Abstract
The medical, agricultural and biotechnological importance of the primitive eukaryotic microorganisms, the Fungi was recognized way back in 1920. Among various groups of fungi, the Aspergillus species are studied in great detail using advances in genomics and proteomics to unravel biological and molecular mechanisms in these fungi. Aspergillus fumigatus, Aspergillus flavus, Aspergillus niger, Aspergillus parasiticus, Aspergillus nidulans and Aspergillus terreus are some of the important species relevant to human, agricultural and biotechnological applications. The potential of Aspergillus species to produce highly diversified complex biomolecules such as multifunctional proteins (allergens, antigens, enzymes) and polyketides is fascinating and demands greater insight into the understanding of these fungal species for application to human health. Recently a regulator gene for secondary metabolites, LaeA has been identified. Gene mining based on LaeA has facilitated new metabolites with antimicrobial activity such as emericellamides and antitumor activity such as terrequinone A from A. nidulans. Immunoproteomic approach was reported for identification of few novel allergens for A. fumigatus. In this context, the review is focused on recent developments in allergens, antigens, structural and functional diversity of the polyketide synthases that produce polyketides of pharmaceutical and biological importance. Possible antifungal drug targets for development of effective antifungal drugs and new strategies for development of molecular diagnostics are considered.
Keywords: Aspergillus species, Allergens, Polyketides
Introduction
Aspergilli are ubiquitous in nature and universal in distribution. The diverse Aspergilli group comprises human, animal and plant pathogens, apart from fungi with a plethora of industrial applications. Aspergillus fumigatus, Aspergillus flavus and Aspergillus niger are known to cause allergic reactions and allergic bronchopulmonary aspergillosis (ABPA) in immuno competent individuals. A. fumigatus represents a major cause of morbidity and mortality in the patients of Allergic bronchopulmonary aspergillosis (ABPA) [1]. A. fumigatus, A. flavus and A. niger are also opportunistic human pathogens in immunocompromised patients such as post transplant cases, HIV etc. where the disease often leads to fatality [2]. A number of novel allergens and antigens of diagnostic and therapeutic importance, multifunctional proteins and toxins have been identified and characterized from Aspergillus species, particularly from A. fumigatus.
The aflatoxin producing A. flavus and ochratoxin producing A. ochraceus are plant pathogens infamous for their ability to affect a wide variety of crops. A. niger and A. oryzae are widely utilized in food industry for citric acid production by fermentation technologies [3, 4]. Lovastatin, the commonly used anticholesterol drug is produced by A. terreus while A. nidulans is being used as a model organism to study cellular physiology and genetics [5].
Currently, only 30% of the genes of Aspergillus species have been characterized leaving the scope for identification of highly diversified biomolecules of human interest [6]. Recent advances in fungal genomics revealed scores of hitherto unknown information of these Aspergillus species. This has opened up new avenues to study biological and molecular mechanisms in host pathogen interactions and also for exploration of unknown secondary metabolites of pharmaceutical and commercial importance. One of the important aspects of Aspergillus biology is the production of secondary metabolites such as polyketides. Very little is known about the structural, molecular and functional aspects of Aspergillus polyketides, their biosynthetic pathways, and the important enzymes involved in these pathways. Polyketide synthase, the key enzyme in polyketide biosynthetic pathways, is a multidomain and multifunctional enzyme.
Recent advances in the understanding of the biomolecules of Aspergillus species and the structural and functional diversity of the polyketide synthases useful for applications are reviewed in the article.
Unique Characteristics of the Aspergillus Species
Aspergillus mycelium forms conidiophores producing large numbers of conidia (asexual spores) which are dispersed through the air and inhaled by humans. These small airborne spores (2–3 μm in diameter) can bypass mucociliary clearance by human host and cause the disease. Aspergillus species can be distinguished from each other by the conidial pigmentation. The conidia of A. fumigatus are bluish to green in color; of A. flavus and A. niger are green and black respectively. The ability to thrive at very high temperature ranges, from 37 to 55°C in soil, mammalian and avian tissues is a unique property of A. fumigatus among Aspergillus species [7]. This may be due to its metabolic adaptation to higher temperature and presence of higher number of heat shock responsive genes compared to other Aspergillus species [7, 8]. To achieve these adaptations, this fungus has evolved distinct mechanisms of stress resistance that may provide basis for its virulence. Apart from its thermotolerant growth, A. fumigatus counters hostile environment while retrieving essential nutrients from the environment whether it is a human host or the decaying organic or plant debris, and thus, adapt well to a broad range of environmental conditions. A. fumigatus secretes number of catabolic enzymes such as peptidases and proteases, to degrade macromolecular polymers for nutrient uptake from the host [9, 10].
Interesting Biomolecules of Aspergilli
Aspergillus species are a continuous source of complex proteins, allergens, antigens and enzymes to the environment. A. fumigatus, A. flavus and A. niger are known to cause Type I and Type III hypersensitivity reactions in humans. A. fumigatus is known to produce multifunctional enzymes and toxins that facilitate the adherence and hydrolysis of the components of the host cell, and complex allergens which cause severe allergic reactions. In view of the clinical importance of the allergens/antigens of Aspergillus species in the allergic and invasive disease, over 20 genes encoding A. fumigatus antigens have been recombinantly expressed and evaluated for their diagnostic importance (Table 1) [11–17]. So far, 34 allergens of A. fumigatus, from Asp f1 to Asp f34, have been designated by IUIS Allergen Nomenclature Committee [18]. Asp f 35 (34 kDa protein with unknown function), Asp f 36 (extracellular arabinase), and Asp f 37 (chitosanase) from A. fumigatus are recently accepted to be included in the list [19]. Asp fl, an 18 kDa allergen/antigen, is a major cytotoxin secreted by A. fumigatus. Multiple roles of this 18 kDa protein such as allergenicity, antigenicity, ribonuclease activity and cytotoxicity have been iterated and this allergen is implicated in pathogenesis [20, 21]. Few more allergens such as 44-kDa allergen, Aspf23 (L3 ribosomal protein), 40 kDa protein disulphide isomerase, 45 and 56kDa glycoprotein antigens and a 34 kDa allergen with protease activity were characterized and were implicated in virulence [22–24]. Among Aspergillusflavus allergens Asp fl 13, an alkaline serine protease and Asp fl 18, vacuolar serine proteases were characterized. These proteases and protease isozymes have been implicated in colonization of animal host. Genes with >90% sequence homology with Asp f 1, Asp f 5, Asp f 12, Asp f 22 and Asp f 23 were mapped on A. flavus genome [25]. However, these allergens are not yet reported from A. flavus and need to be characterized. Alkaline phosphatases of A. flavus are implicated in aggressive colonization of cotton balls, and hydrolases of the same are considered as pathogenic factors for plants [26, 27].
Table 1.
Aspergillus fumigatus allergens/antigens of diagnostic relevance
| Allergen | Biological activity | Use of recombinant allergen for diagnosis |
|---|---|---|
| Asp f 1a (18 kDa) | Allergic, ribonuclease activity, cytotoxicity | IgE reactivity |
| IgG reactivity | ||
| Immunodominant peptides | ||
| Asp f2a (37 kDa) | A fibrinogen-binding protein | IgE reactivity |
| Immunodominant peptides | ||
| Asp f3a (19 kDa) | Peroxisomal membrane proteins (PMP) | IgE reactivity |
| Asp f4a (30 kDa) | Unknown biological function with | IgE reactivity |
| Immunodominant peptides | ||
| Asp f5 (42 kDa) | Metalloprotease (MEP) | IgE reactivity |
| Asp f 6a (23kD) | Manganese superoxide dismutase (MnSOD) | IgE reactivity |
| Immunodominant peptides | ||
| Asp f 8a (11.1 kDa) | A glycoprotein with homology to ribosomal protein P2 | IgE reactivity |
| Asp f 9 (18.8 kDa) | Glycosyl hydrolase | IgE reactivity |
| Asp f 10 (34.4 kDa) | Aspartic proteases (PEP) | IgE reactivity |
| Asp f 11 (95 kDa) | Cyclophilin or a dipeptidyl-peptidase IV | IgE reactivity |
| Asp f 16 (43 kDa) | A protein with unknown function and similarity to Asp f 9. | IgE reactivity |
| Immunodominant peptides | ||
| Asp f 23 (44 kDa) | Ribosomal protein L3 | IgG reactivity |
aAspergillus fumigatus allergens are used in ImmunoCap method for serodiagnosis or skin testing
Advances in the field of genomics and proteomics contributed immensely to better understanding of the host-pathogen factors and their interactions in Aspergillus and other fungal diseases. Some of the promising advances include molecular interaction studies of lung surfactant proteins SP-A, SP-D and Mannan binding lectin (MBL) with A.fumigatus. These studies are based on in vitro cell culture experiments and murine models including knockout mice of ABPA and invasive aspergillosis [28–31]. Therapeutic potential of recombinant human surfactant protein D (rhSP-D) has been established by studies in murine models of lung allergy, hypereosinophilic SP-D gene-deficient mice and also with eosinophils from allergic patients. These studies clearly suggest therapeutic application of rhSP-D for allergic and invasive diseases [32–37]. Single nucleotide polymorphisms (SNPs); have been observed in these genes (SP-A, SP-D and MBL). Such polymorphisms may result in partial or total loss of function and may contribute to the hosts’ susceptibility to aspergillosis. Studies showed association of SNPs in SP-A2 and MBL genes with patients of allergic bronchopulmonary aspergillosis and bronchial asthma with rhinitis [38]. Adhesin proteins of A. fumigatus have been recently identified and characterized. Adhesin protein of A. fumigatus, Extracellular thaumatin domain protein (AfCalA) was recombinantly expressed and specific binding with laminin and murine lung cells was established [39]. This suggests that the mechanism of binding of Afu conidia to the host cells is through adhesin proteins of A. fumigatus. Host-pathogen interaction studies of A. flavus have been carried out with the plant cells using microarray and proteomics technology. However, host–pathogen interactions with respect to A. flavus and A. niger and animal/human host need to be studied in detail.
Virulence of A. fumigatus is not attributed to a single protein/gene and appears to be a multifactorial trait. Cell wall molecules like β-glucan, α-glucan, chitin, galactomannans, galactomannan proteins (Afmp1p, Afmp2p), hydrophobins (rodA/hyp1 and rodB) and DHN-melanin of A. fumigatus are known to interact with the host and alter the immune responses. The genes for β (1-3) glucan synthases (Fks1p), glucanosyltransferase (Gel 1p, Gelp 2p, and Gel3p), β (1-3) endoglucanase (Engl1), α (1-3) glucan synthetase, and chitin synthase (ChsE, ChsG) are studied for their contribution in virulence by gene complementation studies [40]. Genes and proteins/enzymes reported to have a role in the resistance to innate immune response belong to conidial specific catalases (catA, cat1/catB, cat2/katG, catC, and catE), superoxide dismutases (sod1, sod2, sod3/asp f 6, and sod4), fattyacid oxygenases (ppoA–C), glutathione tranferases (gstA–E, afyap1, skn7, and pes1), efflux transporters (mdr1–4, atrF, abcA–E, and msfA–E) and DHN-melanin cluster genes (pksP/alb1, arp1, arp2, abr1, abr2, and ayg1) [41]. Melanin, a pigment (1,8-dihydroxynaphthalene) of A. fumigatus is considered as a virulent factor as it protects the integrity of the conidial cell wall. This also helps in the expression of adhesins at the conidial surface necessary for binding to host cells [42]. Cyclic AMP-dependent Protein Kinase A (PKA) signal cascade is also found to be a critical regulator for conidiation, development, growth, and stress responses [43]. A. fumigatus toxin associated with conidia is fumigaclavine C, an alkaloid metabolite and potent inhibitor of DNA synthesis. It also secretes a number of low molecular weight toxins such as gliotoxins, helvolic acid, verruclogen, a protein toxin Asp f1 that contribute to the virulence and also toxins such as fumagillin, and fumitremorgin A–C (Neurotropic toxins).
Drug Target Pathways/Genes
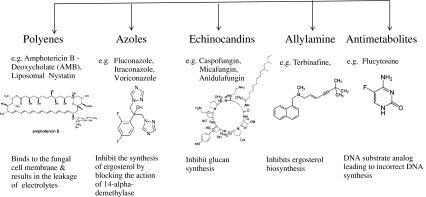
A. fumigatus genome sequencing coupled with the genetic tools for gene manipulation and high-throughput microarray analysis has facilitated the research for novel antifungal drug targets. The drugs currently available for invasive aspergillosis (polyenes, azoles, and echinocandins) target synthesis of cell wall molecules (ergosterol, β-1,3 glucan, and chitin) or molecules involved in the steps for synthesis. This approach has limitations due to the structural homology with cholesterol in the human host and results in toxicity and development of drug resistance by the pathogen (Fig. 1). Unique biochemical pathways and their key enzymes in Aspergillus species which are absent in humans are being explored by various experimental strategies for identification of novel drug targets. The glyoxylate bypass, methylcitrate cycle, lysine biosynthetic pathway and trehalose biosynthetic pathway are few examples (Table 2). New strategies using advanced genomic tools such as conditional promoter replacement (CPR), gene replacement and conditional expression (GRACE) strategies lead to identification of number of essential genes for fungal growth. These can be explored for development of antifungal drug targets [44]. Polysaccharide components of fungal cell wall and its biosynthetic pathway have been attractive targets for selective antifungal drug development [45, 46]. In contrast to yeast cell wall A. fumigatus lacks β-1,6 glucan, which interconnects proteins, chitin, and β1-3 glucans and also lacks cell wall bound homologous GPI-proteins [Glycosylphosphatidylinositol (GPI)-anchored proteins (GPI-Aps)] linked to glucans, important for cell wall organization. Instead, Afu contains hydrophobins that are important for conidial survival and attachment to hydrophobic surfaces [47] that can serve as potential drug targets in A. fumigatus. The methylcitrate cycle, is essential for degradation of toxic propionyl-CoA via methylcitrate synthase enzyme in fungi and some bacteria. A. fumigatus methylcitrate synthase gene transcription was reported during invasion from infected mouse lungs. Virulence of A. fumigatus was attenuated by deletion of methyl citrate synthase gene. Therefore, methylcitrate cycle which is absent in humans may provide a potential drug target [48, 49]. Glyoxylate pathway was implicated in the pathogenicity of other pathogenic organisms such as Candida albicans and Mycobacterium tuberculosis [50]. Mutants of A. fumigatus that do not have isocitrate lyase of glyoxylate pathway retained the same virulence as the wild-type in A. fumigatus [51]. This confirms that glyoxylate cycle is not important in the anaplerotic synthesis of oxaloacetate in A. fumigatus under infectious conditions [52]. Trehalose is a non-reducing disaccharide and functions as a reserve carbohydrate and a stress metabolite which also protects the fungal cell by preventing aggregation of denatured proteins and scavenging free radicals [53, 54]. Trehalose biosynthesis has been linked to virulence in pathogenic fungi and is absent in mammalian cells. Thus, it could be a potential target for antifungal therapy. Trehalose content increases during the life cycle of A. fumigatus and also after heat shock but not in response to other types of stress. Using gene disruption studies, tpsA and tpsB genes of A. fumigatus were found to be involved in the trehalose biosynthesis [55]. Deletion of the two genes led to delayed germination of conidia at 37°C and conidia were susceptible to oxidative stress. Although, trehalose synthesis is related with pathogenicity in Afu, the mechanism of virulence is not clearly understood [56]. Another pathway, alpha-aminoadipate pathway, essential for lysine biosynthesis which is absent in human, has also been suggested as a potential antifungal drug target for A. fumigatus. Deletion mutant of lysine pathway-specific enzyme, homocitrate synthase (HcsA), was found to be attenuating virulence in a corticosteroid-based murine infection model of bronchopulmonary aspergillosis. However, the supply of excess lysine via the drinking water partially restored virulence, implying importance of lysine auxotrophy in virulence [57]. Comparative genomic analysis, manual mining of experimentally confirmed essential genes from fungal pathogen (such as C. albicans or A. fumigatus) and modeling studies proved helpful in drug target discoveries. Four such genes were recently identified as putative drug targets in A. fumigatus with other fungal pathogens such as Candida albicans, Blastomyces dermatitidis, Paracoccidioides brasiliensis, Paracoccidioides lutzii, Coccidioides immitis, Cryptococcus neoformans and Histoplasma capsulatum which were absent in the human genome. These genes are: trr1, that encodes for thioredoxin reductase, rim8, encoding a protein involved in the proteolytic activation of a transcriptional factor, kre2, that encodes for α-1,2-mannosyltransferase and erg6, that encodes for d-(24)-Sterol C-methyltransferase [58]. The validity of these genes as drug targets needs to be experimentally established for development of novel antifungal drugs.
Fig. 1.
Antifungal drugs and the mechanism of action
Table 2.
Biochemical pathways of Aspergillus species targeted for therapy
| Name of the fungus | Biochemical pathway | Important role | +/− in humans | Current drugs | Limitations |
|---|---|---|---|---|---|
| A. fumigatus, C. neoformans | Ergosterol biosynthesis | Integral part of cell membranes | −ve | Amphotericin B, azoles, Echinocandins | Acts also on cholesterols, due to structural similarity |
| A. fumigatus | Methyl citrate cycle | For degradation of toxic propionyl-CoA | −ve | – | |
| A. fumigatus, C. albicans, S. cerevisiae, C. neoformans | Glyoxylate pathway | Serve as carbon sources in gluconeogenesis from fat | −ve | Not essential for survival | |
| A. fumigatus, C. neoformans | Lysine biosynthesis | Essential amino acid | −ve | Lysine biosynthesis may not be required for invasive growth | |
| A. fumigatus, S. cerevisiae | Trehalose biosynthesis | Reserve carbohydrate source | −ve | Virulence mechanism not known clearly | |
| A. fumigatus, C. neoformans | Melanin biosynthesis | Conidial pigment and virulent factor | −ve | Not essential for survival |
Genes and Genomics of Aspergilli
Important Secondary Metabolites: Polyketides
Genome sequencing has facilitated a great insight into the secondary metabolite genes for a comprehensive study. Genome size of important Aspergillus species varies from 30–40 Mega bases (Mb) spanned on eight chromosomes with good synteny among species (50%). Aspergillus species present unique genes which vary from 140 to 500 in numbers for different Aspergilli (Table 3). Currently, only <10% of the 9,000–13,000 open reading frames (ORFs) have been assigned with functions [59–61]. However, a large number of genes are unknown with respect to function. Till date most of the transcriptome of Aspergillus has been predicted based on the bioinformatic approach and needs to be validated in vivo. A. fumigatus (Af293) genome sequencing revealed nine previously unknown allergens, identification of numerous genes involved in the production of specific secondary metabolites, and a set of essential genes that may be potential targets for drug development [62]. Genome-sequencing projects of Aspergillus species and the web-based bioinformatics tool SMURF (Secondary Metabolite Unknown Regions Finder; www.jcvi.org/smurf) have shown that each species of Aspergillus genome contain 30–70 secondary metabolite clusters which includes 15–35 polyketide synthases, 12–30 non ribosomal peptide synthases, and a number of dimethylallyl tryptophan synthases (DMATS) [63, 64] (Table 3).
Table 3.
Genes and genomics of Aspergillus species
| A. fumigatus | A. nidulans | A. oryzae | A. niger | A. flavus | |
|---|---|---|---|---|---|
| Strain | Af293 | FGSC A4 | ATCC 42149 | CBS 513.88 | NRL 3357 |
| Genome size (Mb) | 28.81 | 30.06 | 36.7 | 33.97 | 36.51 |
| Number of chromosomes | 8 | 8 | 8 | 8 | 8 |
| Number of protein coding genes | 9630 | 9541 | 14063 | 14097 | 13515 |
| Annotated proteins (Pfam hits) | 5808 | 4512 | 10416 | 5306 | 5510 |
| Unique genes | 148 | 469 | 331 | 236 | 278 |
| Probable PKS genes | 21 | 24 | 27 | 34 | 25 |
| Secondary metabolite clusters | 28–30 | 46 | 56 | 67 | 55 |
Aspergillus fumigatus shows presence of 28–30 clusters of secondary metabolites. Melanin gene cluster has been identified and characterized from Afu. Several other secondary metabolite gene clusters are characterized in A. fumigatus which produce gliotoxins (immunosuppressive and proapoptotic for mammalian cells) [65], fumigaclavines (ergot alkaloid used for therapeutic use) [66], fumitremorgins (mycotoxin) [67], and siderophores [68]. Gliotoxin, a secondary metabolite produced via gliotoxin biosynthetic pathway using non-ribosomal peptide synthase machinery, has been found in lungs of invasive aspergillosis patients with cancer and also in IA mice model [69]. Other mycotoxins involved in mycosis caused by A. fumigatus are trypacidin, verruculogen and fumigaclavine A. Further metabolomic studies on A. fumigatus suggest that it produces number of extrolites of different chemical nature including polyketides, non ribosomal peptides, terpenes, anthroquinones and compounds of mixed origin with aminoacids, such as siderochromes etc. [70]. Annotation of the A. flavus genome indicates that it has 55 secondary metabolite biosynthetic pathways which may cross link with other pathways to generate various metabolites. So far, only Aflatoxin and cyclopiazonic acid gene clusters have been characterized from A. flavus. Genome sequencing of industrially important species, A. niger revealed presence of highest number of secondary metabolite clusters that is 67, with 34 polyketide synthases and 17 non-ribosomal peptide synthases arranged in clusters [71]. For A. niger a link between genes involved in secondary metabolite synthesis has only been determined for siderophore and spore pigmentation so far [72]. A. niger genome sequencing also showed homology with gene sequences of fumonisin pathway from Gibberella moniliformis indicating its potential to produce mycotoxin such as fumonisin [73]. However, A. niger does not produce this toxin. Some A. niger strains are known to produce ochratoxin but very little is known about the biosynthetic pathway of this mycotoxin in this species. A. oryzae useful for food industry has potential for a remarkably large number of secondary metabolites compared to pathogenic fungus A. fumigatus and A. flavus. It has high number of secondary metabolite genes which are enriched in regions lacking synteny with either A. fumigatus or A. nidulans indicating A. oryzae’s capability to produce specific metabolites. This is also supported by the presence of 56 secondary metabolite clusters and highest no. of cytochrome P450 genes in the A. oryzae genome. Recently, a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene involved in cyclopiazonic acid (CPA) production has been identified from A. oryzae [74]. The presence of putative genes encoding other principal enzymes involved in phenylpropanoid and flavonoid biosynthesis (such as phenylalanine ammonia-lyase, cinnamic acid hydroxylase and p-coumarate CoA ligase) in A. oryzae genome prove the presence of fungal phenylpropanoid-flavonoid metabolite pathway in industrially useful A. oryzae [75]. Although, A. oryzae is not known to produce aflatoxin, aflatoxin biosynthetic clusters genes have been mapped on its genome and these genes appear to be cryptic or silent. Genome sequence of several isolates of A. oryzae when compared with A. flavus, showed presence of aflatoxin biosynthesis genes with deletions, frame shift mutations, and base pair substitutions [76]. These alternations seem to be responsible for the inactive aflatoxin genes in A. oryzae.
Secondary metabolite pathway genes in Aspergillus species are conveniently arranged in gene clusters with transcriptional regulators, mostly at the end of chromosomes, and have been predicted to be involved in the synthesis of various polyketides or non-ribosomal peptides. However, most of these clusters are silent or cryptic, under the laboratory conditions with the exception of few toxins and a virulent factor. To activate expression of silent clusters and to identify the metabolites produced, different strategies based on molecular, epigenetics and cultivation methods are undertaken in Aspergillus field [77, 78]. The novel global regulator of secondary metabolite, LaeA, is recently described in Aspergilli by Keller et al. Loss of this gene eliminates sterigmatocystin, aflatoxin, lovastatin, and gliotoxin production in Aspergillus species and results in an avirulent A. fumigatus mutant strain [79, 80]. LaeA may affect, histone methylation, and this might alter the state of chromatin in metabolite gene clusters. However, the role of this protein in regulation of secondary metabolites has not yet been established fully [81]. In view of the importance of various polyketides, the structural and functional characterization of genes and proteins of the polyketide synthases (PKS), a key enzyme in polyketide biosynthetic pathway in the Aspergillus species needs to be studied in detail.
Aspergillus Polyketides
Polyketides are a large and diverse group of natural products including polyphenols, polyenes, and macrolides with a wide variety of biological activities with antibiotic, antifungal, and anticancer properties. They are chemically distinct group containing multiple –CH2–CO– ketide group synthesized by repetitive condensation or polymerization reactions [82]. Aspergillus species produce a number of secondary metabolites of varied biochemical structures such as terpenes, alkaloids, ergots and polyketides. Aspergillus polyketides are basically naphtho-γ-pyrone and furanocoumarin ring compounds. Important polyketides produced by Aspergillus species are classified as (i) drugs and polyketides of pharmaceutical relevance and (ii) virulent factors and toxins (Table 4). Pharmaceutically relevant polyketides includes cholesterol lowering lovastatins by A. terreus. A. nidulans is also known to produce number of polyketides with antimicrobial and anticancer property [83, 84]. Polyketides as virulent factors and toxins comprise melanin pigments from A. fumigatus, carcinogenic mycotoxins aflatoxins from A. flavus, and A. parasiticus and sterigmatocystin from A. nidulans. Aflatoxin and sterigmatocystin are hepatotoxic, neurotoxic and cause threat to the human health. Mycotoxin contamination of the agricultural crops and agri products leads to great economic losses, as the permeable limit of Aflatoxin (4–20 ppb) for food and food products in international trade has been fixed in view of the WTO stipulations [85]. Important mycotoxins of agricultural crops produced by Aspergillus species include Aflatoxins, ochratoxin and cyclopiazonic acid.
Table 4.
Polyketides and polyketide synthases of Aspergilli and diversity in the KS domain*
 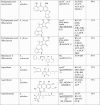
|
aHighly reducing polyketide synthase
bPartially reducing PKs enzyme
cNon reduced PKS enzyme
dNon ribosomal peptide synthase (NRPS) enzyme
eHybrid PKS–NRPS enzyme
f% of KS domain homology with PksA protein (AAS90093.1)
g% of SAT domain homology in afoEa is 27%
* Reference 89
Polyketide Synthases in Aspergillus Species
Polyketide synthases are present in fungi and bacteria. They are the key enzymes in the polyketide biosynthetic pathway. Polyketide synthases are classified in three types based on their structural architect of the domains and their functional use (Table 5). Type I PKS enzymes are large multifunctional proteins of <1000 AA and are encoded by a single gene in fungi and bacteria e.g. PksA of A. flavus. Type II PKS enzymes are dispersed as individual proteins of approximately 500 AA and are found in bacteria e.g. actinorhodin PKS of Streptomyces coelicolor. It is an antibiotic substance from Streptomyces coelicolor [86]. Type III polyketide synthases are comparatively small proteins of 350–500 AA with a single polypeptide chain and are involved in the biosynthesis of precursors for flavonoids like chalcones and stilbenes in plants [87].
Table 5.
Diversity of polyketide synthases in microbes
| Microbes | Type of polyketide synthase | Protein structure | Functional characterization | Types of polyketides produced | Domain organization |
|---|---|---|---|---|---|
| Bacteria | Type I (modular) | Single linear protein with multiple modules | Active site used only once | Macrolide polyketides, such as the erythromycin A and rifamycin | e.g. Saccharopolyspora erythrea KS-AT-ACP-KR-TE = KS-AT-ACP-KR-TE = KS-AT-ACP-KR-TE = KS-AT-ACP-KR-TE |
| Bacteria | Type II (Iterative) | Multiple proteins, with a single mono-functional active site | Active sites may be used only once or repeatedly | Aromatic polyketides, such as actinorhodin and tetracenomycin | e.g. Streptomyces coelicolor KS-AT-ACP-KR-DH-ER |
| Bacteria | Type III (Iterative) | One protein with multiple modules | Active sites are reused repeatedly | Dihydroxyphenylalanine melanins | e.g. bacterial PKS Streptomyces grieseus (RppA) KS-AT-ACP-TE |
| Fungi | Type I (iterative) | Single protein with one module | Active sites are reused repeatedly | Lovastatins, Aflatoxins, Melanin | e.g. polyketide synthase (LovF) from A. terreus KS-AT-DH-MT-(ER)-KR -ACP |
| Fungi | Type III | One protein with multiple modules | Active sites are reused repeatedly | 3,5-dihydroxybenzoic acid | e.g. CsyA from A. oryzae KS-AT-ACP-KR-MT-DH-ER |
| Plants | Type III (Iterative) | One protein with multiple modules | Active sites are reused repeatedly | Flavonoids like chalcones and stilbenes | e.g. Chalcone synthase from Medicago sativa |
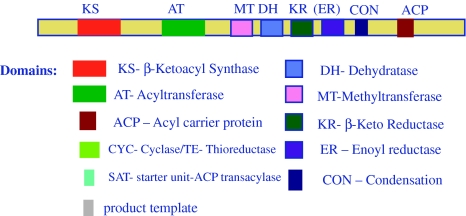
Aspergillus PKS enzymes are Type I and consist of five to nine domains on single polypeptide chain of 1000–3000 AA (Fig. 2). These PKS enzymes use an iterative strategy i.e. each domain is used repeatedly to extend the polyketide chain to build polyketide e.g. Polyketide synthase A (PksA) for Aflatoxin and Polyketide synthase P (PksP) for melanin production. However, Lovastatin diketide synthase (LovF/LDKS) of A. terreus acts non-iteratively like the bacterial modular PKS enzymes [88]. Bacterial PKS enzymes use modular architect, where domains are organized into modules and used for the catalysis of one cycle of polyketide chain elongation non-iteratively as in the case of 6-deoxyerythromycin B synthase (DEBS) for the biosynthesis of erythromycin in streptomyces species [89, 90]. Interestingly, apart from Type I PKS enzymes, recently Type III PKS genes resembling the chalcone synthase genes (CHS) for the production of the flavonoid have been identified in A. flavus and A. oryzae by comparative genome analyses [89, 90].
Fig. 2.
Lovastatin nonaketide synthase(LovB) of Aspergilllus terreus showing general Aspergillus Polyketide synthase domain Architect (*Domains like MT, ER,DH,CON are absent in other Aspergillus PKS enzymes and may have SAT, PT, and TE/CYC domains)
Domain Organization of Aspergillus PKS Enzymes
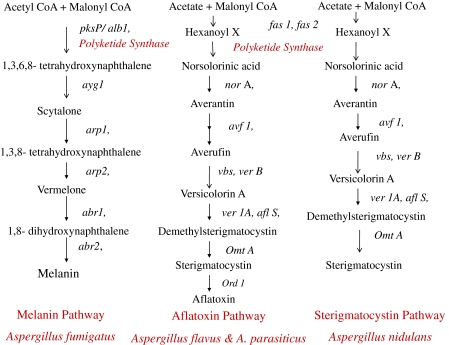
The diversity in polyketide structure and function in Aspergilli is reflected in the structural diversity of PKS enzyme and the Polyketide biosynthetic pathway (Fig. 3). Aspergillus PKS enzymes use substrates such as acetyl-CoA and malonyl-CoA/hexonyl CoA in a similar manner to multi-domain Fatty Acid Synthases but differ by incomplete reduction and/or dehydration steps [91]. A range of domains of the PKS enzyme facilitate different steps in the synthesis of various intermediates of polyketide products. The core domains essential for PKS enzymatic machinery are (i) β-keto synthase (KS) domain for decarboxylative condensation between an acyl CoA and a malonyl CoA, (ii) an acyl carrier protein (ACP) domain for carrying malonyl co-A loading by phosphopantetheinyl arm and (iii) an acyltransferase (AT) domain for carboxylic acid unit, selection and transfer onto the KS. The presence of other domains like β-ketoacyl reductase (KR), enoyl reductase(ER), dehydratases (DH), and methyltrasferase (MT) and CYC/TE is variable in different Polyketide synthases in different Aspergillus species. During polyketide biosynthesis, the newly formed β-ketothiolester is subjected to a series of chemical reactions like reduction by a β-ketoacyl reductase (KR) to a secondary alcohol; dehydration to form an unsaturated thiolester; methylation, using a methyl group from S-adenosylmethionine (SAM) and final enoyl reduction (ER) to form a fully saturated thiolester. Polyketide structural diversity is determined by iterative activity of MT (CMeT), DH and ER domains during each round of extension. However, how those multi-functional enzymes control the product chain length and the site of iteration is largely unknown. In addition, post-translational modifications of the basic polyketide skeleton, by tailoring enzymes such as methyltransferases, esterases, glycosidases and dimerization with other polyketides, also contribute to the diversity of the polyketide compounds. Polyketide synthases are classified as non reducing (NR) and reducing Polyketide snyhthases based on absence of reducing domains [partially reducing (PR) and highly reducing (HR) PKS] [92]. The type of compounds they produce also depend on the functionality of reducing domains like MT, DH, ER, and TE/CLC (thioesterase/Claisen cyclase) present in the enzyme which are classified as Partially reduced or highly reduced PKS producing highly, reduced polyketide compounds such as 6-methylsalicylic acid and lovastatins. Non reduced PKS enzyme produces compounds without reduction reactions. They lack above mentioned reducing domains and instead contain SAT (starter unit-ACP transacylase) and PT (product template) domains which control the structural outcome of non-reduced polyketides compounds such as Aflatoxin and melanin [93]. This diversity in polyketides produced by different Aspergillus species suggests the possible occurrence of variations in the functionality and also molecular structure of the important enzymes in the polyketide biosynthetic pathways. This is evident in the synthesis of melanin, aflatoxin, and lovastatin by A. fumigatus, A. flavus and A. niger respectively.
Fig. 3.
Polyketide biosynthetic pathway in Aspergillus species
Biochemical Pathways and Enzymes
Melanin—Pigment and Virulent Factor
Polyketide Synthase p (PKS P/alb1) of A. fumigatus is well characterized and known to synthesize bluish to green pigment, melanin, by using dihydroxynapthalene (DHN)—melanin pathway [94]. Melanin biosynthesis involves 6 genes, organized in a cluster on chromosome 2. These genes are (i) pigment biosynthesis protein (Ayg1), (ii) polyketide synthase (Alb1/PksP), (iii) THN reductase (Arp2), (iv) scytalone dehydratase (ArP1p), (v) vermelone dehydratase (Abr1p), and (vi) laccase (Abr2P). Melanin synthesis in Afu starts with condensation of acetyl-CoA and malonyl-CoA, which are converted by the products of the genes PksP/Alb1 and Ayg1 into 1,3,6,8 tetrahydroxynaphthalene (THN) [95]. pksP, non reduced polyketide synthase has domains like SAT-KS-AT-PT-ACP-ACP-TE/CLC and it lacks the reducing domains. Conidia lacking pigmentation due to the defective polyketide synthase pks P gene were less resistant to the attack by monocytes in vitro and such conidia showed reduced virulence in a murine model of aspergillosis [96, 97]. pks P, is transcribed in hyphae of germinating conidia isolated from lungs of infected immunosuppressed mice and is also involved in the inhibition of phagosome-lysosome fusion [98]. In addition to a protective role against the host’s immune defenses, melanin is also a structural component of the conidial wall that is required for arrangement of correct assembly of the cell wall layers and for expression of adhesins and virulence factors at the conidial surface [96]. Recently, it was also shown that A. fumigatus produces an alternative type of melanin, designated as pyomelanin, via tyrosine degradation [99]. A. niger produces a black pigment aspergillin; found to be a mixture of a low-molecular weight green pigment hexahydroxyl pentacyclic quinoid (HPQ) and a brown color low-molecular weight melanin pigment [99]. PKS ortholog of alb1 from A. fumigatus is responsible for production of the naphtho-γ-pyrone precursor for the 1,8-dihydroxynaphthalene (DHN) melanin/spore pigment in A. niger too. In contrast to A. fumigatus, the genes for melanin biosynthesis are not clustered in A. niger. However, albA and aygA are present on chromosome 1, they are located on different chromosomal arms [100]. A. nidulans produces yellow spore pigments, made of naphthopyrones which are reported to be important for scavenging reactive oxygen species for protection of conidia against oxidative damage [101]. In contrast to conidial mutant of A. fumigatus where the absence of conidial pigment results in structural changes on the conidial surface, A. nidulans mutant strain (wA) does not show the same structural changes on the conidial surface. The biosynthetic pathways for the pigment formation in the conidia of the two species differ, i.e., A. fumigatus produces the conidial pigment via the 1,8-dihydroxynaphthalene (DHN)-melanin pathway, which appears to be absent in A. nidulans [98]. The pigment synthesis pathway in A. nidulans is still not yet established. Genome sequencing of A. flavus suggest that it has a homolgous pks P/alb1 gene and also additional genes required for pigment biosynthesis. However, these remain to be characterized with respect to pigment formation.
Many A. fumigatus polyketide synthase genes have significant homology with polyketide synthase gene of Mycobacterium tuberculosis (30–40%). Co- infection of A. fumigatus with Mycobacterium tuberculosis is well known as in the case of fungal balls in the post Kochs cavity of the lungs [102]. Genome sequencing information of Mycobacterium tuberculosis H37Rv shows the presence of 22 polyketide synthases of type I and type III, out of which three of the PKS enzymes are characterized and reported to be involved in the production of mycolic acids, phthiocerol dimycocerosate, major virulent factors of the Mycobacterium tuberculosis [103, 104].
Aflatoxins
Extensively studied biosynthetic gene cluster for synthesis of toxins so far is the gene cluster producing Aflatoxin by A. parasiticus and A. flavus. The genes encoding the aflatoxin biosynthetic pathway enzymes are clustered within a 75 kb region in A. flavus genome [105, 106]. Aflatoxin gene cluster includes pksA, a large gene of 6.6 kb encoding for a polyketide synthase that catalyzes the second step, in which hexanoyl tetrahydroxyl anthrone is converted to norsolorinic acid from the hexanoate [107]. The pksA, a non reducing polyketide synthase gene, has SAT, KS, AT, PT, ACP, and TE/CYC domains. A. nidulans has homologous PKS enzyme (pksA) responsible for synthesis of sterigmatocystin, a known intermediate from the Aflatoxin biosynthesis pathway [108]. Annotation of the A. flavus genome indicates the presence of 55 secondary metabolite biosynthetic pathway clusters and highest number of PKS enzymes than any other sequenced Aspergillus species. Apart from Aflatoxin, cyclopiazonic acid biosynthetic clusters and genes encoding for Aflatrem have been identified from A. flavus [109]. Out of 24 putative PKS enzymes only aflC (pks A) is the only gene experimentally characterized as a part of Aflatoxin Biosynthetic cluster. Remaining genes still need to be explored with respect to secondary metabolite production. Recently, A. flavus secondary metabolite gene clusters were examined for their expression over varied experimental conditions to associate gene clusters with known secondary metabolite functions. These clusters have distinctive gene expression profiles where Aflatoxin and cyclopiazonic (CPA) were found to have unique regulation mechanisms [110]. The fact, that the presence of number of PKS enzymes in Aspergillus species exceeds the number of polyketide compounds produced, necessitates further studies of these enzymes using advanced molecular techniques.
Lovastatin
Aspergillus terreus produces non-aromatic, reduced polyketides such as Lovastatin, an HMG-CoA reductase inhibitor. The gene cluster for Lovastatin biosynthesis (over 64 kb) has 18 potential genes as indicated by bioinformatic studies [111]. In the lovastatin pathway, first stable intermediate Dihydromonacolin L is catalyzed by two polyketide synthases LovB and LovF using two acetyl units and eight malonate molecules [112]. LovB/LNKS (lovastatin nonaketide synthase) catalyzes nonketides to a hexahydronaphthalene ring system. The LovB/LNKS, 335 kDa enzyme is a highly reducing iterative type I PKS with KS, AT, DH, MT (ER), KR, and ACP domains. It has a condensation domain (CON) that is similar to the condensation domain of nonribosomal peptide synthetases (NRPSs), which is possibly involved in product transfer or cyclization. It has an inactive ER domain, because of which a separate ER enzyme termed as LovC, carries out the Enoyl reduction function [113]. The other PKS enzyme LovF/LDKS is a simple diketide enzyme, function of which is not characterized fully. The LovF 277 kDa enzyme is similar to LovB and contains domains for KS, AT, DH, ER, and KR. However, its ER domain is active unlike LovB but does not contain the condensation domain (CON). It may encode the enzyme responsible for the biosynthesis of the (2R)-2-methylbutyryl side chain of lovastatin [112, 113]. LovB and LovF lack TE domain required for off loading of growing ketide chain and it is not fully understood how dihydromonacolin L is detached from the enzyme [114]. Lovastatin polyketide synthase (PKS) systems are complex and are of great interest as their genetic engineering can lead to more effective statins with least side effects. Comparative genomic analysis of the Genomes of various Aspergillus species can lead to mapping of complex gene machinery for important polyketides.
Recent Approaches to Study Polyketide Synthases for Applications
A number of gene clusters for secondary metabolites are observed to remain silent in various Aspergillus species. Lack of studies on the environmental signals that trigger the activation of the gene clusters for the synthesis of polyketides are not well established experimentally. The cryptic gene clusters may code for the biosynthesis of important virulent factors, toxins or even drug candidates. New strategies for their activation are urgently needed to make use of this largely untapped reservoir of the bioactive compounds of Aspergillus species. The discovery of new microbial metabolites through genome mining appears to be a promising approach. LaeA, a global regulator of secondary metabolite has proved very helpful in secondary metabolite search [115]. LaeA based gene-mining, deletion and over expression of LaeA in individual Aspergilli, can be helpful in deciphering the secondary metabolites genes and their clusters [116]. The terrequinone A gene cluster from A. nidulans for asterriquinone biosynthetic pathway, was reported using this approach, combined with microarray analyses [84].
Bioinformatic tools have proved helpful in gaining insight into these megaenzymes which are otherwise difficult to handle experimentally due to their large size and lack of advanced technologies for cloning and expression. Variation of the cumulative GC and window–averaged DNA curvature profile of 26 secondary metabolite gene clusters in A. fumigatus genome showed that these clusters are uniquely expressed in Afu and may not have been horizontally transferred. Forty percent of secondary metabolite gene clusters with this conserved pattern were related to regulation by LaeA, transcription factor [117]. Recently, phylogenetic studies were performed for type I polyketide synthases of Ascomycota group fungi, which divides enzymes into eighteen clades indicating the grouping based on presence/absence of reducing and non-reducing domains in PKS, and their linkage with probable polyketide metabolites produced in these fungi [118]. In view of variations in the polyketide biochemical pathways among medically and agriculturally important Aspergillus species, phylogenetic relationship can be explored for assigning functionality to unexplored PKS-enzymes with respect to secondary metabolites.
Invasive Aspergillosis: Molecular Diagnostics
Early diagnosis in ABPA and detection of pathogen in invasive Aspergillosis will be of great relevance today. This is particularly in view of the increasing incidence of Aspergillus induced Asthma and immunocompromised patients. Often early detection of specific pathogen is hampered due to the lack of specific and sensitive diagnostic methods. In the recent past antifungal drug resistance is often reported. Currently available methods of diagnosis for IA are based on Ag detection and PCR tests. These methods are not satisfactory due to cross reactivity and non specificity. Serological tests based on fungal cell wall components, such as galactomannan or (1,3) β-d-glucan is currently used as a diagnostic tool. However, the incidence of false-positive results of galactomannan-based detection tests is the limitation for this test [119]. PCR methods for Aspergillus detection are based on ribosomal DNA (18S rDNA, 28S rDNA, ITS2 and ITS1 regions) but have limitations of genus specificity as they are conserved across a wide range of fungi. PCR based on mycotoxin or polyketide biosynthetic pathway such as melanin or aflatoxin may be of relevance for specific detection of Aspergilli. Diversity of the polyketide synthase enzymes in Aspergillus species can be explored for detection of important Aspergillus species. Conserved and non conserved regions in the functional domains of polyketide synthase gene can be evaluated for their diagnostic utility. Differential detection of clinically important Aspergillus species, such as A. fumigatus, A. flavus and A. niger in a multiplex PCR based on gene validations will be of value for invasive Aspergillosis patients. Apart from clinical applications, these tests will also be useful in specific and differential detection of A. fumigatus, A. flavus and A. niger for the screening of pre and post harvest agricultural products [120, 121].
Aspergillus Research Areas
Current scenario of advancement in the genomic knowledge of Aspergillus species can lead to
New Antifungal drug targets
Novel polyketides of pharmaceutical importance
Molecular diagnostics for fungal diseases : clinical and agricultural screening
Design of new antifungal compounds for human health and new polyketides for antifungal treatment
Synthesis of statins with least side effects.
Acknowledgements
The facilities and the support provided at Division of Plant Pathology, Indian Agricultural Research Institute by Dr. R.K. Jain, Head of the Department and the Director, IARI are highly acknowledged.
References
- 1.Tillie-Leblond I, Tonnel AB. Allergic bronchopulmonary aspergillosis. Allergy. 2005;60(8):1004–1013. doi: 10.1111/j.1398-9995.2005.00887.x. [DOI] [PubMed] [Google Scholar]
- 2.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive Aspergillosis. Clin Microbiol Rev. 2009;22(3):447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett JW. Aspergillus: a primer for the novice. Med Mycol. 2009;47(Suppl 1):S5–S12. doi: 10.1080/13693780802712515. [DOI] [PubMed] [Google Scholar]
- 4.Karaffa L, Sándor E, Fekete E, Szentirmai A. The biochemistry of citric acid accumulation by Aspergillus niger. Acta Microbiol Immunol Hung. 2001;48(3–4):429–440. doi: 10.1556/AMicr.48.2001.3-4.11. [DOI] [PubMed] [Google Scholar]
- 5.Barrios-González J, Miranda RU. Biotechnological production and applications of statins. Appl Microbiol Biotechnol. 2010;85(4):869–883. doi: 10.1007/s00253-009-2239-6. [DOI] [PubMed] [Google Scholar]
- 6.Cleveland TE, Nierman WC. Genomics of industrial Aspergilli and comparison with toxigenic relatives. Food Addit Contam A. 2008;25(9):1147–1151. doi: 10.1080/02652030802273114. [DOI] [PubMed] [Google Scholar]
- 7.Bhabhra R, Askew DS. Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med Mycol. 2005;43(Suppl 1):S87–S93. doi: 10.1080/13693780400029486. [DOI] [PubMed] [Google Scholar]
- 8.Chang YC, Tsai HF, Karos M, Kwon-Chung KJ. THTA, a thermotolerance gene of Aspergillus fumigatus. Fungal Genet Biol. 2004;41(9):888–896. doi: 10.1016/j.fgb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Hohl TM, Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell. 2007;6(11):1953–1963. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beauvais A, Monod M, Wyniger J, Debeaupuis JP, Grouzmann E, Brakch N, Svab J, Hovanessian AG, Latgé JP. Dipeptidyl-peptidase IV secreted by Aspergillus fumigatus, a fungus pathogenic to humans. Infect Immun. 1997;65(8):3042–3047. doi: 10.1128/iai.65.8.3042-3047.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar J, Nigam S, Saxena S, Madan T, Sarma PU. Identification and assignment of function to the genes of Aspergillus fumigatus expressed at 37°C. J Eukaryot Microbiol. 2004;51(4):428–432. doi: 10.1111/j.1550-7408.2004.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 12.Shankar J, Madan T, Basir SF, Sarma PU. Identification and characterization of polyubiquitin gene from cDNA library of Aspergillus fumigatus. Indian J Clin Biochem. 2005;20(1):208–212. doi: 10.1007/BF02893072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautam P, Madan T, Gade WN, Sarma PU. Immunoproteomic analysis of secretory proteins of Aspergillus fumigatus with specific Ig E immunoreactivity. Indian J Clin Biochem. 2006;21(2):12–19. doi: 10.1007/BF02912905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Upadhyay SK, Mahajan L, Ramjee S, Singh Y, Basir SF, Madan T. Expressed sequence tags of Aspergillus fumigatus: extension of catalogue and their evaluation as putative drug targets and/or diagnostic markers. Indian J Clin Biochem. 2009;24(2):131–136. doi: 10.1007/s12291-009-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarma PU, Madan T, Priyadarsini P, Haq W, Katti SB. Polypetides useful for diagnosis of Aspergillus fumigatus and a process for preparing for the same. US patent 6262231.
- 16.Sarma PU, Madan T, Priyadarsini P, Haq W, Katti SB. A process for the preparation of a novel synthetic peptide epitope useful for diagnosis of aspergillosis. Indian patent 184440.
- 17.Sarma PU, Madan T, Saxena S. Method of detection of SP-A2 gene variants useful for prediction of predisposition to aspergillosis. US patent 7288376.
- 18.http://www.allergen.org/search.php?allergensource=Aspergillus+fumigatus.
- 19.Gautam P, Sundaram CS, Madan T, Gade WN, Shah A, Sirdeshmukh R, Sarma PU. Identification of novel allergens of Aspergillus fumigatus using immunoproteomics approach. Clin Exp Allergy. 2007;37(8):1239–1249. doi: 10.1111/j.1365-2222.2007.02765.x. [DOI] [PubMed] [Google Scholar]
- 20.Sarma PV, Purkayastha S, Madan T, Sarma PU. Expression of an epitopic region of AspfI, an allergen/antigen/cytotoxin of Aspergillus fumigatus. Immunol Lett. 1999;70(3):151–155. doi: 10.1016/s0165-2478(99)00140-6. [DOI] [PubMed] [Google Scholar]
- 21.Madan T, Arora N, Sarma PU. Ribonuclease activity dependent cytotoxicity of Asp fl, a major allergen of A. fumigatus. Mol Cell Biochem. 1997;175(1–2):21–27. doi: 10.1023/a:1006822906343. [DOI] [PubMed] [Google Scholar]
- 22.Saxena S, Madan T, Muralidhar K, Sarma PU. cDNA cloning, expression and characterization of an allergenic L3 ribosomal protein of Aspergillus fumigatus. Clin Exp Immunol. 2003;134(1):86–91. doi: 10.1046/j.1365-2249.2003.02257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nigam S, Sarma PV, Ghosh PC, Sarma PU. Characterization of Aspergillus fumigatus protein disulfide isomerase family gene. Gene. 2001;281(1–2):143–150. doi: 10.1016/s0378-1119(01)00794-6. [DOI] [PubMed] [Google Scholar]
- 24.Madan T, Banerjee B, Bhatnagar PK, Shah A, Sarma PU. Identification of 45 kD antigen in immune complexes of patients of allergic bronchopulmonary aspergillosis. Mol Cell Biochem. 1997;166(1–2):111–116. doi: 10.1023/a:1006827126958. [DOI] [PubMed] [Google Scholar]
- 25.Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153(Pt 6):1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 26.St Leger RJ, Screen SE, Shams-Pirzadeh B. Lack of host specialization in Aspergillus flavus. Appl Environ Microbiol. 2000;66(1):320–324. doi: 10.1128/aem.66.1.320-324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellon JE, Cotty PJ, Dowd MK. Aspergillus flavus hydrolases: their roles in pathogenesis and substrate utilization. Appl Microbiol Biotechnol. 2007;77(3):497–504. doi: 10.1007/s00253-007-1201-8. [DOI] [PubMed] [Google Scholar]
- 28.Madan T, Kishore U, Shah A, Eggleton P, Strong P, Wang JY, Aggrawal SS, Sarma PU, Reid KB. Lung surfactant proteins A and D can inhibit specific IgE binding to the allergens of Aspergillus fumigatus and block allergen-induced histamine release from human basophils. Clin Exp Immunol. 1997;110(2):241–249. doi: 10.1111/j.1365-2249.1997.tb08323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madan T, Eggleton P, Kishore U, Strong P, Aggrawal SS, Sarma PU, Reid KB. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect Immun. 1997;65(8):3171–3179. doi: 10.1128/iai.65.8.3171-3179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madan T, Kishore U, Singh M, Strong P, Clark H, Hussain EM, Reid KB, Sarma PU. Surfactant proteins A and D protect mice against pulmonary hypersensitivity induced by Aspergillus fumigatus antigens and allergens. J Clin Invest. 2001;107(4):467–475. doi: 10.1172/JCI10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madan T, Kishore U, Singh M, Strong P, Clark H, Hussain EM, Reid KB, Sarma PU.Surfactant proteins A and D protect mice against pulmonary hypersensitivity induced by Aspergillus fumigatus antigens and allergens J Immunol 2005174116943–6954.15905537 [Google Scholar]
- 32.Madan T, Reid KB, Clark H, Singh M, Nayak A, Sarma PU, Hawgood S, Kishore U. Susceptibility of mice genetically deficient in SP-A or SP-D gene to invasive pulmonary aspergillosis. Mol Immunol. 2010;47(10):1923–1930. doi: 10.1016/j.molimm.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 33.Kaur S, Gupta VK, Thiel S, Sarma PU, Madan T. Protective role of mannan-binding lectin in a murine model of invasive pulmonary aspergillosis. Clin Exp Immunol. 2007;148(2):382–389. doi: 10.1111/j.1365-2249.2007.03351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madan T, Kaur S, Saxena S, Singh M, Kishore U, Thiel S, Reid KB, Sarma PU. Role of collectins in innate immunity against aspergillosis. Med Mycol. 2005;43(Suppl 1):S155–S163. doi: 10.1080/13693780500088408. [DOI] [PubMed] [Google Scholar]
- 35.Madan T, Kaur S, Saxena S, Singh M, Kishore U, Thiel S, Reid KB, Sarma PU. Role of collectins in innate immunity against aspergillosis. Immunobiology. 2002;205(4–5):610–618. doi: 10.1080/13693780500088408. [DOI] [PubMed] [Google Scholar]
- 36.Singh M, Madan T, Waters P, Sonar S, Singh SK, Kamran MF, Bernal AL, Sarma PU, Singh VK, Crouch EC, Kishore U. Therapeutic effects of recombinant forms of full-length and truncated human surfactant protein D in a murine model of invasive pulmonary aspergillosis. Mol Immunol. 2009;46(11–12):2363–2369. doi: 10.1016/j.molimm.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Mahajan L, Madan T, Kamal N, Singh VK, Sim RB, Telang SD, Ramchand CN, Waters P, Kishore U, Sarma PU. Recombinant surfactant protein-D selectively increase apoptosis in eosinophils of allergic asthmatics and enhances uptake of apoptotic eosinophils by macrophages. Int Immunol. 2008;20(8):993–1007. doi: 10.1093/intimm/dxn058. [DOI] [PubMed] [Google Scholar]
- 38.Vaid M, Kaur S, Sambatakou H, Madan T, Denning DW, Sarma PU. Distinct alleles of mannose-binding lectin (MBL) and surfactant proteins A (SP-A) in patients with chronic cavitary pulmonary aspergillosis and allergic bronchopulmonary aspergillosis. Clin Chem Lab Med. 2007;45(2):183–186. doi: 10.1515/CCLM.2007.033. [DOI] [PubMed] [Google Scholar]
- 39.Upadhyay SK, Mahajan L, Ramjee S, Singh Y, Basir SF, Madan T. Identification and characterization of a laminin-binding protein of Aspergillus fumigatus: extracellular thaumatin domain protein (AfCalAp) J Med Microbiol. 2009;58(Pt 6):714–722. doi: 10.1099/jmm.0.005991-0. [DOI] [PubMed] [Google Scholar]
- 40.Abad A, Victoria Fernández-Molina J, Bikandi J, Ramírez A, Margareto J, Sendino J, Luis Hernando F, Pontón J, Garaizar J, Rementeria A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol. 2010;27(4):155–182. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Rementeria A, López-Molina N, Ludwig A, Vivanco AB, Bikandi J, Pontón J, Garaizar J. Genes and molecules involved in Aspergillus fumigatus virulence. Rev Iberoam Micol. 2005;22(1):1–23. doi: 10.1016/s1130-1406(05)70001-2. [DOI] [PubMed] [Google Scholar]
- 42.Pihet M, Vandeputte P, Tronchin G, Renier G, Saulnier P, Georgeault S, Mallet R, Chabasse D, Symoens F, Bouchara JP. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 2009;9:177. doi: 10.1186/1471-2180-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu JH, Mah JH, Seo JA. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot Cell. 2006;5(10):1577–1584. doi: 10.1128/EC.00193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, Jiang B, Roemer T. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 2007;3(3):e24. doi: 10.1371/journal.ppat.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beauvais A, Latgé JP. Membrane and cell wall targets in Aspergillus fumigatus. Drug Resist Updat. 2001;4(1):38–49. doi: 10.1054/drup.2001.0185. [DOI] [PubMed] [Google Scholar]
- 46.Chamilos G, Kontoyiannis DP. Update on antifungal drug resistance mechanisms of Aspergillus fumigatus. Drug Resist Updat. 2005;8(6):344–358. doi: 10.1016/j.drup.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Latge JP, Mouyna I, Tekaia F, Beauvais A, Debeaupuis JP, Nierman W. Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med Mycol. 2005;43:S15–S22. doi: 10.1080/13693780400029155. [DOI] [PubMed] [Google Scholar]
- 48.Ibrahim-Granet O, Dubourdeau M, Latgé JP, Ave P, Huerre M, Brakhage AA, Brock M. Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cell Microbiol. 2008;10(1):134–148. doi: 10.1111/j.1462-5822.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- 49.Maerker C, Rohde M, Brakhage AA, Brock M. Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J. 2005;272(14):3615–3630. doi: 10.1111/j.1742-4658.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 50.Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412(6842):83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 51.Olivas I, Royuela M, Romero B, Monteiro MC, Mínguez JM, Laborda F, Lucas JR. Ability to grow on lipids accounts for the fully virulent phenotype in neutropenic mice of Aspergillus fumigatus null mutants in the key glyoxylate cycle enzymes. Fungal Genet Biol. 2008;45(1):45–60. doi: 10.1016/j.fgb.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Schöbel F, Ibrahim-Granet O, Avé P, Latgé JP, Brakhage AA, Brock M. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect Immun. 2007;75(3):1237–1244. doi: 10.1128/IAI.01416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thevelein JM. Regulation of trehalose mobilization in fungi. Microbiol Rev. 1984;48:42–59. doi: 10.1128/mr.48.1.42-59.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 55.Al-Bader N, Vanier G, Liu H, Gravelat FN, Urb M, Hoareau CM, Campoli P, Chabot J, Filler SG, Sheppard DC. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect Immun. 2010;78(7):3007–3018. doi: 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puttikamonkul S, Willger SD, Grahl N, Perfect JR, Movahed N, Bothner B, Park S, Paderu P, Perlin DS, Cramer RA Jr. Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol Microbiol. 2010 Jun 9. [DOI] [PMC free article] [PubMed]
- 57.Schöbel F, Jacobsen ID, Brock M. Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryot Cell. 2010;9(6):878–893. doi: 10.1128/EC.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abadio AK, Kioshima ES, Teixeira MM, Martins NF, Maigret B, Felipe MS. Comparative genomics allowed the identification of drug targets against human fungal pathogens. BMC Genomics. 2011;12(1):75. doi: 10.1186/1471-2164-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goffeau A. Genomics: multiple moulds. Nature. 2005;438(7071):1092–1093. doi: 10.1038/4381092b. [DOI] [PubMed] [Google Scholar]
- 60.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 61.Rokas A, Payne G, Fedorova ND, Baker SE, Machida M, Yu J, Georgianna DR, Dean RA, Bhatnagar D, Cleveland TE, Wortman JR, Maiti R, Joardar V, Amedeo P, Denning DW, Nierman WC. What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud Mycol. 2007;59:11–17. doi: 10.3114/sim.2007.59.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scazzocchio C. Aspergillus genomes: secret sex and the secrets of sex. Trends Genet. 2006;22(10):521–525. doi: 10.1016/j.tig.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Brakhage AA, Schroeckh V. Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48(1):15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47(9):736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 2005;248(2):241–248. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 66.Coyle CM, Kenaley SC, Rittenour WR, Panaccione DG. Association of ergot alkaloids with conidiation in Aspergillus fumigatus. Mycologia. 2007;99(6):804–811. doi: 10.3852/mycologia.99.6.804. [DOI] [PubMed] [Google Scholar]
- 67.Maiya S, Grundmann A, Li SM, Turner G. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. Chembiochem. 2006;7(7):1062–1069. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 68.Hissen AH, Wan AN, Warwas ML, Pinto LJ, Moore MM. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding l-ornithine N5-oxygenase, is required for virulence. Infect Immun. 2005;73(9):5493–5503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewis RE, Wiederhold NP, Chi J, Han XY, Komanduri KV, Kontoyiannis DP, Prince RA. Detection of gliotoxin in experimental and human aspergillosis. Infect Immun. 2005;73(1):635–637. doi: 10.1128/IAI.73.1.635-637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frisvad JC, Rank C, Nielsen KF, Larsen TO. Metabolomics of Aspergillus fumigatus. Med Mycol. 2009;47(s1):S53–S71. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- 71.Pel HJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25(2):221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 72.Vala AK, Dave BP, Dube HC. Chemical characterization and quantification of siderophores produced by marine and terrestrial aspergilli. Can J Microbiol. 2006;52(6):603–607. doi: 10.1139/w06-012. [DOI] [PubMed] [Google Scholar]
- 73.Mogensen JM, Nielsen KF, Samson RA, Frisvad JC, Thrane U. Effect of temperature and water activity on the production of fumonisins by Aspergillus niger and different Fusarium species. BMC Microbiol. 2009;9:281. doi: 10.1186/1471-2180-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang PK, Horn BW, Dorner JW. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet Biol. 2009;46(2):176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 75.Juvvadi PR, Seshime Y, Kitamoto K. Genomics reveals traces of fungal phenylpropanoid-flavonoid metabolic pathway in the filamentous fungus Aspergillus oryzae. J Microbiol. 2005;43(6):475–486. [PubMed] [Google Scholar]
- 76.Kiyota T, Hamada R, Sakamoto K, Iwashita K, Yamada O, Mikami S. Aflatoxin non-productivity of Aspergillus oryzae caused by loss of function in the aflJ gene product. J Biosci Bioeng. 2011;111(5):512–517. [DOI] [PubMed]
- 77.Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66(3):447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiang YM, Chang SL, Oakley BR, Wang CC. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. 2011;15(1):137–143. doi: 10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3(2):527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Keller NP. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell. 2005;4(9):1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kale SP, Milde L, Trapp MK, Frisvad JC, Keller NP, Bok JW. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet Biol. 2008;45(10):1422–1429. doi: 10.1016/j.fgb.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bentley R, Bennet JW. Construction polyketides: from collie to combinatorial biosynthesis. Annu Rev Microbiol. 1999;53:411–446. doi: 10.1146/annurev.micro.53.1.411. [DOI] [PubMed] [Google Scholar]
- 83.Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131(8):2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76(6):1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.http://www.knowmycotoxins.com/regulations.htm.
- 86.Zhang W, Tang Y. In vitro analysis of type II polyketide synthase. Methods Enzymol. 2009;459:367–393. doi: 10.1016/S0076-6879(09)04616-3. [DOI] [PubMed] [Google Scholar]
- 87.Abe I, Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27(6):809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- 88.Manzoni M, Rollini M. Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Appl Microbiol Biotechnol. 2002;58(5):555–564. doi: 10.1007/s00253-002-0932-9. [DOI] [PubMed] [Google Scholar]
- 89.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14(8):931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seshime Y, Juvvadi PR, Fuji I, Kitamoto K. Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae. Biochem Biophys Res Commun. 2005;331(1):253–60. [DOI] [PubMed]
- 91.Cleveland TE, Yu J, Fedorova N, Bhatnagar D, Payne GA, Nierman WC, Bennett JW. Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 2009;27(3):151–157. doi: 10.1016/j.tibtech.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 92.Chiang YM, Oakley BR, Keller NP, Wang CC. Unraveling polyketide synthesis in members of the genus Aspergillus. Appl Microbiol Biotechnol. 2010;86(6):1719–1736. doi: 10.1007/s00253-010-2525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage AA. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med Microbiol Immunol. 1998;187:79–89. doi: 10.1007/s004300050077. [DOI] [PubMed] [Google Scholar]
- 95.Watanabe A, Fujii I, Tsai HF, Chang YC, Kwon-Chung KJ, Ebizuka Y. Aspergillus fumigatus alb1 encodes naphthopyrone synthase when expressed in Aspergillus oryzae. FEMS Microbiol Lett. 2000;192:39–44. doi: 10.1111/j.1574-6968.2000.tb09356.x. [DOI] [PubMed] [Google Scholar]
- 96.Jahn B, Langfelder K, Schneider U, Schindel C, Brakhage AA. PKSP-dependent reduction of phagolysosome fusion and intracellular kill of Aspergillus fumigatus conidia by human monocyte-derived macrophages. Cell Microbiol. 2002;4:793–803. doi: 10.1046/j.1462-5822.2002.00228.x. [DOI] [PubMed] [Google Scholar]
- 97.Langfelder K, Philippe B, Jahn B, Latgé JP, Brakhage AA. Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect Immun. 2001;69:6411–6418. doi: 10.1128/IAI.69.10.6411-6418.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pihet M, Vandeputte P, Tronchin G, Renier G, Saulnier P, Georgeault S, Mallet R, Chabasse D, Symoens F, Bouchara JP. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 2009;9:177. doi: 10.1186/1471-2180-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, Brakhage AA. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2009;75:493–503. doi: 10.1128/AEM.02077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jørgensen TR, Park J, Arentshorst M, Welzen AM, Lamers G, Vankuyk PA, Damveld RA, Hondel CA, Nielsen KF, Frisvad JC, Ram AF. The molecular and genetic basis of conidial pigmentation in Aspergillus niger. Fungal Genet Biol. 2011;48(5):544–553. doi: 10.1016/j.fgb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 101.Jahn B, Boukhallouk F, Lotz J, Langfelder K, Wanner G, Brakhage AA. Interaction of human phagocytes with pigmentless Aspergillus conidia. Infect Immun. 2000;68(6):3736–3739. doi: 10.1128/iai.68.6.3736-3739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Agarwal R, Singh N, Aggarwal AN. An unusual association between Mycobacterium tuberculosis and Aspergillus fumigatus. Monaldi Arch Chest Dis. 2008;69(1):32–34. doi: 10.4081/monaldi.2008.409. [DOI] [PubMed] [Google Scholar]
- 103.Sankaranarayanan R, Saxena P, Marathe UB, Gokhale RS, Shanmugam VM, Rukmini R. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat Struct Mol Biol. 2004;11(9):894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 104.Chopra T, Gokhale RS. Polyketide versatility in the biosynthesis of complex mycobacterial cell wall lipids. Methods Enzymol. 2009;459:259–294. doi: 10.1016/S0076-6879(09)04612-6. [DOI] [PubMed] [Google Scholar]
- 105.Payne GA, Brown MP. Genetics and physiology of aflatoxin biosynthesis. Annu Rev Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- 106.Yu JJ, Chang PK, Ehrlich KC, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yabe K, Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2004;64(6):745–755. doi: 10.1007/s00253-004-1566-x. [DOI] [PubMed] [Google Scholar]
- 108.Yu JH, Leonard TJ. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol. 1995;177(16):4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nicholson MJ, Koulman A, Monahan BJ, Pritchard BL, Payne GA, Scott B. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl Environ Microbiol. 2009;75(23):7469–7481. doi: 10.1128/AEM.02146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, Woloshuk CP, Yu J, Keller NP, Payne GA. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol Plant Pathol. 2010;11(2):213–226. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326(5952):589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hutchinson CR, Kennedy J, Park C, Kendrew S, Auclair K, Vederas J. Aspects of the biosynthesis of non-aromatic fungal polyketides by iterative polyketide synthases. Antonie Van Leeuwenhoek. 2000;78(3–4):287–295. doi: 10.1023/a:1010294330190. [DOI] [PubMed] [Google Scholar]
- 113.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284(5418):1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 114.Ma SM, Tang Y. Biochemical characterization of the minimal polyketide synthase domains in the lovastatin nonaketide synthase LovB. FEBS J. 2007;274(11):2854–2864. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 115.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 116.Panagiotou G, Andersen MR, Grotkjaer T, Regueira TB, Nielsen J, Olsson L. Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Appl Environ Microbiol. 2009;75(7):2212–2220. doi: 10.1128/AEM.01461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Do JH, Miyano S. The GC and window-averaged DNA curvature profile of secondary metabolite gene cluster in Aspergillus fumigatus genome. Appl Microbiol Biotechnol. 2008;80(5):841–847. doi: 10.1007/s00253-008-1638-4. [DOI] [PubMed] [Google Scholar]
- 118.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA. 2003;100(26):15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Racil Z, Kocmanova I, Lengerova M, Weinbergerova B, Buresova L, Toskova M, Winterova J, Timilsina S, Rodriguez I, Mayer J. Difficulties in using 1, 3-{beta}-d-glucan as the screening test for the early diagnosis of invasive fungal infections in patients with haematological malignancies–high frequency of false-positive results and their analysis. J Med Microbiol. 2010;59(Pt 9):1016–1022. doi: 10.1099/jmm.0.019299-0. [DOI] [PubMed] [Google Scholar]
- 120.Atoui A, Mathieu F, Lebrihi A. Targeting a polyketide synthase gene for Aspergillus carbonarius quantification and ochratoxin A assessment in grapes using real-time PCR. Int J Food Microbiol. 2007;115(3):313–318. doi: 10.1016/j.ijfoodmicro.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 121.Usha Sarma P. Fascinating Potential of Aspergilli. Indian J Clin Biochem. 2010;25(4):331–334. doi: 10.1007/s12291-010-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]