Abstract
Ensuring quality of laboratory services is the need of the hour in the field of health care. Keeping in mind the revolution ushered by six sigma concept in corporate world, health care sector may reap the benefits of the same. Six sigma provides a general methodology to describe performance on sigma scale. We aimed to gauge our laboratory performance by sigma metrics. Internal quality control (QC) data was analyzed retrospectively over a period of 6 months from July 2009 to December 2009. Laboratory mean, standard deviation and coefficient of variation were calculated for all the parameters. Sigma was calculated for both the levels of internal QC. Satisfactory sigma values (>6) were elicited for creatinine, triglycerides, SGOT, CPK-Total and Amylase. Blood urea performed poorly on the sigma scale with sigma <3. The findings of our exercise emphasize the need for detailed evaluation and adoption of ameliorative measures in order to effectuate six sigma standards for all the analytical processes.
Keywords: Six sigma, Coefficient of variation, Total allowable error, Bias, Quality control
Introduction
Quality planning defines quality standards which are the foundation for quality laboratory processes, quality control (QC), quality assessment (QA), and quality improvement. Quality control validation is used to determine the statistical QC procedures appropriate for distinguishing variations critical for clinical interpretation of the test [1].
Quality requirement differs significantly between various analytes. Serum electrolyte levels are stringently regulated and make even a small change to be clinically significant.
Whereas cholesterol levels show a greater variability; therefore much larger rise is obligatory to cause a clinically considerable change that necessitates further investigation or treatment.
Sigma (σ) is the mathematical symbol for standard deviation (SD) [2]. Six sigma is a process quality measurement and improvement program developed by Motorola in the early 1980s. Sigma methodology can be applied wherever an outcome of a process is be measured. A poor outcome is counted as an error or defect. This is quantified as defects per million (DPM). Six sigma provides a more quantitative frame work for evaluating process performance with evidence for process improvement and describes how many sigma fit within the tolerance limits [3]. Quality is assessed on the σ scale with a criterion of 3 σ as the minimum allowable sigma for routine performance and a sigma of 6 being the goal for world-class quality [4]. The present study was undertaken to evaluate the quality of the analytical performance of clinical chemistry laboratory of a tertiary care hospital based in Delhi, India on sigma scale.
Materials and Methods
We aim to present the sigma metrics observed in our clinical chemistry laboratory in GB Pant hospital during a period of 6 months. Our clinical biochemistry laboratory caters to a 600-bedded tertiary hospital. Internal statistical QC data was extricated from the Olympus biochemistry analyzer [AU 400, Tokyo, Japan] data for the period of 6 months from July 2009 to December 2009. Control materials were obtained from Randox, Antrim, UK. Both normal (L2) and pathological (L3) levels of QC materials were assayed before commencing reporting of patient samples every day. Next QC scheduled event was undertaken after running 50 patient samples (bracketed QC).
Various parameters scrutinized were glucose, urea, Creatinine, cholesterol, triglycerides, High Density Lipoprotein (HDL), total bilirubin, alkaline Phosphatase (ALP), alanine amino transferase (ALT), aspartate amino transferase (AST), total protein, creatine kinase total (CKT), amylase, sodium and potassium.
Validation of quality control of our lab was done by calculating 6 months mean from the data of internal QC and External Quality Assurance Scheme (EQAS) to establish the CV and bias respectively, for each analyte.
The sigma metrics for the various analytes was calculated by the following equation
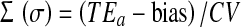 |
[TEa—total allowable error, CV—coefficient of variation]
TEa values of various parameters were taken from the Clinical Laboratories Improvement Act (CLIA) guidelines [5].
Bias was computed from the external Quality assurance records using the following formula:
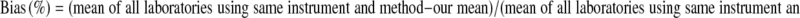 |
CV was determined from the calculated laboratory mean and calculated standard deviation procured from the internal QC data over the last 6 months:
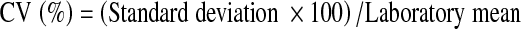 |
Results
Table 1 illustrates the designated mean, laboratory mean and the calculated standard deviation values of the two levels namely normal (L2) and pathological (L3) quality controls run in our laboratory for the different parameters. The laboratory mean and designated target mean were similar (<2% variation) for both the levels of internal QC in the case of urea, triglycerides, SGOT, SGPT, ALP and total bilirubin. Cholesterol, HDL, sugar and sodium showed negligible variation between the lab mean and the designated value for normal internal quality control (IQC); whereas in the case of the pathological level, lab mean for cholesterol, sugar and sodium was approximately 3% higher than the designated value and HDL was found to be 8.6% lower than the assigned target mean. The estimated laboratory mean of L3 in case of total protein and CK-total was similar to the allocated target value, however L2 was 3.7% higher in case of protein and 8% lower in case of CK total. For creatinine, amylase and potassium, the computed mean values for both the levels of IQC were higher than the stipulated target means (6.6, 10; 2.8, 3.9 and 3, 2% respectively).
Table 1.
Comparison of designated and calculated laboratory mean for different parameters for both levels of quality control
| Parameter | L2 | L3 | ||||
|---|---|---|---|---|---|---|
| Designated mean | Lab mean | SD | Designated mean | Lab mean | SD | |
| Sugar (mg/dl) | 110 ± 8.35 | 110.36 | 1.87 | 288 ± 21 | 295.02 | 4.06 |
| Urea (mg/dl) | 45 ± 3.4 | 46.28 | 1.43 | 124 ± 9.1 | 120.12 | 4.33 |
| Creatinine (mg/dl) | 1.5 ± 0.14 | 1.61 | 0.14 | 3.8 ± 0.36 | 4.22 | 0.08 |
| Cholesterol (mg/dl) | 157 ± 11.5 | 152.4 | 3.38 | 327 ± 24.5 | 317.3 | 8.42 |
| Triglycerides (mg/dl) | 99 ± 8.15 | 97.9 | 2.5 | 265 ± 21.5 | 261.4 | 8.53 |
| HDL (mg/dl) | 30.6 ± 5.2 | 28.9 | 1 | 58 ± 6.4 | 52.7 | 3.93 |
| SGOT (IU/l) | 35 ± 3.5 | 34.66 | 0.93 | 151 ± 15.5 | 156.2 | 3.49 |
| SGPT (IU/l) | 43 ± 4.5 | 41.2 | 1.77 | 147 ± 14.5 | 151.7 | 4.74 |
| ALP (IU/l) | 220 ± 16.5 | 220 | 15.5 | 459 ± 34.5 | 459 | 30 |
| T bilirubin (mg/dl) | 1.8 ± 0.2 | 1.76 | 0.06 | 5.12 ± 0.53 | 5.05 | 0.18 |
| T protein (g/dl) | 5.91 ± 0.27 | 6.13 | 0.116 | 4.7 ± 0.46 | 4.75 | 0.098 |
| CPK T (IU/l) | 235 ± 21 | 216 | 6.53 | 548 ± 49.5 | 549.5 | 15.7 |
| Amylase (IU/l) | 81 ± 6 | 83.3 | 1.86 | 330 ± 24.5 | 343.7 | 7.2 |
| Sodium (mEq/l) | 144 ± 4.5 | 144 | 1.77 | 160 ± 5 | 165 | 2.17 |
| Potassium (mEq/l) | 3.98 ± 0.17 | 4.1 | 0.06 | 6.08 ± 0.25 | 6.24 | 0.1 |
Bias was calculated from data of external quality assurance program provided by Randox (RIQAS) for the months of July to December 2009 for the different parameters and the average calculated. This is tabulated in Table 2. Average bias was <3 for 5 chemistries (cholesterol, triglycerides, protein, sodium, potassium), 3.1–6.0 for 6 chemistries (sugar, urea, SGOT, SGPT, CK-total, amylase) and >6 for 4 chemistries (creatinine, HDL, ALP bilirubin).
Table 2.
Percentage bias calculated from RIQAS results for a period of 6 months
| Parameter | Month 1 | Month 2 | Month 3 | Month 4 | Month 5 | Month 6 | Average |
|---|---|---|---|---|---|---|---|
| Sugar | 2.36 | 2.7 | 4.2 | 2.8 | 0 | 9.4 | 3.5 |
| Urea | 0.6 | 2.85 | 6.7 | 5.5 | 0 | 4 | 4.2 |
| Creatinine | 6.5 | 2.0 | 11 | 3.2 | 0.6 | 7.1 | 6.3 |
| Cholesterol | 2.7 | 3.5 | 2.1 | 3.2 | 3 | 1.1 | 2.6 |
| Triglycerides | 1.96 | 4.3 | 4.4 | 2.1 | 2.1 | 2.5 | 2.9 |
| HDL | 6.8 | 12 | 8.1 | 16 | 1.9 | 5.2 | 8.3 |
| SGOT | 0.06 | 1.7 | 9 | 2.8 | 2.9 | 10.5 | 4.5 |
| SGPT | 2.8 | 3.5 | 6.1 | 2.4 | 2.7 | 2.9 | 3.4 |
| ALP | 10.2 | 4.3 | 3.4 | 7.9 | 5.5 | 15 | 7.7 |
| T bilirubin | 10 | 4.08 | 16 | 5.8 | 0.6 | 11 | 7.9 |
| T protein | 0.2 | 0.14 | 2.41 | 6.25 | 1.7 | 4.9 | 2.6 |
| CPK T | 0.82 | 3.4 | 8.4 | 0.53 | 8.4 | 3.5 | 4.75 |
| Amylase | 3.6 | 4.27 | 1.05 | 1 | 6 | 5.3 | 5.4 |
| Sodium | 1.65 | 0.3 | 1.91 | 1.3 | 2.1 | 4.2 | 1.91 |
| Potassium | 2.0 | 0.6 | 1.82 | 1.8 | 2.4 | 2.3 | 1.82 |
Table 3 highlights TEa, average bias, coefficient of variation (CV) and sigma values of the two levels of quality control for the different parameters. TEa is less for the sodium and potassium signifying the criticality of these analytes and the need for stringent quality control conformity. Amylase, ALP, CKT and HDL have been assigned a higher TEa of 30% as mentioned in CLIA guidelines. The coefficient of variation (CV) varied from 0.86 (creatinine) to 3.08 (urea) for quality control L2 and 1.3 (sodium) to 7.45 (HDL) for quality control L3.
Table 3.
Table showing the average calculated bias %, TEa %, CV and sigma values for a period of 6 months
| Parameter | TEa (%) | Average bias | L2 | L3 | ||
|---|---|---|---|---|---|---|
| CV | σ | CV | σ | |||
| Sugar | 10 | 3.5 | 1.7 | 3.8 | 1.37 | 4.7 |
| Urea | 9 | 4.2 | 3.08 | 1.5 | 3.6 | 1.3 |
| Creatinine | 15 | 6.3 | 0.869 | 10.0 | 1.89 | 4.6 |
| Cholesterol | 10 | 2.6 | 2.22 | 3.3 | 2.65 | 2.79 |
| Triglycerides | 25 | 2.9 | 2.55 | 8.6 | 3.25 | 6.8 |
| HDL | 30 | 8.3 | 3.4 | 6.3 | 7.45 | 2.9 |
| SGOT | 20 | 4.5 | 2.6 | 5.9 | 2.2 | 7.0 |
| SGPT | 20 | 3.4 | 4.2 | 3.9 | 3.12 | 5.3 |
| ALP | 30 | 7.7 | 7 | 3.2 | 6.5 | 3.4 |
| T bilirubin | 20 | 7.9 | 3.4 | 3.5 | 3.5 | 3.4 |
| T protein | 10 | 2.6 | 1.89 | 3.9 | 2.06 | 3.5 |
| CPK T | 30 | 4.75 | 3.02 | 8.3 | 2.8 | 9.0 |
| Amylase | 30 | 5.4 | 2.2 | 11.2 | 2.09 | 11.7 |
| Sodium | 5 | 1.91 | 1.23 | 2.5 | 1.3 | 2.3 |
| Potassium | 6 | 1.82 | 1.46 | 2.8 | 1.6 | 2.6 |
The sigma value >6 was observed for triglycerides, CPK-Total and amylase for both the levels of QC. Creatinine and HDL depicted a sigma value of >6 for the normal levels of quality control and 4.6 and 2.9 for the L3 of quality control respectively. The sigma values for L2 and L3 were 5.9 and 7 respectively for SGOT. We have achieved sigma metrics of the range 3.1–5.9 for 5 parameters namely sugar, SGPT, ALP, bilirubin and total protein. Cholesterol showed a sigma value of 3.3 for L2 and <3 (2.79) for L3. A sigma value of <3 for both the levels of QC was observed for urea, sodium and potassium (Table 3).
Table 4 depicts RIQAS results for one of the 6 months of the study period. Total Score (TS) allows the laboratories to assess their performance. TS relates the percentage difference between the lab results and the mean for comparison to a target CV. The target CV (TCV) is the precision target of any analyte which is calculated from state of art historical performance and are reviewed annually.
Table 4.
RIQAS results of 1 month to demonstrate lab mean, average mean and bias
| Parameter | Our result | Mean for comparison | Total score | Bias |
|---|---|---|---|---|
| Sugar | 101 | 104 | 83 | 2.8 |
| Urea | 38 | 36 | 89 | 5.5 |
| Creatinine | 1.2 | 1.24 | 100 | 3.2 |
| Cholesterol | 158 | 153 | 86 | 3.2 |
| Triglycerides | 94 | 92 | 120 | 2.1 |
| HDL | 57 | 50 | 65 | 16 |
| SGOT | 36 | 35 | 120 | 2.8 |
| SGPT | 45 | 44 | 120 | 2.4 |
| ALP | 190 | 176 | 85 | 7.9 |
| T bilirubin | 1.8 | 1.7 | 94 | 5.8 |
| T protein | 5.1 | 4.8 | 69 | 6.25 |
| CPK T | 187 | 188 | 120 | 0.53 |
| Amylase | 100 | 99 | 120 | 1 |
| Sodium | 137 | 135 | 96 | 1.3 |
| Potassium | 3.7 | 3.77 | 87 | 1.8 |
Discussion
Attainment of six sigma is envisaged as the gold standard for defining world class measure of quality. Six sigma concentrates on regulating a process to 6 SDs, which represents 3.4 DPM opportunities [6]. Functioning at the 3-sigma level is regarded as the minimum acceptable level of quality. The six sigma idea asserts an association between the numbers of product defects, wasted operating costs and levels of customer satisfaction. It can be inferred that as sigma increases, the consistency and steadiness of the test improves, thereby reducing the operating costs.
Laboratory performance can be appraised with the application of six sigma in laboratory functions [7]. When the method sigma is ≥6, stringent internal QC rules need not be adopted. In such cases, false rejections can be minimized by relaxing control limits up to 3 s. A method sigma below 3 calls for the adoption of a newer and better method as quality of the test cannot be assured even after repeated QC runs [8].
Employing six sigma in laboratory involves quantifying the performance of the test using standard QC methods; specifying the quality requirements for the test (TEa); scrutinizing the data and computing a six sigma value (sigma (σ) = [TEa − bias)/CV]); recuperating the process based on the results of analysis; and required follow up [9].
Total allowable error (TEa) refers to the degree of change that needs to be detected in an analyte for a clinically important decision to be made with regard to further investigation or treatment [5]. Bias or inaccuracy, emphasizes lack of agreement among methods being compared. Systematic error is detected as positive or negative bias for a given analytical method. Coefficient of variation (CV) is used to describe the variation of a test. The CV expresses the variation as a percentage of the mean [5]. In the laboratory functions, the CV is preferred mode of variance determination when the SD increases in proportion to concentration. The CV also provides a general perception about the performance of a method. CVs of 5% or less generally denotes a good method performance, whereas CVs of 10% and higher implies unsatisfactory performance.
QC materials are used for monitoring the performance of analytical methods. When applying any criteria (including Westgard rules) for acceptability of control data, determination of probability for rejection is paramount importance [10]. The term probability of false rejection (Pfr) is used signifies a situation where there are no analytical errors present except for the inherent imprecision or random error of the method. Probability of error detection (Ped) is the term used to describe where an analytical error occurs in addition to the inherent random error. It has been observed that a high probability of error detection and a low probability of false rejection are desirable [11].
We obtained sigma >6 for TG, CPK-Total and Amylase for both the levels of QC. This implies that the analytical method in use is appropriate for detecting both low and high values. The QC strategies that should be implemented in such cases need not be draconian and we can release the patient results immediately. The parameters which demonstrated wide variation in the sigma values for both the levels of QC should be evaluated with discretion. The methodology should be re evaluated. There is also a need to strictly follow Westgard multi rules as well as increase the number of QC runs so as to abolish this discrepancy. Blood urea, sodium and potassium being the worst performers in our laboratory, diverting special attention to them is mandatory for revamping performance. It is of utmost importance to explore urea method performance and practice stringent maintenance of ISE unit to alleviate inaccuracies resulting in poor performance of ISE module. Most of other parameters demonstrated sigma metrics 3.1 to 5.9 signifying acceptable laboratory performance with a scope for improvisation.
The practice of correlating the results with clinical features and results of other related analytes will aid to overcome the limitations that we have confronted during the interpretation of QC and corresponding sigma metrics. We also propose the custom of critical appraisal of the sigma values of all the parameters on a regular basis to achieve exceptional quality.
The main limitation in our work is the lack of knowledge about the corresponding Pfr and Ped for the different analytes due to lack of appropriate software as a result of financial constraints. This would have made our results and interpretation more explicit and ultra precise.
Conclusion
The six sigma motive is to minimize both variance and quality control processes to guarantee compliance with the critical specifications. Sigma metrics will also facilitate the inculcation of ideal analytical methodologies in order to augment laboratory performance. We, clinical biochemists affirm that we should nurture realistic quality goals for the laboratories keeping in mind the inherent random errors and performance capability of biochemistry analyzers. It is also imperative to implement appropriate QC strategies in order to augment the judicious use of QC.
References
- 1.Westgard JO, Barry PL. Beyond quality assurance: committing to quality improvement. Lab Med. 1989;20:241–247. [Google Scholar]
- 2.Nevalainen D, Berte L, Kraft C, Leigh E, Picaso L, Morgan T. Evaluating laboratory performance on quality indicators with six sigma scale. Arch Pathol Lab Med. 2000;124(4):516–519. doi: 10.5858/2000-124-0516-ELPOQI. [DOI] [PubMed] [Google Scholar]
- 3.Coskun A. Six sigma and calculated laboratory tests. Clin Chem. 2006;52:770–771. doi: 10.1373/clinchem.2005.064311. [DOI] [PubMed] [Google Scholar]
- 4.Harry M, Schroeder R. Six sigma: the breakthrough management strategy revolutionizing the world’s top corporations. New York, NY: Currency; 2000. [Google Scholar]
- 5.Westgard JO, Klee GG. Quality management. In: Burtis CA, Ashwood ER, Bruns DE, editors. Tietz textbook of clinical chemistry. 4. Philadelphia: Saunders; 2006. [Google Scholar]
- 6.Revere L, Black K. Integrating six sigma with total quality management: a case example for measuring medication errors. J Healthc Manag. 2003;48:377–391. [PubMed] [Google Scholar]
- 7.Tetrault G. Evaluating laboratory performance with the six sigma scale. Arch Pathol Lab Med. 2000;124(12):1748–1749. doi: 10.5858/2000-124-1748c-ELPWTS. [DOI] [PubMed] [Google Scholar]
- 8.Westgard JO, Westgard SA. The quality of laboratory testing today an assessment of σ metrics for analytic quality using performance data from proficiency testing surveys and the CLIA Criteria for Acceptable Performance. J Clin Pathol. 2006;125:343–354. [PubMed] [Google Scholar]
- 9.Westgard JO. Quality control. How labs can apply six sigma principles to quality control planning. Clin Lab News. 2006;32:10–12. [Google Scholar]
- 10.Westgard JO. Internal quality control: planning and implementation strategies. Ann Clin Biochem. 2003;40:593–611. doi: 10.1258/000456303770367199. [DOI] [PubMed] [Google Scholar]
- 11.Westgard JO, Groth T, Aronsson T, Falk H, Verdier CH. Performance characteristics of rules for internal quality control: probabilities for false rejection and error detection. Clin Chem. 1977;23:1857–1867. [PubMed] [Google Scholar]


