Abstract
The chicken karyotype consists of 39 chromosomes of which 33 are classed as microchromosomes (MICs). MICs contain about one third of genomic DNA. The majority of mapped chicken genes are assigned to macrochromosomes (MACs), but a recent study indicated that CpG islands (CGIs), which are associated with most vertebrate genes, map predominantly to MICs. The present work establishes that chicken genes are concentrated on MICs by several criteria. Acetylated (lysine 5) histone H4, which is strongly correlated with the presence of genes, is highly enriched on MICs by immunocytochemistry. In addition, detailed analysis of chicken cosmids shows that CGI-like fragments are approximately six times denser on MICs than on MACs. Published mapping of randomly chosen genes by fluorescent in situ hybridization (FISH) also shows a significant excess of microchromosomal assignments. Finally, the finding that MICs replicate during the first half of S phase is also compatible with the suggestion that MICs represent gene-rich DNA. We use the cosmid data to predict that ∼75% of chicken genes are located on microchromosomes.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AJ001643 and AJ001644.]
Avian karyotypes invariably comprise microchromosomes (MICs) and macrochromosomes (MACs) (Bloom et al. 1993). In the chicken, MICs 11–39 constitute approximately one-quarter of the genome and are cytologically indistinguishable from each other because of their small size (Bloom et al. 1993). MICs contain GC-rich DNA (Auer et al. 1987), are enriched for repetitive sequences (Stefos and Arrighi 1974; Matzke et al. 1990), and contain heterochromatin by the criterion of C banding (Schmid et al. 1989). About two-thirds of the chicken genome (65%) is found on MACs 1–6 (Stubblefield and Orro 1982), which are the only chicken chromosomes larger than the smallest human chromosomes. Chromosomes 7–10 are intermediate in size and have been arbitrarily designated as larger MICs (Bloom et al. 1993). In contrast to the MIC group, MACs show only faint C banding caused by heterochromatin in centromeric and telomeric regions. However, R bands, which are associated with transcriptionally active DNA, are also faint on MACs, having lower intensity and contrast than those of mammalian chromosomes (Schmid et al. 1989). MACs contain much AT-rich DNA (Auer et al. 1987) and, unlike MICs, are relatively rich in (CA)n microsatellite repeats (Primmer et al. 1997). MICs were originally thought to be genetically inert (Newcomer 1955), but are now known to be bona fide chromosomes that are maintained at a constant number and have conserved telomere sequences (Solovei et al. 1994). Many genes have recently been assigned microchromosomal locations (Bloom and Bacon 1985; Dominguez-Steglich et al. 1990, 1992a, 1992b, 1993; Jones et al. 1997). Nevertheless, published gene-mapping data remains biased in favor of macrochromosomal assignments (Burt et al. 1995).
As in other vertebrates, the majority of genes in chicken have CpG islands (CGIs) (Cooper et al. 1983; McQueen et al. 1996). CGIs are associated with the promoters of genes and can be differentiated from bulk DNA by their high GC content, lack of depletion of the dinucleotide CpG, and lack of methylation (for review, see Cross and Bird 1995). It is possible to exploit these characteristics for purification of CGIs from bulk DNA (Cross et al. 1994). In situ hybridization with a chicken CGI library constructed by this technique indicated that CGIs are concentrated on MICs. MACs by contrast were relatively CGI-poor by this criterion (McQueen et al. 1996). The implication that the majority of genes are localized on MICs was in contrast to mapping data and suggested that mapping data was biased towards MACs for technical reasons. The results led us to predict that gene density on chicken MICs is similar to that of the pufferfish Fugu rubripes. Fugu has proven to be an excellent model for the study of vertebrate gene organization because of its compact genome (Elgar 1996). Should chicken MICs reveal a similarly high concentration of genes, then they may also serve as a useful model system for the study of larger vertebrate genomes. Reduced intron size of chicken genes relative to mammalian homologs, as seen for Fugu genes, is compatible with this idea (Hughes and Hughes 1995; Riegert et al. 1996).
In this study we have analyzed the distribution of acetylated histone H4 in the chicken genome. Increased acetylation of the amino-terminus of histone H4 is observed in transcriptionally active regions (for review, see Turner 1993; Wade et al. 1997). For example, the distribution of acetylated H4 in human and hamster chromosomes has been shown to be nonrandom, with hyperacetylation of gene-rich R bands (Jeppesen et al. 1992), and hypoacetylation of heterochromatic domains (Jeppesen and Turner 1993). Histone H4 in CGI chromatin is known to be hyperacetylated compared with bulk chromatin (Tazi and Bird 1990). Acetylation studies therefore provide a method for visualizing regions of high gene content that is independent of sequence characteristics.
Our study also includes a reinvestigation of the time of MIC replication during S phase. In mammals, transcriptionally active DNA is early-replicating (Holmquist 1987) and CGI-rich regions have been shown to replicate early in human chromosomes (Craig and Bickmore 1994). Assuming that transcription and early replication are also correlated in the chicken, it is expected that microchromosomal DNA should be early-replicating. Previous studies give an inconsistent picture with respect to MICs. Schmid et al. (1989) concluded that MIC replication occurs predominantly in the second half of S phase, whereas Ponce de Leon et al. (1992) concluded that MICs replicate during both early and late S phase. We have therefore conducted replication timing experiments with particular attention to MICs.
Finally, we have estimated CGI density on microchromosomal DNA relative to macrochromosomal DNA by counting CGI-like sequences on cosmids whose genomic origin is known. We find a significant enrichment of CGIs on MICs.
RESULTS
Microchromosomes Are Selectively Enriched for Acetylated Histone H4
The histone H4 acetylation profile of chicken chromosomes was determined by immunofluorescence with an antiserum raised against histone H4 acetylated at lysine 5 (a gift from Bryan Turner; Turner and Fellows 1989). For comparison, Figure 1a shows hybridization of the CGI library against chicken chromosomes (McQueen et al. 1996). The acetylation pattern for the chicken (Fig. 1b) is strikingly similar to the CGI localization in that MICs are strongly labeled. MICs aggregate somewhat under the exceptionally mild fixation conditions that are required to detect histone immunofluorescence and we cannot, therefore, be certain whether all MICs are stained with equal intensities. There is some evidence for heterogeneity of hybridization between MICs with the CGI probe (Fig. 1a), but one would need to compare the same MIC on different photomicrographs to be sure that the variation is real. This is impossible because MICs are indistinguishable. MACs are less stained than MICs by the antibody, but there is evidence for an uneven pattern of acetylated histone H4 along their length. The sharp distinction between hyperacetylated and hypoacetylated regions of the chicken genome appears to be more exaggerated than found in mammalian autosomes, although the inactive X chromosome of mammals is dramatically hypoacetylated (Jeppesen et al. 1992; Jeppesen and Turner 1993).
Figure 1.
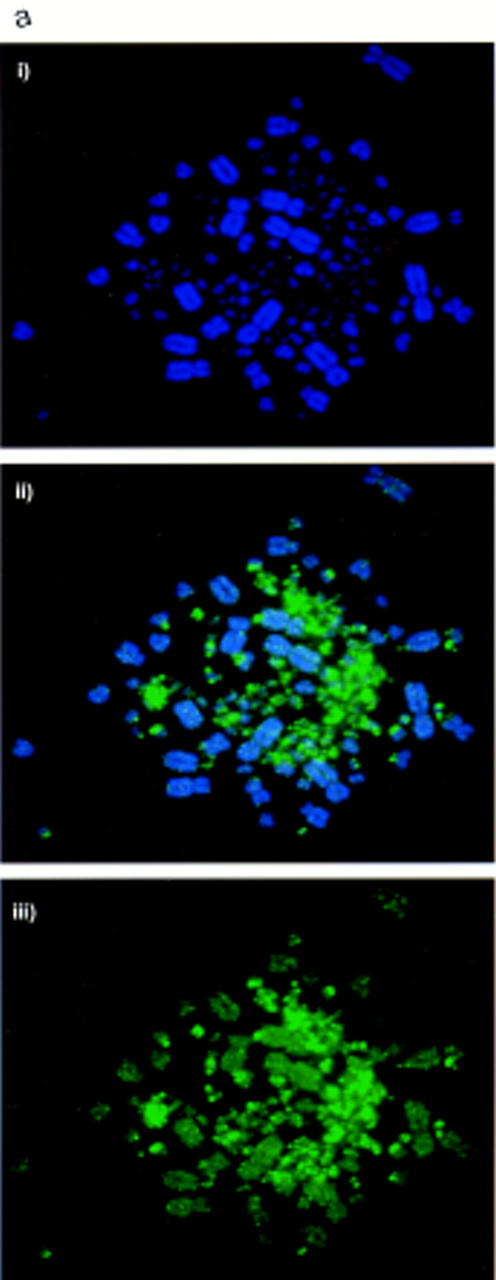
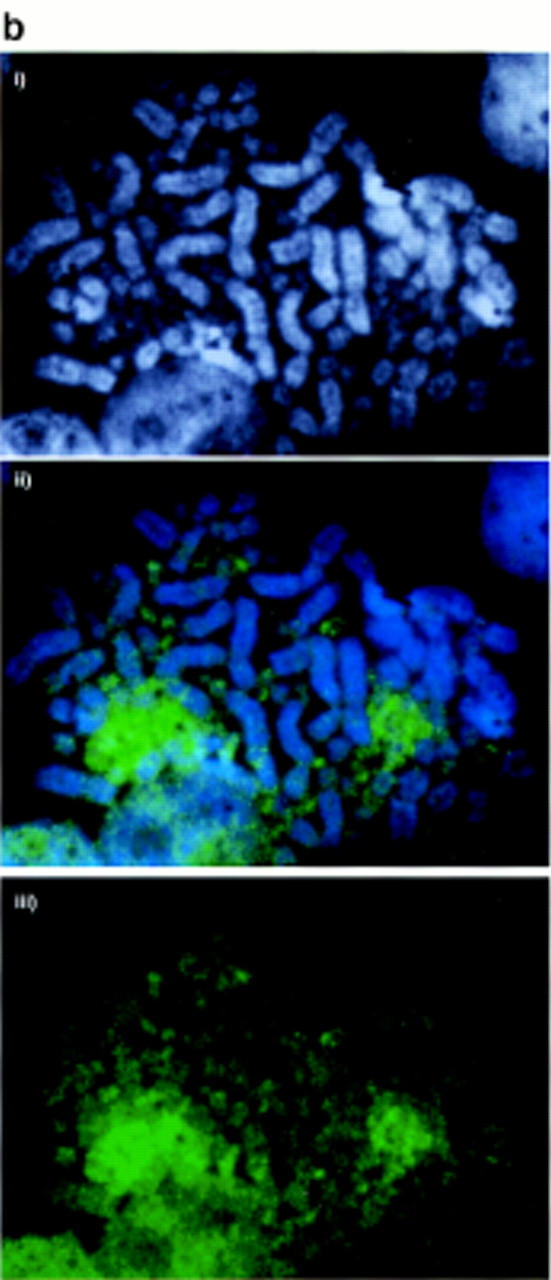
Acetylated histone H4 and CGIs are concentrated on MICs. (a) CGI–FISH. (i) DAPI-stained DT40 chromosomes. (ii) CGI library probe (green) prepared and used as in McQueen et al. (1996), showing strong signal in MICs. (iii) CGI library probe without counterstain showing the contrast in signal intensity between the MIC and MAC groups. (b) H4 acetylation immunolabeling. (i) DAPI-stained DT40 chromosomes. (ii) H4 acetylation immunostaining (green) showing strong staining over MIC clusters. (iii) H4 acetylation immunostaining without counterstain showing the contrast in signal intensity between the MIC and MAC groups.
Microchromosomes Replicate During the First Half of S Phase
Replication timing of chicken chromosomes was assessed by labeling unsynchronized DT40 cells with bromodeoxyuridine (BrdU) for between 1 and 10 hr before arresting with colcemid. Incorporated BrdU was detected in arrested metaphases with a fluorescein-conjugated antibody. Labeled cells that reached metaphase a short time after the pulse must have been in late S phase when the BrdU pulse occurred. Conversely, labeled cells reaching mitosis 10 hr after the pulse must have started to incorporate BrdU during early S phase. We estimated S phase to last no more than 10 hr for this lymphoblastoid culture, as complete labeling of the chromosome set was observed when label was present during the 10 hr prior to mitosis (Fig. 2). Chromosomes that have incorporated BrdU for 10, 7, 4, and 2 hr prior to mitosis are shown in Figure 2. Little or no BrdU incorporation into MICs was noted during the last 4 hr of S phase, although parts of the MACs were strongly banded by replication during this period of mid-to-late S phase. In contrast, virtually all of the microchromosomal BrdU incorporation occurs within the first 2–3 hr of S phase. These results were confirmed by detection of microchromosomal incorporation before arrest with methotrexate at mid-S phase, but not after release from the block (data not shown).
Figure 2.
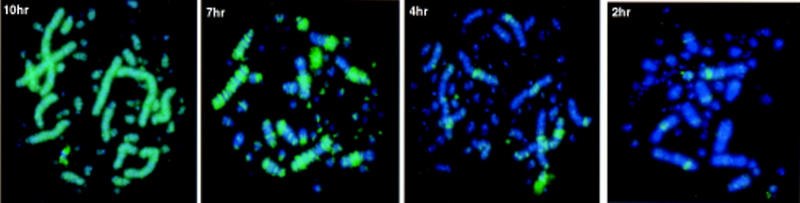
BrdU incorporation patterns on chicken metaphase chromosomes after labeling with BrdU for 10, 7, 4, and 2 hr prior to mitosis. BrdU incorporation is stained green. Little or no microchromosomal incorporation is detected throughout the last 4 hr of S phase, whereas some regions of MACs are replicated at later and some specifically at earlier time points.
Microchromosomal Cosmids Are Enriched for CGI-Like Fragments
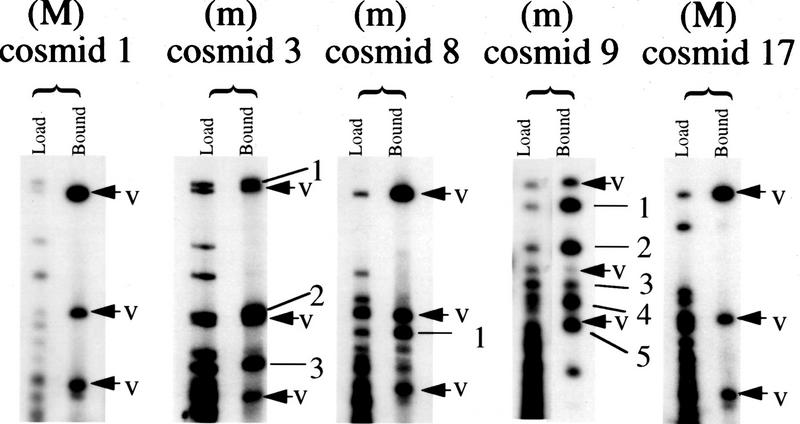
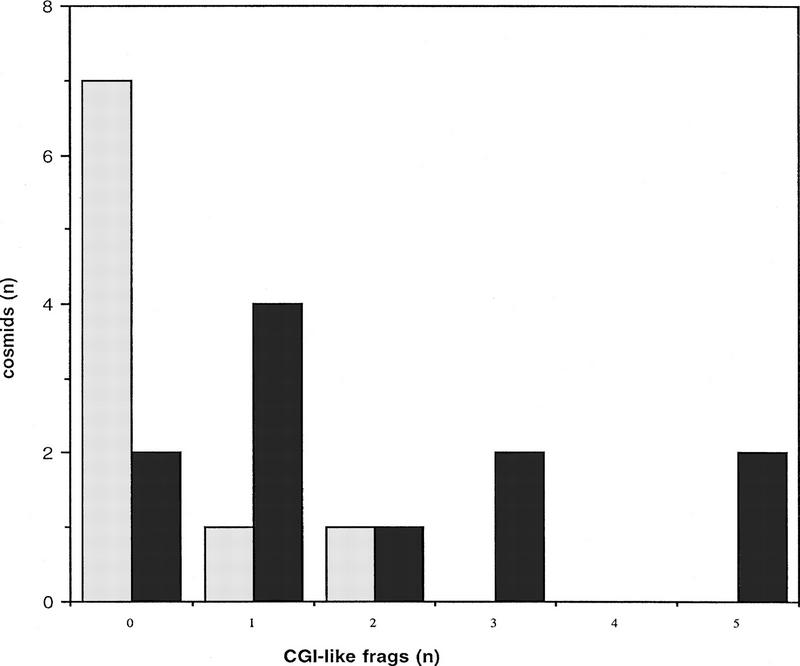
To compare the density of CGIs on MICs and MACs, 20 chicken cosmids were selected for analysis. The presence of insert was confirmed for all 20 by digestion with NotI and HindIII. All 20 cosmids were labeled with either biotin or digoxigenin and the resulting probes were used for FISH against chicken metaphase spreads. Cosmids that hybridized to one of the 6 largest chromosomes were designated macrochromosomal, whereas those mapping to one of the 33 smallest chromosomes were termed microchromosomal. Of the 20 cosmids, 11 were microchromosomal, 1 of these (cosmid 9) originating from an intermediate MIC, and 9 were macrochromosomal (Table 1). To estimate the number of CGIs, cosmids were cut with the enzyme MseI to leave most CGIs intact (Cross et al. 1994; McQueen et al. 1996) and were methylated to completion. By passing the end-labeled fragments over methyl-CpG binding domain columns (MBD columns; Cross et al. 1994), it was possible to separate strongly bound CGI DNA from weakly bound bulk genomic DNA. Weakly bound fragments were eluted from the column with 0.5 m NaCl, and strongly bound fragments were eluted at 0.7–1.0 m NaCl. The total MseI fragment pattern was visualized by autoradiography and compared with the pattern of bound CGI-like fragments. In this way we were able to identify and number CGI-like fragments, as indicated for five examples in Figure 3. The cosmid vector (pWE15) has four MseI fragments >500 bp in size that have an average G + C content of 53.0%, the greatest being 57.0%, with no apparent under-representation of the CpG dinucleotide. Because of the CGI-like nature of their sequences, these fragments bound tightly to the MBD column after de novo methylation, and their presence within the bound fraction therefore served as an internal control for the functioning of the MBD column. This was particularly valuable in cases in which no CGI was present in the cosmid insert (see Fig. 3). The average G + C content for chicken CGI MseI fragments is 68% (McQueen et al. 1996). As a result, chicken CGI DNA was expected to bind to MBD columns at least as tightly as vector fragments. Any band of weaker intensity than similar sized vector was therefore discounted, as was any band less than 500 bp in size. The results for each of the 20 cosmids are summarized in Table 1 and Figure 4. Seven of the nine macrochromosomal cosmids did not have any CGI-like fragments according to our assay, and only one macrochromosomal cosmid had as many as two CGI-like fragments (Fig. 4). In contrast, the numbers of CGI-like fragments in microchromosomal cosmids ranged from zero to five and averaged two. The methylation status of CGI-like fragments of cosmid 23 was tested by using the gel-purified fragments as radioactive probes on Southern blots of chicken genomic DNA. The DNA was cut with MseI alone, and MseI plus HpaII or MspI (methylation-sensitive and methylation-insensitive, respectively). All three fragments were part of unmethylated tracts (data not shown) as expected of CGI DNA.
Table 1.
Cytogenetic Location and CGI-Like Fragment Number for Each of 20 Cosmids
| Cosmid | Insert size (kb) | Locationa | CGI-like fragments |
|---|---|---|---|
| 1 | 23 | Ma | 0 |
| 5 | 23 | Ma | 2 |
| 10 | 30 | Ma | 0 |
| 15 | 43 | Ma | 0 |
| 17 | 40 | Ma | 0 |
| 18 | 32 | Ma | 0 |
| 19 | 33 | Ma | 1 |
| 24 | 31 | Ma | 0 |
| 25 | 32 | Ma | 0 |
| 9 | 34 | I. mi | 5 |
| 2 | 32 | mi | 1 |
| 3 | 33 | mi | 3 |
| 6 | 30 | mi | 2 |
| 7 | 34 | mi | 5 |
| 8 | 30 | mi | 1 |
| 16 | 39 | mi | 1 |
| 20 | 36 | mi | 0 |
| 21 | 27 | mi | 0 |
| 22 | 35 | mi | 1 |
| 23 | 29 | mi | 3 |
(Ma) Macrochromosomal; (I. mi) located on an intermediate MIC; (mi) microchromosomal.
Figure 3.
Binding of CGI-like fragments from 5 cosmids to MBD columns. For each cosmid the fragment profile of MseI-digested DNA that was loaded onto the column is shown beside the tightly bound fragments that eluted at high salt. The vector bands present in the eluate are arrowed and labeled V. Bound insert fragments for each cosmid are numbered. The profiles refer to 2 macrochromosomal cosmids (M) with no CGI-like chicken fragments and 3 microchromosomal examples (m) with 1, 3, and 5 CGI-like fragments.
Figure 4.
Graphic summary of the number of microchromosomal (dark shading) and macrochromosomal (light shading) cosmids having 0, 1, 2, 3, 4, and 5 CGI-like fragments. The majority of macrochromosomal cosmids have no CGI-like fragments, whereas most microchromosomal cosmids have one or more CGI-like fragments.
Column-Purified CGI-Like Fragments Were Associated with Genes
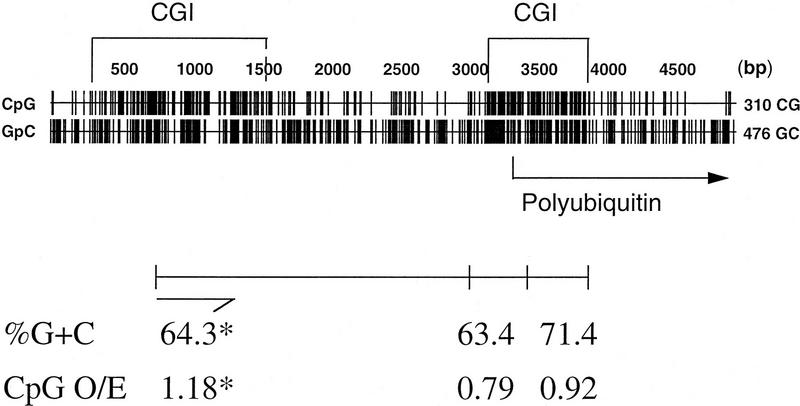
To extend the observed enrichment of CGI-like fragments on MICs to bona fide genes, the CGI-like fragments of cosmid 7 were cloned and sequenced. The CGI-like nature of these fragments was confirmed with G + C contents ranging from 63% to 71% and CpG densities (expressed as the ratio of observed over expected CpGs) of up to 1.18. Three fragments showed identity to genomic chicken DNA in the EMBL database: two MseI fragments from the CGI of the polyubiquitin gene and a fragment from a second CGI which lies ∼2 kb upstream to polyubiquitin and probably belongs to a separate gene. The positions and GC characteristics of these three fragments are shown in Figure 5. The predicted protein encoded by a fourth fragment from this cosmid contained a 42-amino-acid region of protein that was 64%–66% identical to a region found in the human Cla-1 membrane glycoprotein and in scavenger receptor proteins of mouse and Chinese hamster. This suggests that a gene related to human Cla-1 and rodent scavenger receptor genes resides, with polyubiquitin, on cosmid 7 although further data is required to confirm the identity of this chicken gene. Cla-1 and polyubiquitin are known to be syntenic on human chromosome 12. Thus, CGI-like fragments selected by the column can correspond to CGI-associated genes. As cosmid 7 is of microchromosomal origin, our data establishes a microchromosomal location for the previously unmapped polyubiquitin gene.
Figure 5.
Genomic context of 3 CGI-like fragments purified from cosmid 7. Occurrences of the dinucleotides CpG and GpC are plotted throughout this genomic sequence, which was assembled by overlapping two genomic fragments from the polyubiquitin gene with EMBL database accession nos. Z14958 and X58195. The positions of 2 CGIs and the start site for the polyubiquitin gene are indicated. The positions and GC characteristics of 3 sequenced cosmid 7 fragments, which contain CGIs, are shown below. (*) Characteristics pertain to the 504-bp sequenced end of this fragment as indicated.
DISCUSSION
Preferential staining of MICs with antibody against acetylated H4 provides strong evidence for elevated gene density. The antibody, in mammals, detects only the most highly acetylated H4 isoforms and is specific for lysine 5 in the H4 tail. Acetylation of amino-terminal lysines 8 and 12 correlates with acetylation of lysine 5 in histone H4 (Belyaev et al. 1996). Therefore the results indicate that microchromosomal H4 is acetylated at multiple lysine residues. Histone acetylation is a feature of gene-dense regions of the genome (Hebbes et al. 1991; Brownell et al. 1996), and is therefore a strong indicator that genes are concentrated on MICs. At first sight it is paradoxical that the smallest MICs have been described as heterochromatic (Schmid et al. 1989), as our data suggest that most, if not all MICs, including the smallest subset, are heavily acetylated at H4. The results can be reconciled if we propose that MICs contain both constitutive heterochromatin (most probably near the centromeres) and active euchromatin. Although the smallest MICs are near the size limit of light microscopy at metaphase, they are estimated to contain at least 7 Mb of DNA (Bloom et al. 1993). There is therefore room for regions of activity and regions of silence. Chicken MICs were at one time equated with plant B chromosomes, but in contrast to MIC hyperacetylation as described here, underacetylation of the B chromosomes of the plant Brachycome dichromosomatica has been demonstrated recently with the same antibody (Houben et al. 1997).
In calculating the frequency of CGIs on cosmids, we assumed that fragments retained by the MBD-column represent genuine CGI DNA rather than the generally GC-rich DNA that has been detected in MICs by chromomycin staining (Auer et al. 1987). The validity of this assumption rests on several observations. Firstly, the MBD protein binds methylated DNA according to the number of CpG dinucleotides, not the overall G + C content. Thus GC-rich DNA would not be retained by the column unless it had an undepleted CpG frequency. Previous studies have shown that GC-rich DNA with the expected frequency of CpGs is invariably derived from CGIs (Matsuo et al. 1993) and CpG frequency therefore appears to be a reliable means to distinguish CGI from non-CGI DNA. Sequencing of retained fragments from cosmid 7 confirmed that they are CGI-like in GC content and CpG frequency and in their association with genes (Fig. 5). In contrast, bulk genomic DNA of chicken has an average CpG frequency (observed over expected) of 0.27 (Setlow 1976). Methylation analysis was also used to confirm the CGI-like nature of fragments retained by the column.
Do the number of retained fragments accurately reflect the number of CGIs on a cosmid? Errors in CGI counting may be expected due to multiple large fragments derived from the same island (leading to an overestimate), or multiple small fragments that are below the 0.5-kb threshold for scoring by our assay (leading to an underestimate). However, plots of MseI fragments around 14 chicken CGIs showed 11 to contain only one fragment larger than 0.5 kb, two to contain multiple large fragments, and one to be cut into fragments, all of which were <0.5 kb (Table 2). From this analysis we consider that our method for CGI counting is reasonably accurate.
Table 2.
Cytogenetic Assignments for Chicken Genes with and without CGIs, and Number of MseI Fragments per CGI
| Gene | Accession no. | CGI | Assignment | CGI MseI fragments (kb) |
|---|---|---|---|---|
| MHC class II β chain | M26306 | + | MIC 16 | 1 frag > 0.5 |
| MHC class II β chain | M26307 | + | MIC 16 | 1 frag > 0.5 |
| MHC class I α chain | M31012 | + | MIC 16 | 1 frag > 0.5 |
| Polyubiquitin Ub II | X58195 | + | MIC (results) | 0 frags > 0.5 |
| Adenylate kinase I | D00251 | + | MIC > 16 | 1 frag > 0.5 |
| Conalbumin | Y00407 | − | MIC 10–12 | – |
| α-A and α-D globin | X59989 | − | MIC 10–15 | – |
| Dmd | X06294 | − | MIC 10 | – |
| Transforming GF β3 | X58127 | + | MAC 5 | 1 frag > 0.5 |
| Transforming GF β2 | X58071 | + | MAC 3 | 2 frags > 0.5 |
| GAPDH | M11213 | + | MAC 1 | 1 frag > 0.5 |
| Acetylcholine receptor | J05218 | + | – | 1 frag > 0.5 |
| Retinoblastoma | X72217 | + | – | 1 frag > 0.5 |
| Heat shock protein 70 | J02579 | + | – | 1 frag > 0.5 |
| U plasminogen activator | J05188 | + | – | 1 frag > 0.5 |
| Lipoprotein lipase | X60547 | + | – | 2 frags > 0.5 |
| G protein α-subunit | L24550 | + | – | 1 frag > 0.5 |
All assignments from ChickGBase at http://www.ri.bbsrc.ac.uk except α-globin genes (Hughes et al. 1979), and the polyubiquitin gene (see Results). (U) Urokinase.
Based on our estimates of CGI density, we have calculated the approximate number of genes expected on microchromosomal versus macrochromosomal DNA. Comparing the 22 CGIs on 11 microchromosomal cosmids with the 3 CGIs found in 9 macrochromosomal cosmids gives a sixfold enrichment for CGIs in microchromosomal DNA. A database survey suggests the proportion of genes with CGIs to be roughly similar in chicken and humans (McQueen et al. 1996). Importantly, we find that genes with CGIs can be found on both MICs and MACs, and that genes without CGIs are present on the MICs (Table 2). Thus, there is no evidence for genomic segregation of CGI-genes from non-CGI genes. Assuming 60% of genes to have CGIs, we calculate that there is a gene on average every 10 kb of microchromosomal DNA. This is similar to the average gene density of one gene every 7 kb in Fugu. Because the MIC group, including those of intermediate size, constitutes 35% of the chicken genome (Stubblefield and Orro 1982), we estimate a total of 42,000 genes in the microchromosomal compartment and 13,000 genes in the macrochromosomal compartment. The derived total number of genes in the chicken genome of 55,000 must be regarded as very approximate. The estimate is somewhat lower than previous estimates of 80,000 (Antequera and Bird 1993), 64,000 (Fields et al. 1994), and 60,000 (Elgar 1996) for gene number in vertebrates. Given the assumptions involved, and the relatively small sample of genomic DNA on which the chicken estimate is based, we do not consider the difference significant at this stage.
A discrepancy remains between the predominantly macrochromosomal location of mapped chicken genes (Burt et al. 1995) and our finding that over 75% of genes reside on MICs. We suggest that the chicken genome map has become biased for two reasons. Firstly, physical mapping of microchromosomal genes has been technically difficult because of the intrinsic difficulty of identifying specific MICs; this would have been even more difficult when in situ hybridization used predominantly radioactive probes, as microchromosomal signal and random noise would often have been indistinguishable (Dominguez-Steglich et al. 1991). Secondly, the liberal use of randomly isolated microsatellite markers for the building of the chicken genetic map may have prejudiced against the mapping of microchromosomal genes, since MICs are poorly populated by at least (CA)n microsatellites (Primmer et al. 1997). An independent and direct assessment of gene density can be derived from assignments of randomly selected genes by FISH. A survey of the recent literature showed that of 17 genes apparently chosen at random, 12 were located on MICs and 5 were on MACs (Dominguez-Steglich et al. 1990, 1992a,b,c,d; Li et al. 1995; Riegert et al. 1996; Nanda et al. 1996, 1997; Marienfeld et al. 1997). The results of this limited data set agree well with our CGI analysis, as does the sequence scanning approach of M.S. Clark et al. (pers. comm.).
Altogether, the results of several distinct experimental approaches consistently show that chicken MICs are unusually gene-rich. The asymmetric distribution of genetic activity within the chicken may have implications for our understanding of vertebrate genome evolution and for efforts to construct the chicken genome map. It also suggests that chicken may complement Fugu as a system for gene discovery and sequencing.
METHODS
Immunofluorescence with Acetylated H4-Specific Antiserum
DT40 cells were arrested in metaphase with colcemid. After 75 mm KCl hypotonic solution treatment, 106 cells were spun gently onto each glass slide and permeabilized by immersion in KCM consisting of 120 mm KCl, 20 mm NaCl, 10 mm Tris-HCl (pH 8.0), 0.5 mm EDTA, and 0.1% Triton X-100. The primary antibody, R41/5, was a rabbit serum raised against histone H4 acetylated at Lys-5, and was a gift from Bryan Turner (University of Birmingham, UK). This was detected using TRITC conjugated anti-rabbit immunoglobulins (Sigma). Slides were fixed in paraformaldehyde, mounted in antifadent containing DAPI, and examined with a fluorescence microscope.
BrdU Incorporation for Replication Timing
DT40 cells were grown continuously in the presence of 0.1 μm BrdU for 1-, 2-, 3-, 4-, 5-, 6-, 7-, 8-, 9-, and 10-hr periods, and those cells having reached mitosis were immediately arrested in 0.1 μg/ml colcemid. Metaphase chromosomes, fixed in 3:1 methanol/acetic acid, were prepared on slides from each culture. Slides were denatured in 0.07 m NaOH/EtOH and blocked in 1% skim milk before applying FITC conjugated anti-BrdU antibody (Boehringer). Slides were mounted and examined as for FISH.
FISH
Chicken metaphase chromosomes were prepared from DT40 cells according to standard procedures. Cosmids were selected, 17 at random and 3 for the presence of CGIs, from a male adult leghorn library (Clontech). Cosmid DNA was prepared using Qiagen plasmid columns and following recommendations for low-copy plasmid purification. Biotin 16–dUTP (Boehringer) and digoxigenin 11–dUTP (Boehringer) were incorporated into cosmid DNA by nick translation and labeled probes were concentrated by precipitation. One hundred nanograms of probe was ethanol precipitated in the presence of 5 μg of salmon sperm DNA as a carrier and 2 μg of sonicated chicken genomic DNA as competitor. The pellet was resuspended in 15 μl of hybridization mix, denatured, and preannealed for 15 min at 37°C to block repetitive sequences. After RNase treatment and denaturation, slides were hybridized at 37°C overnight. Biotinylated probes were detected with avidin-FITC or avidin-Texas Red followed by biotinylated anti-avidin (all from Vector Laboratories) and an additional layer of avidin-FITC or avidin-Texas Red. Digoxigenin-labeled probes were detected with antidigoxigenin antibody conjugated with fluorescein (Boehringer). Slides were mounted in antifadent containing DAPI and examined with a fluorescence microscope using filter blocks for FITC, Texas Red, and DAPI. Images were collected with a Photometrics CCD camera and Digital Scientific software.
CGI Purification from Chicken Cosmids
For each cosmid, 2.5 μg of DNA (and of vector DNA) was cut to completion with the enzyme MseI, methylated de novo with SssI methylase (NEB), and after phosphatase treatment, was end labeled with [γ32P]rATP using polynucleotide kinase. Unincorporated nucleotides were removed by passing labeled DNA over G50 Sephadex columns. MBD columns were prepared according to a previously described method (Cross et al. 1994) except that in this instance only 100 μl of nickel agarose-coupled MBD protein was loaded into the reservoir of a Wizard minicolumn (Promega) for each cosmid. DNA was loaded onto the column, washed, and eluted in 20 mm HEPES (pH 7.9), 10% glycerol, 0.1% Triton X-100, 0.5 mm PMSF, and NaCl concentrations of up to 1.0 m. DNA was loaded and washed on the columns at 0.5 m salt to remove unbound fragments before eluting at 1.0 m. Collected samples were diluted back to 0.5 m and reloaded onto the column for a second round of purification. CpG-rich fragments were eluted at 0.7–1.0 m salt and were precipitated, counted on a scintillation counter, and loaded onto 1.5% agarose gels. Gels were run at 80–125 V for 4–5 hr before drying and exposing to X-ray film.
Cloning and sequence analysis were carried out using methods and reagents previously described (McQueen et al. 1996). Sequences from both ends of an ∼1-kb fragment of cosmid 7 were submitted to the EMBL database and were given accession numbers AJ001643 and AJ001644, where the latter sequence shows partial homology to Cla-1.
Acknowledgments
We are very grateful to Bryan Turner for the gift of the anti-acetylated histone H4 antibody. We also thank Joan Davidson and Aileen Greig for technical help, Vicky Clark for DNA sequencing, and Sally Cross for critical reading of the manuscript. This work was funded by the Biotechnology and Biological Sciences Research Council, and a Medical Research Council studentship to G.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL heather.mcqueen@ed.ac.uk; FAX 0131 650 5379.
REFERENCES
- Antequera F, Bird AP. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer H, Mayr B, Lambrou M, Schleger W. An extended chicken karyotype, including the NOR chromosome. Cytogenet Cell Genet. 1987;45:218–221. doi: 10.1159/000132457. [DOI] [PubMed] [Google Scholar]
- Belyaev ND, Keohane AM, Turner BM. Histone H4 acetylation and replication timing in Chinese hamster chromosomes. Exp Cell Res. 1996;225:277–285. doi: 10.1006/excr.1996.0177. [DOI] [PubMed] [Google Scholar]
- Bloom SE, Bacon LD. Linkage of the major histocompatability (B) complex and the nucleolar organiser in the chicken. J Hered. 1985;76:146–154. [PubMed] [Google Scholar]
- Bloom SE, Delany ME, Muscarella DE. Constant and variable features of avian chromosomes. In: Etches RJ, Gibbons AMV, editors. Manipulation of the avian genome. Boca Raton, FL: CRC Press; 1993. pp. 39–59. [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Burt DW, Bumstead N, Bitgood JJ, Ponce De Leon AF, Crittenden LB. Chicken genome mapping: A new era in avian genetics. Trends Genet. 1995;11:190–194. doi: 10.1016/s0168-9525(00)89042-3. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Taggart MH, Bird AP. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Bickmore WA. The distribution of CpG islands in mammalian chromosomes. Nature Genet. 1994;7:376–382. doi: 10.1038/ng0794-376. [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nature Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Devel. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Meng G, Bettecken T, Muller C, Schmid M. The dystrophin gene is autosomally located on a microchromosome in chicken. Genomics. 1990;8:536–540. doi: 10.1016/0888-7543(90)90041-r. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Auffray C, Schmid M. Linkage of the chicken MHC to the nucleolus organizer region visualised using non-isotopic in situ hybridisation. J Hered. 1991;82:503–505. doi: 10.1093/oxfordjournals.jhered.a111138. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Lichter P, Carrier A, Auffray C, Schmid M. Mapping the bNGF gene in situ to a microchromosome in chicken. Genomics. 1992a;12:829–832. doi: 10.1016/0888-7543(92)90318-m. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Jeltsch J-M, Nakazawa A, Schmid M. Microchromosomal assignment of the chicken ovotransferrin and adenylate kinase genes. Cytogenet Cell Genet. 1992b;61:155–157. doi: 10.1159/000133396. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Jeltsch JM, Garnier JM, Schmid M. In situ mapping of the chicken progesterone receptor gene and the ovalbumin gene. Genomics. 1992c;13:1343–1344. doi: 10.1016/0888-7543(92)90064-y. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Carrier A, Auffrey C, Schmid M. Assignment of the chicken tyrosine hydroxylase gene to chromosome 6 by FISH. Cytogenet Cell Genet. 1992d;60:138–139. doi: 10.1159/000133324. [DOI] [PubMed] [Google Scholar]
- Dominguez-Steglich M, Robbins J, Schmid M. Mapping of the chicken N-CAM gene and a myosin heavy chain gene: Avian microchromosomes are not genetically inert reserves of DNA. J Exp Zool. 1993;265:295–300. doi: 10.1002/jez.1402650310. [DOI] [PubMed] [Google Scholar]
- Elgar G. Quality not quantity: The pufferfish genome. Hum Mol Genet. 1996;5:1437–1442. doi: 10.1093/hmg/5.supplement_1.1437. [DOI] [PubMed] [Google Scholar]
- Fields C, Adams MD, White O, Venter JC. How many genes in the human genome? Nature Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Clayton AL, Crane-Robinson C. Histone acetylation and globin gene switching. Nucleic Acids Res. 1991;20:1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist GP. Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet. 1987;40:151–173. [PMC free article] [PubMed] [Google Scholar]
- Houben A, Belyaev ND, Leach CR, Timmis JN. Differences of histone acetylation and replication timing between A and B chromosomes of Brachycome dichromosomatica. Chromosome Res. 1997;5:233–237. doi: 10.1023/B:CHRO.0000032297.10876.86. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MK. Small genomes for better flyers. Nature. 1995;377:391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- Hughes SH, Stubblefield E, Payvar F, Engel JD, Dodgson JB, Spector D, Cordell B, Schimke RT, Varmus HE. Gene localization by chromosome fractionation: Globin genes are on at least two chromosomes and three estrogen-inducible genes are on three chromosomes. Proc Natl Acad Sci. 1979;76:1348–1352. doi: 10.1073/pnas.76.3.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P, Mitchell A, Turner B, Perry P. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Jones CT, Morrice DR, Paton IR, Burt DW. Gene homologs on human chromosome 15q21-q26 and a chicken microchromosome identify a new conserved segment. Mamm Genome. 1997;8:436–440. doi: 10.1007/s003359900463. [DOI] [PubMed] [Google Scholar]
- Li H, Grenet J, Valentine M, Lahti JM, Kidd VJ. Structure and expression of chicken protein kinase PITSLRE-encoding genes. Gene. 1995;153:237–242. doi: 10.1016/0378-1119(94)00801-x. [DOI] [PubMed] [Google Scholar]
- Marienfeld R, Nanda I, Zoller B, Schmid M, Rebbert M, Jungwirth C. Cloning of chicken interferon regulatory factor-2 (IRF-2) cDNA: Expression and mapping of the IRF-2 gene. J Interferon Cytokine Res. 1997;17:219–227. doi: 10.1089/jir.1997.17.219. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Clay O, Takahashi T, Silke J, Schaffner W. Evidence for erosion of mouse CpG islands during mammalian evolution. Somatic Cell Mol Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Varga F, Berger H, Schernthaner J, Schweizer D, Mayr B, Matzke AJ. A 41–42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma. 1990;99:131–137. doi: 10.1007/BF01735329. [DOI] [PubMed] [Google Scholar]
- McQueen HA, Fantes J, Cross SH, Clark VH, Archibald AL, Bird AP. CpG islands of chicken are concentrated on microchromosomes. Nature Genet. 1996;12:321–324. doi: 10.1038/ng0396-321. [DOI] [PubMed] [Google Scholar]
- Nanda I, Tanaka T, Schmid M. The intron-containing ribosomal protein-encoding genes L5, L7a and L37a are unlinked in chicken. Gene. 1996;170:159–164. doi: 10.1016/0378-1119(95)00828-4. [DOI] [PubMed] [Google Scholar]
- Nanda I, Peters MA, Taparowsky EJ, Sperling K, Schmid M. Assignment of the chicken MAX gene to chromosome 5p by fluorescent in situ hybridisation. Cytogenet Cell Genet. 1997;76:229–232. doi: 10.1159/000134556. [DOI] [PubMed] [Google Scholar]
- Newcomer EH. Accessory chromosomes in the domestic fowl. Genetics. 1955;40:587–588. [Google Scholar]
- Ponce de Leon FA, Li Y, Weng Z. Early and late replicative chromosomal banding patterns of Gallu domesticus. J Hered. 1992;83:36–42. doi: 10.1093/oxfordjournals.jhered.a111154. [DOI] [PubMed] [Google Scholar]
- Primmer CR, Raudsepp T, Chowdhary BP, Moller AP, Ellegren H. Low frequency of microsatellites in the avian genome. Genome Res. 1997;7:471–482. doi: 10.1101/gr.7.5.471. [DOI] [PubMed] [Google Scholar]
- Riegert P, Andersen R, Bumstead N, Dohring C, Dominguez-Steglich M, Enberg J, Salomonsen J, Schmid M, Schwager J, Skjodt K, Kaufman J. The chicken beta 2-microglobulin gene is located on a non-major histocompatability complex microchromosome: A small G+C-rich gene with X and Y boxes in the promoter. Proc Natl Acad Sci. 1996;93:1243–1248. doi: 10.1073/pnas.93.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Enderle E, Schindler D, Schempp W. Chromosome banding and DNA replication patterns in bird karyotypes. Cytogenet Cell Genet. 1989;52:139–146. doi: 10.1159/000132864. [DOI] [PubMed] [Google Scholar]
- Setlow P. Nearest neighbour frequencies in deoxyribonucleic acids. In: Fasman GD, editor. CRC Handbook of biochemistry and molecular biology: Nucleic acids. Vol. 2. Boca Raton, FL: CRC Press; 1976. pp. 312–318. [Google Scholar]
- Solovei I, Gaginskaya ER, Macgregor HC. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 1994;2:460–470. doi: 10.1007/BF01552869. [DOI] [PubMed] [Google Scholar]
- Stefos K, Arrighi F. Repetitive DNA of gallus domesticus and its cytological locations. Exp Cell Res. 1974;83:9–14. doi: 10.1016/0014-4827(74)90681-8. [DOI] [PubMed] [Google Scholar]
- Stubblefield E, Orro J. The isolation of specific chicken macrochromosomes by zonal centrifugation and flow sorting. Cytometry. 1982;2:273–281. doi: 10.1002/cyto.990020502. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Turner BM, Fellows G. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur J Biochem. 1989;179:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- Wade PA, Pruss D, Wolfe AP. Histone acetylation: Chromatin in action. Trends Biochem Sci. 1997;4:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]