorf ME, Mimami M, Makino S, Mariwaki K, Kuzuya H, Imura H, et al. The NOD mouse: Recessive diabetogenic gene within the major histocompatibility complex. Science. 1986;231:733–735. doi: 10.1126/science.3003909. [DOI] [PubMed] [Google Scholar]
- Hattori M, Yamato E, Fukuda M, Petruzelli M, Weaver A, MacMurray A, Lander E, Chapman VM. Type-I (insulin-dependent) and type-II (non-insulin-dependent) diabetes mellitus in BC1[(NODxMus Spretus)F1xNOD] mice. Autoimmunity: Experimental aspects. NATO ASI Series H : Cell Biol. 1993;80:275–297. [Google Scholar]
- Homo-Delarche F, Boitard C. Autoimmune diabetes, the role of the islets of Langerhans. Immunol Today. 1996;17:456–460. doi: 10.1016/0167-5699(96)10053-8. [DOI] [PubMed] [Google Scholar]
- Ikegami H, Makino S. Genetic susceptibility to insulin-dependent diabetes mellitus: from NOD mice to humans. In: Shafrir E, editor. Lessons from animal diabetes. London, UK: Smith-Gordon; 1993. pp. 39–50. [Google Scholar]
- Ikegami H, Makino S, Yamato E, Kawaguchi Y, Ueda H, Sakamoto T, Takegawa K, Ogihara T. Identification of a new susceptibility locus for insulin-dependent diabetes mellitus by ancestral haplotype congenic mapping. J Clin Invest. 1995;96:1936–1942. doi: 10.1172/JCI118239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HJ, Pettersson A, Wilson D, Mao Y, Lernmark A, Lander ES. Genetic dissection of autoimmune type I diabetes in the BB rat. Nature Genet. 1992;2:56–60. doi: 10.1038/ng0992-56. [DOI] [PubMed] [Google Scholar]
- Julier C, Hyer RN, Davies J, Merlin F, Soularue P, Briant L, Catelineau G, Deschamps I, Rotter JL, Froguel P, et al. Insulin-IGF2 region on chromosome 11p encodes a gene implicated in HLA-DR4-dependant diabetes susceptibility. Nature. 1991;354:155–159. doi: 10.1038/354155a0. [DOI] [PubMed] [Google Scholar]
- Julier C, Lucassen A, Villedieu P, Delepine M, Levy-Marchal C, Danze PM, Bianchi F, Boitard C, Froguel P, Bell J, et al. Multiple DNA variant association analysis: Application to the insulin gene region in type I diabetes. Am J Hum Genet. 1994;55:1247–1254. [PMC free article] [PubMed] [Google Scholar]
- Julier C, Hashimoto L, Lathrop M. Genetics of insulin-dependent diabetes mellitus. Curr Opin Genet Dev. 1996;6:354–360. doi: 10.1016/s0959-437x(96)80014-1. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nature Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lucassen AM, Juliet C, Beressi JP, Boitard C, Froguel P, Lathrop M, Bell JI. Susceptibility to insulin dependent diabetes mellitus maps to a 4.1 kb segment of DNA spanning the insulin gene and associated VNTR. Nature Genet. 1993;4:305–310. doi: 10.1038/ng0793-305. [DOI] [PubMed] [Google Scholar]
- Lucassen AM, Bell JI. Genetics of insulin-dependent diabetes. In: Leslie RDG, Robins DC, editors. Diavets: Clinical science in practice. Cambridge, MA: Cambridge University Press; 1995. [Google Scholar]
- Luo D-F, Buzetti R, Rotter JI, Maclaren NK, Thomson G, She J-Y. Affected-sib-pair mapping of a novel susceptibility gene to insulin-dependent diabetes mellitus (IDDM8) on chromosome 6q25-q27. Hum Mol Genet. 1995;57:911–919. [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Bohlander SK, Hopes EA, Montague CT, Hill NJ, Prins J-B, Rejilian RJ, Peterson LB, Wicker LS, Todd JA, Denny P. Mapping the diabetes polygene Idd3 on mouse chromosome 3 by use of novel congenic strains. Mamm Genome. 1995;6:563–570. doi: 10.1007/BF00352359. [DOI] [PubMed] [Google Scholar]
- Makino S, Kumimoto R, Muraoka Y, Mizushima Y, Katagiri K, Tokino Y. Breeding of a non-obese diabetic strain of mice. Exp Anim. 1980;29:1–13. doi: 10.1538/expanim1978.29.1_1. [DOI] [PubMed] [Google Scholar]
- Martin R, McFarland HF, McFarlin DE. Immunological aspects of demyelinating diseases. Annu Rev Immunol. 1992;10:153–187. doi: 10.1146/annurev.iy.10.040192.001101. [DOI] [PubMed] [Google Scholar]
- McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powel EE, Rodrigues NR, Prins J-P, Serreze DV, et al. Crosses of the NOD mouse with the related NON strain: A polygenic model for type 1 diabetes. Diabetes. 1995;44:1186–1195. doi: 10.2337/diab.44.10.1186. [DOI] [PubMed] [Google Scholar]
- McCoy KL, Neilson K, Clagett J. Spontaneous production of colony-stimulating activity by splenic MAC-1 antigen-positive cells from autoimmune motheaten mice. J Immunol. 1984;132:272–276. [PubMed] [Google Scholar]
- Melanitou E, Simmler M-C, Heard E, Rougeulle C, Avner P. Selected methods related to the mouse as a model system. Methods Mol Genet. 1996;8:439–469. [Google Scholar]
- Mimura G. Present status and future view of the genetic study of diabetes mellitus. In: Mimura G, et al., editors. Clinico-genetic genesis of diabetes mellitus. Amsterdam, The Netherlands: Excerpta Medica; 1982. pp. >xiii–xxviii. [Google Scholar]
- Morahan G, McClive PJ, Huang D, Little P, Baxter A. Genetic and physiological association of diabetes susceptibility with raised Na+/H+ exchange activity. Proc Natl Acad Sci. 1994;91:5898–5902. doi: 10.1073/pnas.91.13.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penha-Gonçalves C, Leijon K, Persson L, Holmberg D. Type 1 diabetes and the control of dexamethasone-induced apoptosis in mice maps to the same region on chromosome 6. Genomics. 1995;28:398–404. doi: 10.1006/geno.1995.1167. [DOI] [PubMed] [Google Scholar]
- Plutzky J, Neel BG, Rosenberg RD, Eddy RL, Byers MG, Jani-Sait S, Shows TB. Chromosomal localization of an SH2-containing tyrosine phosphatase (PTPN6) Genomics. 1992;13:869–872. doi: 10.1016/0888-7543(92)90172-o. [DOI] [PubMed] [Google Scholar]
- Podolin PL, Denny P, Lord CJ, Hill NJ, Todd JA, Peterson LB, Wicker LS, Lyons PA. Cogenic mapping of the insulin-dependent diabetes (Idd) gene Idd10, localizes two genes mediating the Idd10 effect and eliminates the candidate Fcgr1. J Immunol. 1997;159:1835–1843. [PubMed] [Google Scholar]
- Risch N, Ghosh S, Todd JA. Statistical evaluation of multiple locus linkage data in experimental species and relevance in human studies: Application to murine and human IDDM. Am J Hum Genet. 1993;53:702–714. [PMC free article] [PubMed] [Google Scholar]
- Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH. Use of recombinant inbred and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J Exp Med. 1994;180:1553–1558. doi: 10.1084/jem.180.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudweeks JD, Todd JA, Blankenhorn EP, Wardell BB, Woodward SR, Meeker ND, Estes SS, Teuscher C. Locus controlling Bordetella pertussis-induced histamine sensitization (Bphs), an autoimmune disease-susceptibility gene, maps distal to T-cell receptor β-chain gene on mouse chromosome 6. Proc Natl Acad Sci. 1993;90:3700–3704. doi: 10.1073/pnas.90.8.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contribution to insuline-dependant diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- Todd JA, Aitman JT, Cornall JR, Ghosh S, Hall RSJ, Hearne MC, Knight AM, Love JM, McAleer MA, Prins J-B, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Todd JA. Genetic analysis of autoimmune disease. Cell. 1996;85:311–318. doi: 10.1016/s0092-8674(00)81110-1. [DOI] [PubMed] [Google Scholar]
- Wicker LS, Miller BJ, Coker LZ, McNally SE, Scott S, Mullen Y, Appel MC. Genetic control of diabetes and insulitis in the nonobese diabetic (NOD) mouse. J Exp Med. 1987;165:1639–1654. doi: 10.1084/jem.165.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker LS, Appel MC, Dotta F, Pressey A, Miller BJ, Delarato NH, Fischer PA, Boltz RC, Jr, Peterson LB. Autoimmune syndromes in major histocompatibility (MHC) congenic strains of non-obese diabetic (NOD) mice. The NOD MHC is dominant for insulitis and cyclophosphamide-induced diabetes. J Exp Med. 1992;176:67–77. doi: 10.1084/jem.176.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker LS, Todd JA, Prins J-B, Podolin P L, Renjilian R J, Peterson LB. Resistance alleles at two non-major histocompatibility complex-linked insulin-dependent diabetes loci on chromosome 3, Idd3 and Idd10, protect diabetic mice from diabetes. J Exp Med. 1994;180:1705–1713. doi: 10.1084/jem.180.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]