Abstract
Background
Peanut allergy affects 1% of the population and causes the most fatal food-related anaphylactic reactions. The protein Ara h 2 is the most potent peanut allergen recognized by 80–90% of peanut allergic patients.
Methods
The crystal structure of the major peanut allergen Ara h 2 was determined for the first time at 2.7 Å resolution using a customized MBP-fusion system. IgE antibody binding to the MBP fusion construct versus the natural allergen was compared by ELISA using sera from peanut allergic patients.
Results
The structure of Ara h 2 is a five helix bundle held together by four disulfide bonds and related to the prolamin protein superfamily. The fold is most similar to other amylase and trypsin inhibitors. The MBP-Ara h 2 fusion construct was positively recognized by IgE from 76% of allergic patients (25/33). Two populations of patients could be identified. Sub-population 1 (n=14) showed an excellent correlation of IgE antibody binding to natural versus recombinant Ara h 2. Sub-population 2 (n=15) showed significantly reduced IgE binding to the MBP fusion protein. Interestingly, about 20% of the IgE binding in sub-population 2 could be recovered by increasing the distance between MBP and Ara h 2 in a second construct.
Discussion
The reduced IgE binding to the MBP-Ara h 2 of sub-population 2 indicates that the MBP molecule protects an immunodominant epitope region near the first helix of Ara h 2. Residues involved in the epitope(s) are suggested by the crystal structure. The MBP-Ara h 2 fusion constructs will be useful to further elucidate the relevance of certain epitopes to peanut allergy.
Keywords: Peanut, Allergy, Ara h 2, Immunotherapy, Structure
Introduction
The prevalence of food allergy is estimated to be 6% in young children and 3.7% in adults (1). Most children grow out of common allergies to milk or eggs, but allergies to peanuts generally persist, affecting approximately 1% of the population (2). Peanut allergies are of particular concern due to the extreme hypersensitivity of some individuals (less than 100 μg dose (3)) and adverse reactions to peanuts are the most frequent type of fatal anaphylaxis among food allergens (4). Ara h 2 is the most potent peanut allergen recognized by >90% of peanut allergic patients (5–7). Studies in children demonstrated that Ara h 2 and the homologous Ara h 6 (59% identity) are the most commonly recognized allergens and IgE reactivity to these proteins is a risk factor for the most serious reactions (8, 9).
Currently, patients are advised to strictly avoid peanut consumption. In traditional immunotherapy treatments for allergy, patients are exposed to small but escalating doses of protein (10). Studies with peanuts have demonstrated initial promise, but still use extremely small doses of peanut protein in order to avoid serious side effects and, at present, utilize only oral administration due to safety concerns (10, 11). It has been proposed that a safer alternative would be to design hypoallergenic variants of the major allergens, which could avoid the serious side effects, allow for higher doses, and still generate tolerance or desensitization (5, 12). There have been many attempts to modify inhalant and food allergens (12, 13), however this approach seems particularly appropriate for peanut allergy since the adverse reactions can be severe.
Herein, we present the first empirically determined crystal structure of Ara h 2 at 2.7 Å, which we have used to analyze IgE antibody binding using sera from peanut allergic patients. Antibody epitopes usually extend 600–900 Å2 in surface area and, except in special cases, interact with discontinuous elements of the primary structure (14). Indeed, Albrecht et al demonstrated that peptides derived from Ara h 2 could not inhibit IgE binding to the native allergen, and unfolded Ara h 2 had significantly reduced IgE binding capacity (15). While mapping antibody epitopes with peptides is expedient and may provide some useful information, the full structure can provide detailed information about the complete interacting surface.
Materials and Methods
Crystallization and Structure Determination
A codon-optimized gene of Ara h 2.01 was obtained from GenScript (Piscataway, NJ) and used as a template for PCR to amplify the DNA to be inserted into the pMALX_E plasmid (16) using the NotI and EcoRI restriction sites. The pMALX_E plasmid contains the MBP mutations D82A, K83A, E172A, N173A, K239A, and E359A as well as changes in the C-terminal helix as previously described to improve the likelihood of crystallization (16). Several constructs with different N-terminal truncations of Ara h 2 were tested for expression of soluble protein (data not shown). The two constructs discussed in this paper are rMBP-Ara h 2-N19 and rMBP-Ara h 2-N28, which connect to the C-terminus of MBP-pMALX_E via the N-terminus of Ara h 2 residues 19 or 28, respectively, following the numbering of Stanley et al (5). Origami B cells were serially transformed with a pACYCDuet-1 plasmid encoding thioredoxin, followed by the pMALX_E Ara h 2 plasmids. For large scale purification, cells were grown in 12 L of LB media containing 100 mg/ml ampicillin, 35 mg/ml chloramphenicol, 12.5 mg/ml tetracycline, and 50 mg/ml kanamycin in twelve 2.8 L Fernbach flasks at 37° C. When the A600 reached 0.75, the temperature of the incubator was set to 18° C. When the incubator temperature reached 18° C, IPTG was added to a final concentration of 500 μM and the cells were allowed to grow overnight. Cells were pelleted by centrifugation and lysed by sonication in 25 mM Tris pH 7.5, 500 mM NaCl and 1 mM EDTA containing Complete protease inhibitors (Roche). Soluble protein was separated from debris by centrifugation and loaded in batch onto amylose resin at 4° C. The resin was washed with sonication buffer followed by elution with sonication buffer containing 40 mM maltose. Protein was concentrated then loaded onto a 16/60 Superdex200 column pre-equilibrated with elution buffer. Peaks containing pure MBP-Ara h 2 protein were pooled and dialyzed against 25 mM Tris pH 7.5, 75mM NaCl, and 5mM maltose. Protein was then concentrated to 38 mg/ml and used for crystallization trials.
Diffraction quality crystals of MBP-Ara h 2-N28 were obtained using the vapor diffusion hanging drop method, mixing 1 μl of protein with 1 μl of reservoir containing 0.1 M sodium citrate pH 5.5 and 1.7 M ammonium sulfate. Resulting crystals were used for micro-seeding into similar conditions containing a reservoir of 0.1 M sodium citrate pH 6.5 and 1.8 M of ammonium sulfate. For data collection, a small, thin plate was transferred to 0.1M sodium citrate pH 6.5, 100 mM NaCl, 2.2 M ammonium sulfate, 5 mM maltose, and 15% ethylene glycol and then flash frozen in liquid nitrogen. Data were collected at APS to 2.7 Å on the ID22 SER-CAT beamline. Data were processed using HKL2000 (17). CCP4i was used to obtain initial phases (18). MolRep (19) was used to determine the position of the MBP using coordinates from pdb code 2DMO followed by initial refinement in Refmac (20). Coot was used to build Ara h 2 into the electron density (21). The structure of Ara h 6 was manually placed into the electron density and used as a visual guide for model building. The structure was further refined through iterative cycles of model building in Coot and refinement in Phenix (21, 22). All MBP residues are present in the density, as well as residues 28-56 and 84-148 of Ara h 2. Note that in the PDB file (3OB4), the Ara h 2 residues are numbered 1028–1148 to differentiate them from the N-terminal MBP fusion. In addition, density for eleven residues found in a lattice contact with an MBP molecule have been modeled with residues 59-68 of Ara h 2. The overall quality of the model has been assessed using MolProbity and is in the 97th percentile for structures at 2.7 Å +/− 0.25 Å (Table 1) (23).
Table 1.
Crystallographic data statistics
| data set | MBP-Ara h 2 (28–148) |
| unit cell | a=68.85Å, b=87.38Å, c=113.14Å; α=γ=90°, β =103.93° |
| Space Group | C2 |
| Resolution (Å) | 50.0 - 2.7 |
| # of observations | 56,256 |
| unique reflections | 16,353 |
| Rsym(%)(last shell)1 | 10.4 (32.1) |
| I/σI (last shell) | 7.9 (2.3) |
| Mosaicity range | 0.55–0.95 |
| completeness(%) (last shell) | 91.7 (50.0) |
| Refinement statistics | |
| Rcryst(%)2 | 18.3 |
| Rfree(%)3 | 25.6 |
| # of waters | 69 |
| Overall Mean B value (Å) | 39.8 |
| Average for MBP | 37.8 |
| Ara h 2 | 47.0 |
| maltotriose | 26.1 |
| water | 31.6 |
| r.m.s. deviation from ideal values | |
| bond length (Å) | 0.005 |
| bond angle (°) | 0.8 |
| dihedral angle (°) | 18.7 |
| Ramachandran Statistics4 | |
| residues in: | |
| favored (98%) regions (%) | 95.5 |
| allowed (>99.8%) regions (%) | 99.8 |
| MolProbity Score | 2.01, 97th percentile |
Rsym = Σ(|Ii − <I>|)/Σ(Ii) where Ii is the intensity of the ith observation and <I> is the mean intensity of the reflection.
Rcryst = Σ||Fo|−|Fc||/Σ|Fo| calculated from working data set.
Rfree was calculated from 5% of data randomly chosen not to be included in refinement.
Ramachandran results were determined by MolProbity.
ELISA for measuring IgE antibodies to Ara h 2
IgE binding to natural Ara h 2 and the recombinant constructs rMBP-Ara h 2-N19 and rMBP-Ara h 2-N28 was analyzed by a chimeric ELISA previously described (24). The plates were coated overnight at 4°C with anti-Ara h 2 mAb 1C4. Plates were blocked with PBS-Tween-1% BSA, and in the following step, they were incubated with 5 mg/mL of either natural Ara h 2 or one of the two recombinant allergen constructs (100 μl per well) at room temperature. Natural Ara h 2 was purified from defatted peanut extract by affinity chromatography using the Ara h 2 specific mAb 1C4. Proteins were quantified by Advance Protein Assay (Cytoskeleton, Denver, CO). Sera from peanut allergic patients (CAP values from 0 to >100 kUA/L; average 26 ± 33 kUA/L for n = 21) were obtained from Bioreclamation (Westbury, NY), which operates in full compliance with Food and Drug Administration guidelines, or kindly provided by Dr. Peter Heymann (University of Virginia). Thirty-three sera were added at 1:2 and 1:10 dilutions, or further diluted for IgE antibody binding to fall in the linear range of the standard curves. Two sera without peanut reactivity were used as negative controls. Anti-Der p 2 mAb αDpX, purified natural Der p 2 at 500 ng/ml, and chimeric Ab 2B12-IgE were used to perform IgE standard curves. Bound IgE was detected using biotinylated goat anti-human IgE (KPL, Gaithersburg, MD) at 1:1,000 dilution. Streptavidin peroxidase was added at 250 ng/ml, followed by development with 1mM ABTS and 0.03% hydrogen peroxide as substrate. IgE antibody binding was quantified by measuring the OD at 405 nm. OD values for negative control sera were subtracted from the values for peanut allergic sera. Relative values of IgE binding were calculated by multiplying OD 405 nm by the dilution at which the sera were tested. As an aside, CD spectra of the two rAra h 2-MBP constructs are nearly identical within experimental error (data not shown), indicating that Ara h 2 is similarly folded in both constructs.
Results
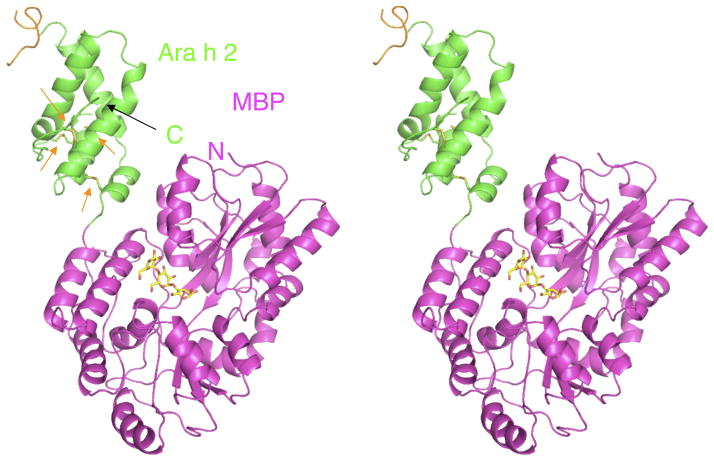
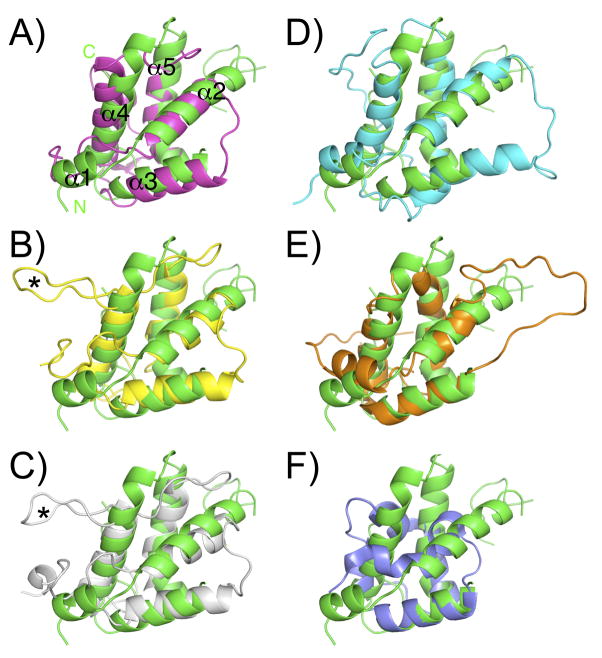
Ara h 2 was crystallized by the MBP fusion/surface entropy reduction technique that uses an engineered MBP as a carrier protein (Figure 1), to aid in crystal lattice formation (16). The construct that crystallized was rMBP-Ara h 2-N28, which eliminated the 21 residue signal sequence and 6 residues predicted to be disordered based on the NMR structure of Ara h 6 (25). Ara h 2 has five helices and four disulfide bonds with at least one disulfide bond connecting each helix to another (orange arrows Figure 1). The fold of Ara h 2 belongs to the prolamin protein superfamily, which usually consists of a 4–5 helix bundle held together by 4–5 disulfide bonds. Figure 2 shows superpositions of Ara h 2 with other members of the prolamin family. Using the web server DALI (26), the structure of Ara h 2 was ranked most similar to the proteinase/alpha-amylase inhibitors with very high Z scores ranging from 8 to 9.3. The top match is an alpha-amylase inhibitor (AI, Figure 2A) from wheat kernel (27), the second best match is the corn hageman factor inhibitor (CHFI, Figure 2B) (28), and the third best match is the RAGI-bifunctional inhibitor (RBI, Figure 2C) from Indian finger millet (29, 30). A significant structural difference between Ara h 2 and RBI or CHFI, is that the bifunctional inhibitors have a short fourth helix interrupted by a structured extended loop (asterisk in Figure 2) that interacts with the target amylase. Ara h 2 and AI have a longer helix (α4) in a structurally similar position (Figure 2A), so Ara h 2 is structurally most similar to AI.
Figure 1.
Stereo view of MBP-Ara h 2 fusion protein. The colors are: Ara h 2 (green and orange (residues 56-69)), MBP (magenta), maltotriose (yellow). Orange arrows indicate the disulfide bonds.
Figure 2.
Superpositions of Ara h 2 and other prolamin family members. Ara h 2 is green in all panels. A) AI, magenta B) CHFI, yellow C) RBI, white D) Ara h 6, cyan E) SFA-8, orange F) nsLTP, blue. The asterisk indicates a structured loop in the bifunctional inhibitors RBI and CHFI that interacts with an amylase.
Other structural matches to Ara h 2 include Ara h 6, which is the fourth highest Z-score (Figure 2D) and has the highest sequence identity (59%). Previous studies have also compared Ara h 2 with the 2S albumins such as SFA-8 from sunflower (Figure 2E) and the castor bean allergen Ric c 3 (31). However, the low DALI scores clarify that these are more distantly related. Additionally, the lower Z-scores and a visual inspection confirm that Ara h 2 is more similar to the proteinase/alpha-amylase inhibitors and the 2S albumins, than it is to the nsLTP family as shown with the example in Figure 2F (32).
The extended 31 residue loop between helices 2 and 3 of Ara h 2 was demonstrated to be sensitive to proteolysis (25) and is mostly unobserved in the crystal structure. The loop may also be cleaved in the crystal, however, mass spectrometry of the sample used for the crystallization found no evidence of proteolysis (data not shown) suggesting that the residues are simply disordered. The few residues of this loop that are observed (orange color Figure 1) are significantly displaced from comparable residues in Ara h 6, but this region is not well conserved and is known to be flexible (25). The sequence of this disordered loop differentiates Ara h 2.01 and Ara h 2.02, hence the structure here can be considered a model for the core structure of either isoform of Ara h 2.
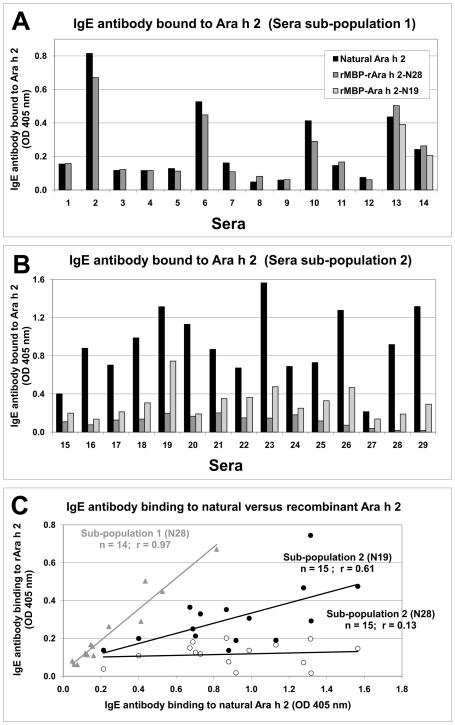
In order to assess the correct folding of Ara h 2 in the MBP fusion construct, IgE antibody binding to the natural allergen was compared to the recombinant constructs (Figure 3). Twenty-nine of thirty-three sera samples from peanut allergic patients reacted to natural Ara h 2, representing an 88% (29/33) prevalence of IgE antibody binding to this allergen. The overall percentage of sera reacting to rMBP-Ara h 2-N28 was 76% (25/33) with a clear distinction between two sub-populations that had previously reacted positively to the natural allergen. Sub-population 1 (n=14) showed an excellent correlation of IgE antibody binding to natural versus the rMBP-Ara h 2-N28 construct, indicating the recombinant Ara h 2 used in this crystallographic study is properly folded (Figure 3A and 3C). In contrast, sub-population 2 (n=15) showed low IgE antibody binding to rMBP-Ara h 2-N28 with an average binding reduction of 90% compared to the natural allergen. To test whether the MBP molecule was protecting an important IgE epitope region, the IgE antibody binding of 15 sera from subpopulation 2 was tested against rMBP-Ara h 2-N19, a construct with 9 more amino acids between MBP and the first helix of Ara h 2 (Figure 3B). The IgE antibody binding was on average 19% higher to the rMBP-Ara h 2-N19 than the binding to the rMBP-Ara h 2-N28 construct (Figure 3B and 3C). The construct with longer linker between the molecules should allow more access of IgE to the protected epitope(s). Additionally, the range of relative IgE antibody binding (OD 405 nm times sera dilution) was similar for natural and rMBP-Ara h 2-N28 allergen for sub-population 1 (mean absorbance of 6.0 ± 11.9 and 6.7 ± 14.0 OD units, respectively). In contrast, for sub-population 2, the range was larger for IgE antibody binding to natural allergen (mean absorbance of 91.3 ± 88.1) than for rMBP-Ara h 2-N28 (5.3 ± 4.8) or rMBP-Ara h 2-N19 (20.1±18.6) (Figure 3C).
Figure 3.
IgE binding to MBP-Ara h 2 fusion constructs and natural Ara h 2 measured by ELISA. A) IgE antibody binding to natural and recombinant Ara h 2 using 14 sera in sub-population 1. B) IgE antibody binding to natural and recombinant Ara h 2 using sera from sub-population 2. Sera #26–29 were considered negative for IgE binding to rMBP-Ara h 2-N28 due to values below the non-allergic control. In panels A & B, black bars represent natural Ara h 2, grey bars rMBP-Ara h 2-N28, and light grey rMBP-Ara h 2-N19. Panel C shows the correlation of IgE antibody binding to natural Ara h 2 versus recombinant fusion constructs. Grey triangles represent sub-population 1 comparing natural versus rMBP-Ara h 2-N28. Open circles show sub-population 2 comparing natural versus rMBP-Ara h 2-N28. Filled circles represent sub-population 2 comparing natural versus rMBP-Ara h 2-N19. Results are reported as OD 405 nm measured for the sera at dilutions that fall in the linear range of the standard curve.
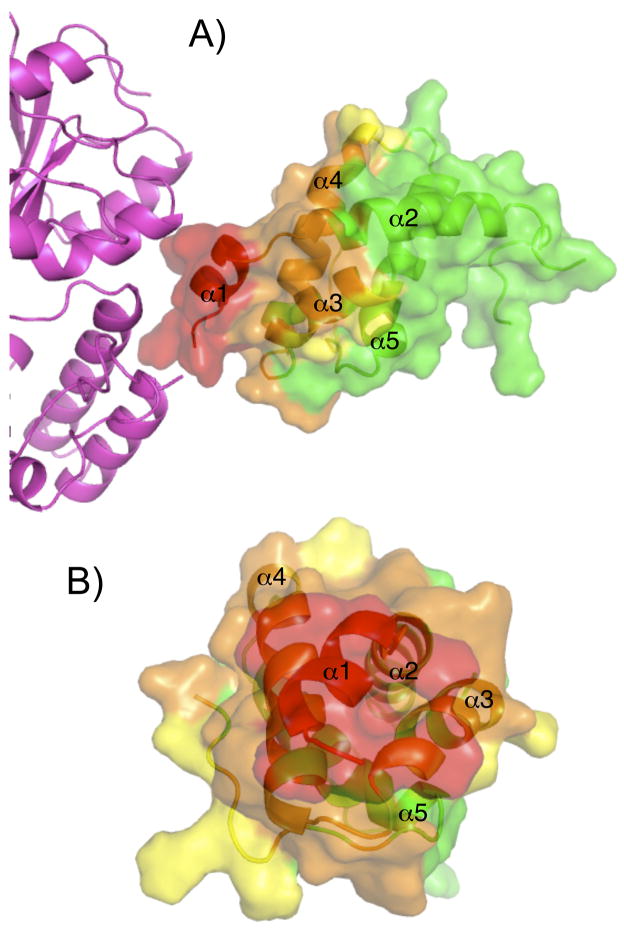
In examining the structure of the fusion protein for residues likely to be blocked by MBP-Ara h 2-N28, residues were classified into to three categories using different sized spherical probes to estimate access to Ara h 2 by IgE. The radii of the probes were: 1.4 Å (similar to solvent and very likely obscured by MBP), 10 Å (an intermediate value), and 17 Å (a circle with equivalent radius would be similar in size to an antibody epitope). Figure 4 shows the residues colored by estimated relative accessibility according to the spherical probes. The same color scheme is reiterated in Figure 5 to specify exactly what residues are potentially protected by MBP. Broadly speaking, it is very likely that residues in helix 1 are obscured by MBP, moderately likely that residues in helix 4 are obscured by MBP, and possible that residues at the C-terminus and in helix 3 would be difficult to access with a bulky antibody based on this analysis. It is important to state that this analysis is extremely qualitative and it is unknown if there is any flexibility between MBP and Ara h 2 in solution.
Figure 4.
Proposed surface of Ara h 2 that is inaccessible to IgE due to MBP. Spheres of various sizes (1.4, 10, and 17 Å) were used to estimate the relative accessibility of residues. MBP is magenta in panel A and absent in panel B. The surface and ribbon diagram of Ara h 2 are colored green for accessible, yellow for possibly accessible, orange for unlikely to be accessible, and red for most likely to be inaccessible. The perspective in panel B is looking from the MBP molecule.
Figure 5.
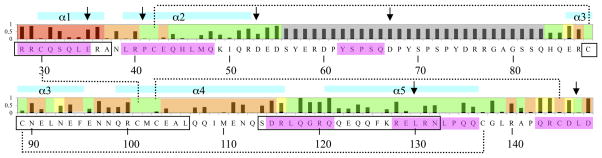
Ara h 2 Epitope Data and Fractional Side Chain Accessibility. Black bars plot the fractional side chain accessibility determined by VADAR (38). Overlayed on the plot, residues that are inaccessible due to MBP measured with a 1.4, 10, and 17 Å sphere are highlighted red, orange, and yellow, respectively. Other residues are highlighted green. Other highlights indicate: disordered residues and estimated side chain accessibility (grey); Helices (cyan); potentially important peptide IgE epitopes in (5) (magenta); T-cell epitopes (37) (black box). Arrows indicate the positions of a multiple alanine-substituted mutant of Ara h 2 with reduced IgE binding (12). Dotted lines indicate disulfide bonds.
Discussion
The fold of Ara h 2 belongs to the prolamin superfamily, which is subdivided into three families (33), the non-specific lipid transfer proteins (nsLTP), the proteinase/alpha-amylase inhibitors, and the seed storage 2S albumins. The nsLTP’s have a hydrophobic cleft for binding lipid substrates, which is not present in Ara h 2. Based on the structural data presented here, Ara h 2 belongs in the proteinase/alpha-amylase inhibitor family. The initial motivation for the development of the MBP constructs that are tightly packed against Ara h 2 was to aid in crystallization. To date, 21 different proteins have been successfully crystallized using MBP as a carrier protein (16). The MBP fixed-arm constructs, such as the MBP-Ara h 2, have been used to solve the structures of other unrelated proteins, including another allergen, Der p 7 (34). There are two main lines of evidence that suggest the structure of Ara h 2 determined here is not significantly perturbed by MBP. First, the structure is similar to other members of the prolamin family (Figure 2) including the proper connectivity of the disufide bonds, as predicted from sequence homology (31). And second, in 42% (14/33) of the sera from peanut allergic patients there is excellent correlation between IgE binding to the natural allergen and the fusion protein indicating that MBP does not alter the overall structure (Figure 3).
Additionally, the MBP-Ara h 2 fusion constructs have proven useful tools for identifying areas in Ara h 2 protected from antibody access by MBP. These protected areas appears to contain immunodominant IgE epitopes for a sub-population of peanut allergic patients. In this sera sub-population 2 (n=15), the IgE antibody binding to rMBP-Ara h 2-N28 was strongly reduced (by 90%) compared to the natural allergen, and the correlation between IgE antibody binding to both allergens was low (r = 0.13) (Figure 3C). Interestingly, with a longer linker (construct rMBP-Ara h 2-N19) a fraction of the patient response (19%) could be recovered and the correlation between IgE antibody binding to natural versus the construct rMBP-Ara h 2-N19 is positive (r = 0.61) compared to the rMBP-Ara h 2-N28 construct. From these results, MBP appears to block IgE antibody binding to an immunodominant epitope region for the set of sera from sub-population 2, rather than altering the rAra h 2 structure. In agreement with these results, one of the previously reported immunodominant linear epitopes of Ara h 2 corresponded to residues 27-36 (5), which is significantly blocked by MBP. It is intriguing that sub-population 2 appears to have a main immunodominant epitope region near helix 1 (Figures 4 and 5). A positive correlation between the number of IgE epitopes recognized and clinical sensitivity has been reported (35, 36). Therefore, the use of MBP-Ara h 2 fusion constructs in future studies should continue to elucidate the relevance of these Ara h 2 epitopes in peanut allergy.
The structure also provides information of potential value for the design of possible hypoallergenic mutants as described below. Hypoallergens seek to improve the safety and efficacy of immunotherapy by reducing IgE binding while maintaining important T-cell epitopes (5, 11, 12, 37). Figure 5 graphs the fractional surface exposure of each residue, and highlights in magenta important residues for IgE binding as identified with peptide mapping (5). These studies suggested that residues R28, R29, Q31 and E35 were crucial for IgE binding based on alanine substitutions (5). All of these residues are significantly surface exposed in the structure so the alanine scanning nicely matches the relative surface area for the residues where there is structural data. Based on the mapping, a mutant Ara h 2 with multiple residues changed to alanine (arrows Figure 5) was created that demonstrated reduced IgE binding (12). The structure presented here indicates that the mutations P41A, L130A, and L148A are likely of limited value in reducing IgE binding because of low surface exposure, whereas the highly solvated E35 and D53 may represent important antigenic features for IgE binding. Based on Figure 5, future design could focus on surface residues such as Q48, Q51, R52, E86, E111, Q135, Q143, R144, D146, and/or D148 for mutational studies to reduce IgE binding. These mutations avoid key T-cell epitopes (Fig. 5 black box), thought to be pivotal for the efficacy of selective allergen immunotherapy (11, 37). We anticipate that the structural data presented here will provide useful information for the rational design of hypoallergenic Ara h 2 molecules.
Additional bioinformatic analysis of epitopes from the structure and comparisons of Ara h 2 and possible Ara h 6 epitopes can be found in the Supplementary Material.
Supplementary Material
Acknowledgments
We are grateful to Dr. Robert Petrovich in the Protein Expression Core Facility for use of the CD spectrometer and Dr. Jason Williams in the Protein Microcharacterization Facility for mass spectrometric analysis. We thank Dr. Michael Fessler and Dr. Donald Cook for a critical reading of the manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. AP and SW were funded in part by grant R01AI077653 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health. X-ray diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID (or 22-BM) beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Abbreviations
- MBP
maltose binding protein
- RMSD
root mean squared deviation
Footnotes
Conflict of Interest: The authors confirm that they have no conflict of interest to report.
Contributions: GAM conceived of the study, performed structural analysis, and wrote the manuscript. RAG refined the crystallographic coordinates. AP and SW designed and executed the IgE studies and analysis. AFM contributed to the production of sample. REL conceived of the study and edited the manuscript. LCP produced the sample, collected data, and refined the coordinates. All authors contributed to the editing of the document.
References
- 1.Sampson HA. Update on food allergy. J Allergy Clin Immunol. 2004 May;113(5):805–19. doi: 10.1016/j.jaci.2004.03.014. quiz 20. [DOI] [PubMed] [Google Scholar]
- 2.Sicherer SH, Munoz-Furlong A, Sampson HA. Prevalence of peanut and tree nut allergy in the United States determined by means of a random digit dial telephone survey: a 5-year follow-up study. J Allergy Clin Immunol. 2003 Dec;112(6):1203–7. doi: 10.1016/s0091-6749(03)02026-8. [DOI] [PubMed] [Google Scholar]
- 3.Wensing M, Penninks AH, Hefle SL, Koppelman SJ, Bruijnzeel-Koomen CA, Knulst AC. The distribution of individual threshold doses eliciting allergic reactions in a population with peanut allergy. J Allergy Clin Immunol. 2002 Dec;110(6):915–20. doi: 10.1067/mai.2002.129235. [DOI] [PubMed] [Google Scholar]
- 4.Bock SA, Munoz-Furlong A, Sampson HA. Further fatalities caused by anaphylactic reactions to food, 2001–2006. J Allergy Clin Immunol. 2007 Apr;119(4):1016–8. doi: 10.1016/j.jaci.2006.12.622. [DOI] [PubMed] [Google Scholar]
- 5.Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, et al. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch Biochem Biophys. 1997 Jun 15;342(2):244–53. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- 6.Koppelman SJ, Wensing M, Ertmann M, Knulst AC, Knol EF. Relevance of Ara h1, Ara h2 and Ara h3 in peanut-allergic patients, as determined by immunoglobulin E Western blotting, basophil-histamine release and intracutaneous testing: Ara h2 is the most important peanut allergen. Clin Exp Allergy. 2004 Apr;34(4):583–90. doi: 10.1111/j.1365-2222.2004.1923.x. [DOI] [PubMed] [Google Scholar]
- 7.Palmer GW, Dibbern DA, Jr, Burks AW, Bannon GA, Bock SA, Porterfield HS, et al. Comparative potency of Ara h 1 and Ara h 2 in immunochemical and functional assays of allergenicity. Clin Immunol. 2005 Jun;115(3):302–12. doi: 10.1016/j.clim.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Flinterman AE, van Hoffen E, den Hartog Jager CF, Koppelman S, Pasmans SG, Hoekstra MO, et al. Children with peanut allergy recognize predominantly Ara h2 and Ara h6, which remains stable over time. Clin Exp Allergy. 2007 Aug;37(8):1221–8. doi: 10.1111/j.1365-2222.2007.02764.x. [DOI] [PubMed] [Google Scholar]
- 9.Pastorello EA, Pompei C, Pravettoni V, Brenna O, Farioli L, Trambaioli C, et al. Lipid transfer proteins and 2S albumins as allergens. Allergy. 2001;56(Suppl 67):45–7. doi: 10.1034/j.1398-9995.2001.00914.x. [DOI] [PubMed] [Google Scholar]
- 10.Jones SM, Pons L, Roberts JL, Scurlock AM, Perry TT, Kulis M, et al. Clinical efficacy and immune regulation with peanut oral immunotherapy. J Allergy Clin Immunol. 2009 Aug;124(2):292–300. e1–97. doi: 10.1016/j.jaci.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolland JM, Gardner LM, O’Hehir RE. Allergen-related approaches to immunotherapy. Pharmacol Ther. 2009 Mar;121(3):273–84. doi: 10.1016/j.pharmthera.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 12.King N, Helm R, Stanley JS, Vieths S, Luttkopf D, Hatahet L, et al. Allergenic characteristics of a modified peanut allergen. Mol Nutr Food Res. 2005 Oct;49(10):963–71. doi: 10.1002/mnfr.200500073. [DOI] [PubMed] [Google Scholar]
- 13.Linhart B, Valenta R. Molecular design of allergy vaccines. Curr Opin Immunol. 2005 Dec;17(6):646–55. doi: 10.1016/j.coi.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Benjamin DC, Berzofsky JA, East IJ, Gurd FR, Hannum C, Leach SJ, et al. The antigenic structure of proteins: a reappraisal. Annu Rev Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- 15.Albrecht M, Kuhne Y, Ballmer-Weber BK, Becker WM, Holzhauser T, Lauer I, et al. Relevance of IgE binding to short peptides for the allergenic activity of food allergens. J Allergy Clin Immunol. 2009 Aug;124(2):328–36. 36, e1–6. doi: 10.1016/j.jaci.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 16.Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010 May;19(5):901–13. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994 Sep 1;50(Pt 5):760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997 Dec 1;30:1022–5. [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997 May 1;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004 Dec;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–35. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 23.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Research. 2007 Jul;35:W375–W83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuurman J, Perdok GJ, Lourens TE, Parren PW, Chapman MD, Aalberse RC. Production of a mouse/human chimeric IgE monoclonal antibody to the house dust mite allergen Der p 2 and its use for the absolute quantification of allergen-specific IgE. J Allergy Clin Immunol. 1997 Apr;99(4):545–50. doi: 10.1016/s0091-6749(97)70083-6. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann K, Schweimer K, Reese G, Randow S, Suhr M, Becker WM, et al. Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions. Biochem J. 2006 May 1;395(3):463–72. doi: 10.1042/BJ20051728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008 Dec 1;24(23):2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oda Y, Matsunaga T, Fukuyama K, Miyazaki T, Morimoto T. Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution. Biochemistry. 1997 Nov 4;36(44):13503–11. doi: 10.1021/bi971307m. [DOI] [PubMed] [Google Scholar]
- 28.Behnke CA, Yee VC, Trong IL, Pedersen LC, Stenkamp RE, Kim SS, et al. Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution. Biochemistry. 1998 Nov 3;37(44):15277–88. doi: 10.1021/bi9812266. [DOI] [PubMed] [Google Scholar]
- 29.Strobl S, Maskos K, Betz M, Wiegand G, Huber R, Gomis-Ruth FX, et al. Crystal structure of yellow meal worm alpha-amylase at 1.64 A resolution. J Mol Biol. 1998 May 8;278(3):617–28. doi: 10.1006/jmbi.1998.1667. [DOI] [PubMed] [Google Scholar]
- 30.Strobl S, Maskos K, Wiegand G, Huber R, Gomis-Ruth FX, Glockshuber R. A novel strategy for inhibition of alpha-amylases: yellow meal worm alpha-amylase in complex with the Ragi bifunctional inhibitor at 2.5 A resolution. Structure. 1998 Jul 15;6(7):911–21. doi: 10.1016/s0969-2126(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 31.Radauer C, Breiteneder H. Evolutionary biology of plant food allergens. J Allergy Clin Immunol. 2007 Sep;120(3):518–25. doi: 10.1016/j.jaci.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Hoh F, Pons JL, Gautier MF, de Lamotte F, Dumas C. Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding. Acta Crystallogr D Biol Crystallogr. 2005 Apr;61(Pt 4):397–406. doi: 10.1107/S0907444905000417. [DOI] [PubMed] [Google Scholar]
- 33.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995 Apr 7;247(4):536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 34.Mueller GA, Edwards LL, Aloor JJ, Fessler MB, Glesner J, Pomes A, et al. The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins. Journal of Allergy and Clinical Immunology. 2010 Apr;125(4):909–17. doi: 10.1016/j.jaci.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flinterman A, Shreffler W, Lencer D, Bardina L, den Hartog JS, Lin J, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitised children in relation to severity of peanut allergy. Allergy. 2008;63:32. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 36.Lewis SA, Grimshaw KEC, Warner JO, Hourihane JOB. The promiscuity of immunoglobulin E binding to peanut allergens, as determined by Western blotting, correlates with the severity of clinical symptoms. Clinical and Experimental Allergy. 2005 Jun;35(6):767–73. doi: 10.1111/j.1365-2222.2005.02252.x. [DOI] [PubMed] [Google Scholar]
- 37.Glaspole IN, de Leon MP, Rolland JM, O’Hehir RE. Characterization of the T-cell epitopes of a major peanut allergen, Ara h 2. Allergy. 2005 Jan;60(1):35–40. doi: 10.1111/j.1398-9995.2004.00608.x. [DOI] [PubMed] [Google Scholar]
- 38.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003 Jul 1;31(13):3316–9. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.