Abstract
Glycosaminoglycans (GAGs) are anionic polysaccharides, which participate in key processes in the extracellular matrix by interactions with protein targets. Due to their charged nature, accurate consideration of electrostatic and water-mediated interactions is indispensable for understanding GAGs binding properties. However, solvent is often overlooked in molecular recognition studies. Here we analyze the abundance of solvent in GAG-protein interfaces and investigate the challenges of adding explicit solvent in GAG-protein docking experiments. We observe PDB GAG-protein interfaces being significantly more hydrated than protein–protein interfaces. Furthermore, by applying molecular dynamics approaches we estimate that about half of GAG-protein interactions are water-mediated. With a dataset of eleven GAG-protein complexes we analyze how solvent inclusion affects Autodock 3, eHiTs, MOE and FlexX docking. We develop an approach to de novo place explicit solvent into the binding site prior to docking, which uses the GRID program to predict positions of waters and to locate possible areas of solvent displacement upon ligand binding. To investigate how solvent placement affects docking performance, we compare these results with those obtained by taking into account information about the solvent position in the crystal structure. In general, we observe that inclusion of solvent improves the results obtained with these methods. Our data show that Autodock 3 performs best, though it experiences difficulties to quantitatively reproduce experimental data on specificity of heparin/heparan sulfate disaccharides binding to IL-8. Our work highlights the current challenges of introducing solvent in protein-GAGs recognition studies, which is crucial for exploiting the full potential of these molecules for rational engineering.
Electronic supplementary material
The online version of this article (doi:10.1007/s10822-011-9433-1) contains supplementary material, which is available to authorized users.
Keywords: Glycosaminoglycan-protein interfaces, GRID, Docking, Interfacial solvent, Molecular dynamics, SCOWLP
Introduction
Glycosaminoglycans (GAGs) represent a class of negatively charged heteropolysaccharides containing repeating disaccharides units. Each repeating unit consists of a hexose or a hexuronic acid linked to a hexosamine, while hydroxyl groups of hexose and hexosamine can be sulfated at different positions. Being localized in the extracellular matrix, GAGs participate in cell proliferation, regeneration, lipids metabolism, angiogenesis, and metastatis [1] by interactions with proteins such as growth factors [2–4], antithrombin [5], cytokines [6–8], cell adhesion molecules [9], and phospholipase A2 [10]. Natural and modified GAGs are of high interest for the design of biomaterials to be used to promote bio-specific cell behaviour in skin and bone tissue regeneration [11].
Computational approaches to study GAG-protein interactions face similar challenges as studies of protein interactions with other classes of saccharides because of high conformational flexibility of these molecules, indispensability of solvent for their interactions, lack of specialized tools for their molecular modelling and simulation, and due to a scarce availability of structural data on GAG-protein complexes. Although there are some general computational approaches applicable for identification of saccharide binding sites on proteins, the state of the art has not been developed as for protein-peptide recognition [12, 13]. Currently existing docking techniques, though so far not tuned for saccharides, seem to be promising for prediction and analysis of saccharide-protein interactions [14, 15], especially if they are combined with experimental data obtained from NMR [16]. Training a scoring function on a dataset of non-charged saccharides [17] and its implementation within a docking program (BALLDock/SLICK) have been shown to improve significantly the results of docking experiments for saccharides [18]. However, this program does not consider parameters for sulfate moieties, which are abundant in GAGs and, therefore cannot be yet used for docking GAGs and other sulfated saccharides. Another challenge for GAGs docking is their high symmetry in terms of orientations of reducing and non-reducing termini [19]. Besides that, due to their high conformational flexibility, only relatively short GAGs (up to tetramers) could be so far docked reliably without applying constraints or using experimental data for post-processing filtering of docking results. To be able to dock longer GAGs, docking of mono- and disaccharides could be useful as a first step in obtaining hints about possible binding poses [14]. Another important issue for docking GAGs to proteins is electrostatics. Due to the charged nature of GAGs, electrostatic interactions are especially important for GAG-protein recognition [20], which makes impact of GAGs hydration and water-mediated interactions crucial in their analysis by computational means [21]. Several studies have found tight interconnections between saccharides conformations and dynamical behaviour of the solvent surrounding them [22–24]. For instance, Sheehan’s group succeeded in explaning specific structural properties of GAGs by analyzing interactions of free GAGs with solvent [25–28]. In general, docking results for some other classes of ligands were shown to improve when crystal water molecules were explicitly included in the docking experiments as a part of the receptor [29, 30] or when water molecules were added by a Monte Carlo based solvated docking approach [31].
In this work, we perform a detailed analysis of solvent in GAG-protein interfaces at three different levels. Firstly, we analyze the abundance of solvent-mediated saccharides-protein and GAG-protein interactions in the PDB and compare them with similar available data on protein–protein interfaces. For this we use the SCOWLP database, which is based on the SCOP protein classification and contains detailed data on all protein interfaces from the PDB, including interfacial solvent (www.scowlp.org) [32]. In our previous work we performed statistical analysis of water-mediated interactions in protein interfaces based on the SCOWLP definition [32, 33] and characterized them from a dynamic and energetic point of view [34], showing indispensability of water-mediated interactions for a complete description of protein–protein interactions. The interacting solvent data obtained from SCOWLP also assisted to improve protein contact predictions [35]. We apply molecular dynamics (MD) approaches in order to further analyze the significance of water-mediated interactions in GAG-protein interfaces. Our findings emphasize the high abundance of water-mediated GAG-protein interactions and its significance for molecular recognition of GAGs. Then, we perform docking experiments with four different methods and compare the results obtained with and without explicit water molecules in the GAGs binding sites. In our studies we compare the docking results obtained by using solvent crystallographic data and the ones obtained by using de novo predicted water positions in the binding site. In general, we observe improvement in the docking results by inclusion of explicit water molecules at the protein binding site. The results we obtain underline the challenges for positioning of water molecules in the GAG binding sites and for the application of docking approaches for reproducing experimental data on short GAGs binding. Our work contributes to a better understanding of the GAG-protein interactions and of the role of solvent in GAG-protein recognition.
Methods
Analysis of GAG-protein and other saccharide-protein complexes
We used the SCOWLP database (www.scowlp.org) to extract structural data on saccharides-protein interfaces available in the PDB. SCOWLP consists of a SCOP-based classification of protein binding regions that takes into account interfacial solvent as a descriptor of protein interfaces. In SCOWLP all interfacial residues are divided into three classes: dry (direct interaction), dual (direct and water-mediated interactions), and wet spots (residues interacting only through one water molecule). The following types of interactions are defined in SCOWLP: hydrogen bonds, with distance donor/acceptor atom ≤3.6 Å; salt bridges, with charged atom distance ≤4 Å; van der Waals, with hydrophobic atoms at distance ≤4.5 Å [32].
Molecular dynamics simulations of GAG-protein complexes
We used experimental structures of two GAG-protein complexes: CD44 with heptameric hyaluronan (PDB ID: 2JCQ, 1.25 Å) and cathepsin K with hexameric chondroitin sulfate (PDB ID: 3C9E, 1.80 Å). MD simulations for these GAG-protein complexes were carried out with the AMBER 10.0 package [36] using ff03 force field parameters [37] for protein and GLYCAM06 for GAGs, respectively. Sulfate charges were obtained from the work of Huige et al. [38], and the corresponding to hyaluronan, chondroitin sulfate and heparin/heparan sulfate monosaccharide libraries were created using the LEaP tool of AMBER. GAG-protein complexes were solvated in a truncated octahedron periodic box filled with TIP3P water molecules and neutralized by counterions. MD simulations were preceded by two energy-minimization steps: 500 cycles of steepest descent and 103 cycles of conjugate gradient with harmonic force restraints on protein atoms, then 3 × 103 cycles of steepest descent and 3 × 103 cycles of conjugate gradient without constraints. This was followed by heating the system from 0 to 300 K for 10, and a 30 ps MD equilibration run at 300 K and 106 Pa in isothermal isobaric ensemble (NPT). Following the equilibration procedure, 10 ns of productive MD runs were carried out in periodic boundary conditions in NPT ensemble with Langevin temperature coupling with collision frequency parameter γ = 1 ps−1 and Berendsen pressure coupling with a time constant of 1.0 ps. The SHAKE algorithm was used to constrain all bonds that contain hydrogen atoms. A 2 fs time integration step was used. An 8 Å cutoff was applied to treat non-bonded interactions, and the Particle Mesh Ewald (PME) method was introduced for long-range electrostatic interactions treatment. The scaling parameters SCEE and SCNB were set to 1 as required for the use of the GLYCAM06 force field within AMBER [22]. Pyranose rings of β-d-glucuronic acid were restrained to be in 4C1 chair conformation since our tests MD simulations (data not shown) with unrestrained β-d-glucuronic acid demonstrated that the GLYCAM06 force field is unable to reproduce experimentally observed prevalence of 4C1 chair conformation [39].
While analyzing the trajectories we used the SCOWLP definition of residue interactions based on physico-chemical and distance criteria between atoms (described in previous section). Each frame of the trajectory was processed so that each residue in the system was described in terms of the relative time fractions (TFs) of total, dry, dual and wet spot interactions (TFT, TFD,TFd, Tfws) that they were establishing during the simulation [34]. The total interaction per residue was defined as the sum of all three defined interaction types. A residue was considered interacting when the total time of interaction was at least 5% of the simulation time. Energetic post-processing of the trajectories was done in a continuous solvent model as implemented in the AMBER 10.0 MM-GBSA (Molecular Mechanics- Generalized Born Surface Area) module. The snapshots for these calculations were chosen as described by Lafont and coworkers [40].
GAG-protein complexes docking
For our docking experiments we selected eleven GAG-protein interfaces from ten structures (the structure 3IN9 contains two different GAG-protein interfaces) from the PDB based on resolution criteria (≤2.2 Å) and size of the ligand (not longer than a tetramer). From the structures with the same protein we choose the one with the highest resolution as the representative to avoid redundancy (Table 1). We define binding site as all protein residues with at least one atom within a distance cutoff of 4.5 Å to the ligand in the crystal structure. Prior to docking calculations, ligands were extracted from the complex structure and minimized using the default procedure in MOE [41] with the Amber99 force field. Then, we calculated positions for water molecules in the protein binding sites. As reference, docking calculations were first carried out by taking into account the information about solvent placement from the crystal structure. Furthermore, we performed docking calculations without using this information and, therefore, de novo positioning solvent in the binding site:
Table 1.
GAG-protein complex structures used for the reference docking runs
| PDB ID | Res. (Å) | Description | GAG length | Crystal waters | GRID waters | Overlapc |
|---|---|---|---|---|---|---|
| 1DBO | 1.70 | Chondroitinase B + CSa | Dimer | 8 | 2 | 1 |
| 1OJN | 1.60 | Hyaluronate lyase + CSa | Dimer | 24 | 7 | 8 |
| 1RWH | 1.25 | Chondroitin lyase AC + CSa | Tetramer | 30 | 4 | 16 |
| 1G5N | 1.90 | Annexin V + HEb | Tetramer | 15 | 1 | 6 |
| 1T8U | 1.95 | 3-O-Sulfotransferase3 + HEb | Tetramer | 15 | 2 | 6 |
| 3E7J | 2.10 | Heparinase II + HEb | Tetramer | 17 | 2 | 5 |
| 2HYU | 1.42 | Annexin 2A + HEb | Tetramer | 35 | 1 | 9 |
| 2BRS | 2.20 | EMBP + HEb | Dimer | 3 | 2 | 0 |
| 1BFB | 1.90 | FGF2 + HEb | Tetramer | 0 | 5 | 0 |
| 3IN9_1 | 2.00 | Heparin lyase 1 + HEb | Dimer | 7 | 4 | 3 |
| 3IN9_2 | 2.00 | Heparin lyase 1 + HEb | Dimer | 7 | 5 | 3 |
aChondroitin sulfate
bHeparin/heparan sulfate
cThis number shows how many GRID-generated water molecules were discarded because of the overlap with crystal water molecules
1. Reference docking experiments: crystallographic water molecules in the binding site were considered part of the protein and were left for the docking calculations. We then used the GRID program [42] in order to account for additional water molecules that could be missing in the binding site. After running a GRID water probe on the protein surface, the grid points with the most negative energy values were chosen for placing a water oxygen. The GRID-generated water molecules with oxygen atoms closer than 2.8 Å to any of the crystal waters or to the ligand atoms were discarded. All the remained water molecules within a distance of 4.5 Å from ligands were considered explicitly for the docking calculations. This procedure, clearly biased due to the a priori knowledge of the crystal structures, is used in order to estimate to which extent the correct positioning of the water molecules in the binding site improves docking in comparison to docking with de novo positioning of solvent and also without explicit solvent.
2. Docking experiments with de novo positioning of solvent: in order to avoid biases due to the knowledge of the crystal structure, and as it is the case of de novo docking experiments, we removed all crystallographic water molecules and minimized the binding site using the AMBER 99 force field as implemented in MOE (combination of steepest descent, conjugate gradient and truncated Newton methods using 0.01 Å RMSD for convergence criteria). We then used the GRID program to position water molecules within the binding site. In addition, we used a GRID carbon sp3 probe (Csp3) as an approximation to account for solvent exclusion upon ligand binding. We discarded those predicted water molecules that were positioned within 1.5 Å to the minima of the Csp3 energy grid. This procedure is independent of the previous knowledge about positions of crystallographic water molecules. Furthermore, it considers certain relaxation in all atoms in the binding site upon ligand removal.
To perform docking experiments we have used the following methods: Autodock 3 [43], eHiTs [44], MOE docking [41], and FlexX [45]. In each of them, the adjustable parameters were optimized for GAG-protein complexes in the following way:
– Autodock 3. Atomic potential grid was calculated by autogrid3 with a 0.375 Å spacing in a box of size 15 × 15 × 15 Å for disaccharides and 18.75 × 18.75 × 18.75 Å for tetrasaccharides. Initially, the box was centered on the center of mass of the bound ligand in each of the crystal complexes, and then translated towards the binding surface to minimize its intersection with protein core residues and to enhance the sampling of the available space for ligand placement. Docking simulations were done with autodock3 using the genetic algorithm with 2.5 × 107 energy evaluations for disaccharides and 2.5 × 108 energy evaluations for tetrasaccharides.
– eHiTs. All default parameters were used except for: accuracy = 6; optimized for GAGs weights in scoring function: desol, depth were increased by 2, Lelec, Coulo were increased by 3 in order to favour electrostatic interactions.
– MOE docking. All default parameters with Triangle Matcher, retaining 105 poses and Alpha HB rescoring (with the equal weights for Hydrogen bonds and Alpha parameter) were used.
– FlexX. All default parameters for the FlexX 3.1 version with type 1 and type 3 water molecules were used. In comparison to type 1 water molecules, which presence and coordinates are user-defined, in case of use of FlexX type 3 water molecules, a receptor without explicit crystallographic and GRID-generated water molecules in the binding site was used, and the water molecules were added by the placement algorithm implemented in FlexX.
The docking experiments were firstly performed without explicit solvent. Additionally, new docking calculations were carried out with explicit solvent by using the predicted water molecules positioned prior to docking as described above. In both cases, the best ranked 50 solutions were used for further analysis. The results were described in terms of the best pose, amount of poses that qualitatively reproduced the crystal structure (defined by visual criteria and described as ‘correct pose’) and the pose with the closest RMSD to the crystal structure.
Comparison of docking scores with experimental data on GAGs disaccharides bound to IL-8
For comparison of the obtained GAGs docking results with experimental binding (Kd) data from isothermal fluorescence titration experiments [46] for IL-8 with heparin/heparan sulfate disaccharides (Idu(2S)-GlcNAc(6S); Idu-GlcNS(6S); Idu-GlcNS; Idu-GlcNAc; Idu(2S)-GlcNS; Idu-GlcN(6S)), which experimental crystal structures are not known, we did the following. We built the structures of these ligands using the crystal structure of heparin (PDB ID: 1HPN) as template, and prior to docking runs we minimized them using the default option in MOE with the Amber99 force field. The structure of monomeric IL-8 (PDB ID: 3IL8, 2.00 Å) was used as receptor, and the binding site was defined by H23, K25, R65, K69, K72, R73 residues according to literature mutagenesis data on the IL-8 heparin binding site [47]. To position explicit solvent in the binding site, crystallographic water molecules within 7 Å distance from the residues H23, K25, R65, K69, K72, R73 were taken into account together with the water molecules predicted by GRID. Then, we discarded those GRID-generated water molecules that were overlapping with crystallographic water molecules, and also all water molecules that were ovelapping with the energy minima points of the Csp3 grid. This resulted in a total of 15 water molecules added in the binding site (5 crystallographic and 10 GRID-generated). Using Autodock 3, we obtained the energies of the top scoring poses and the mean energy from the 150 top scoring poses calculated with and without explicit solvent. The mean energies were calculated as weighed means with the weights proportional to the probabilities of the energetic states of docking solutions, and were compared with the experimentally obtained Kd values.
Data analysis and its graphical representation were done with the use of the R-package [48].
Results
GAG-protein and saccharides-protein complexes in the PDB
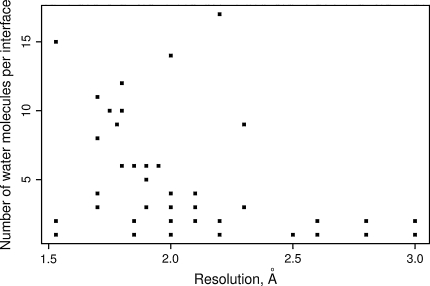
Using SCOWLP we have obtained 1,910 saccharide-protein (excluding GAG-protein) interfaces represented by 715 crystal structures, and 57 GAG-protein interfaces from 31 crystal structures. The hydration level of these GAG-protein interfaces versus their crystal structure resolution is plotted in Fig. 1. In order to describe quantatively if the hydration of these interfaces is different to other types of interfaces, we normalize the number of water molecules by interface area. When comparing GAG-protein interfaces and the rest of saccharides-protein interfaces (Table 2), we do not find significant differences (t Test), though GAG-protein interfaces are expected to be more hydrated because of their charged nature. This absence of differences is possibly due to the low number of currently available GAG-protein structures in the PDB. The comparison with the data obtained for a manually curated protein–protein interfaces dataset with resolution <2.00 Å [33] shows that both GAG-protein and other saccharides-protein interfaces are significantly more hydrated.
Fig. 1.
Dependence of water molecules abundance in GAG-protein interfaces on crystal structure resolution
Table 2.
Hydration of GAG-protein, other saccharide-protein and protein–protein interfaces
| Interfaces dataset | Number of interfaces | Water molecules/interface area (1/1000 Å2) |
|---|---|---|
| GAG-protein | 57 | 10.8 |
| Saccharide–protein (not GAGs) | 1,910 | 9.5 |
| Protein–protein [33] | 176 | 3 |
MD analysis of protein-GAGs complexes
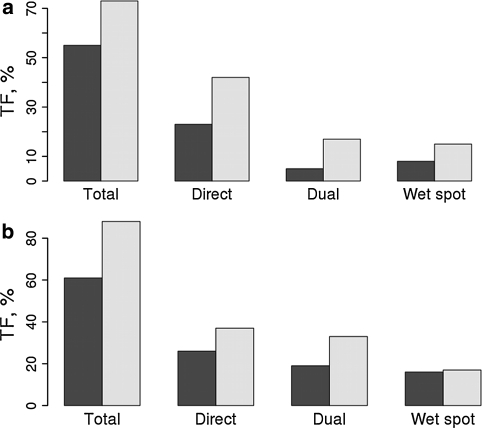
We have run two MD simulations to define the abundance of water-mediated interaction in GAG-protein interfaces from a dynamical point of view. Time fractions of interactions obtained for CD44-hyaluronan and cathepsin K-chondroitin sulfate complexes are shown in Fig. 2. Half of the interactions in the first system and even more in the second one are water-mediated, which is qualitatively similar to the time fractions of interactions found previously for protein–protein interfaces [34]. However, when analyzing the corresponding structures in SCOWLP, we find that the amount of dry, dual and wet spot interactions are 19, 0, 3 and 8, 7, 9 for 2JCQ and 3CE9, respectively. This example illustrates the importance of dynamics-based studies of hydration of the protein interfaces.
Fig. 2.
Time fractions of interactions per protein (dark grey) and per GAGs (light grey) residues for CD44—HA (a); Cathepsin K—CS (b) complexes
From the energetic point of view these two complexes represent very different binding modes. According to MM-PBSA free energy calculations, cathepsin K demostrates strong electrostatically driven binding with the ratio between electrostatic and van der Waals component of 40, while the electrostatic component in vacuo for CD44 is positive and close to the van der Waals component by absolute value. At the same time, both complexes have very similar van der Waals as well as GB surface energies (Table 3). Therefore, despite these substantial differences in the electrostatics participation in binding observed for these two complexes, the amount of water-mediated interactions is similar. This suggests that solvent plays an active role in GAG-protein interfaces, even for the interfaces where total electrostatic contribution is not the driving force for complex formation. We assume that besides direct electrostatic impact on the intermolecular interaction, water molecules can still play an important role by at least two other mechanisms. First, tightly bound water molecules may contribute to define the binding site geometry and, hence have an effect on binding. In addition, water molecules reorganization upon ligand binding greatly affects the entropic component of the binding energy, which is not explicitly considered by continuum solvent approaches as MM-PBSA and, therefore is not detectable in these calculations.
Table 3.
MM-PBSA free energy decomposition for CD44—HA and Cathepsin K—CS complexes
| Complex/component | Free energy (kcal/mol) | |||||||
|---|---|---|---|---|---|---|---|---|
| ELE | VDW | GAS | PBSUR | PB | PBSOL | PBELE | PBTOT | |
| CD44—HA | 34.2 | −35.7 | −1.5 | −3.1 | −14.1 | −17.3 | 20.0 | −18.8 |
| Cathepsin K—CS | −1399.2 | −35.0 | 0.0 | −5.1 | 1398.9 | 1393.7 | −0.4 | −40.5 |
Energy components: ELE electrostatic, VDW van der Waals, GAS full energy in gas phase (ELE + VDW), PBSUR hydrophobic contribution to solvation, PB reaction field calculated by PB, PBSOL full solvation (PBSUR + PB), PBELE sum of electrostatic energy in vacuo and reaction field energy (PB + ELE), PBTOT total energy (PBSOL + GAS)
Docking of GAGs to “dry” and “wet” binding sites of proteins
We compared the results obtained in our docking experiments performed with four different methods with and without explicit solvent (see details in the Methods section) in terms of the following docking quality parameters: RMSD between the experimental structure and the top scoring pose; the lowest RMSD obtained in any of the poses; the rank of the pose with the lowest RMSD; and the number of correct poses found within 50 and 10 top scoring solutions.
One of the most important current challenges in docking with explicit solvent is the correct placement of the water molecules in the binding site. This is due to the fact that two important aspects need to be taken into account. First, some solvent molecules are displaced from the binding site upon ligand binding. Second, some solvent molecules play a bridging role between ligand and protein. In de novo docking it is very challenging to predict and position these water molecules a priori. As reference, we first performed docking experiments using crystallographic information on solvent positioning to discard predicted waters in the binding site. In the studied complexes, some of the GRID-predicted waters (total of 99) could be rejected because of their overlapping with crystal waters (a total of 35), and some others because of their overlapping with the ligand (a total of 17). Although it is clear that favourable positions for solvent in the binding site in a complex and in an unbound protein differ, the considerable proportion of mutual water overlapping obtained justifies the use of GRID to predict some of the important waters that form the “ligand water bed” in the binding site (Table 1). As for the overlapping of predicted waters with positions that are eventually occupied by the ligands in the complexed structures, the approximation we used for taking into account possible “solvent exclusion” upon ligand binding (energy grids obtained with a Csp3 probe; see Methods section for details) shows that a substantial amount of these waters can be detected (total of 12). Therefore, these observations define the Csp3 grids as a valid approximation for exclusion of ligand-overlapping water molecules in the studied cases.
In these reference docking experiments, Autodock 3 performed very well (Supplementary Table 1). The average RMSD for the top scoring poses are 1.94 and 1.60 Å for the binding sites without and with explicit water molecules in the binding site, respectively (Fig. 3a). The lowest RMSD poses are very close in ranks and values independently of water molecules presence (Fig. 3b, c). Improvement in the results by inclusion of solvent is observed for the number of correct poses within the 50 top scoring poses, and even more evident for number of correct poses within 10 top scoring poses (Fig. 3d, e). eHiTs performed significantly worse than Autodock 3 (Supplementary Table 2) in terms of RMSD for the top scoring poses (Fig. 3b). The lowest RMSD values are still mainly ≤3 Å, though the ranks of the corresponding poses are lower than in case of Autodock 3 (Fig. 3b, c). Also the number of correctly found poses is significantly lower than for Autodock 3 in both 50 and 10 top scoring poses (Fig. 3d, e). Addition of water molecules to the binding site does not change RMSD of the top scoring poses but slightly improves the performance in terms of the lowest RMSD and number of correctly found poses. This improvement is the most evident for the ranking of the lowest RMSD pose. MOE docking did not yield good results in terms of the RMSD of the top scoring pose (Supplementary Table 3; Fig. 3a). In terms of other docking quality parameters, MOE docking had similar performance to eHiTs with the exception that the number of correctly found poses within the 10 top scoring solutions was in particular low for docking without explicit solvent. MOE docking results clearly improved when the water molecules were added to the binding site, especially the ranking of the pose with the lowest RMSD. FlexX performed significantly worse than the other tested programs (Supplementary Table 4; Fig. 3). Only in three complexes out of eleven it found correct poses within the 50 top solutions when no water molecules were added, and only in two when FlexX type 3 water molecules were used. This suggests that use of the explicit water molecules implemented in FlexX does not improve the results for GAG-protein systems. Yet FlexX improves its performance almost up to the level of the performance of eHiTs and MOE when explicit crystallographic and GRID-generated water molecules are used. The fact that FlexX performs worse for GAG-protein complexes than other tested programs could be explained by its so-called ‘anchor-and-grow’ algorithm of ligand’s placement, in which first a part of a ligand is placed in its energetical minimum pose, and then the rest of ligand is grown using the already docked part as an anchor. This strategy could be expected to be unsuccessful in cases where the interactions are electrostatics-driven and the ligand is symmetric and repetitive, as it is in the case of GAGs.
Fig. 3.
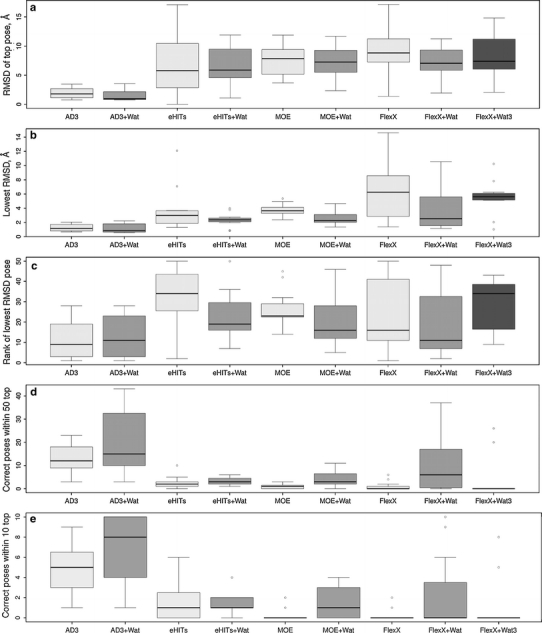
Comparison of the reference docking experiments of Autodock 3, eHiTs, MOE docking and FlexX for 11 complexes: a RMSD of top scoring pose; b Lowest RMSD within 50 top poses; c Rank of the pose with the lowest RMSD in 50 top poses; d Number of correct poses in 50 top poses; e Number of correct poses in 10 top poses. ‘+Wat’ relates to the runs with explicit water molecules. ‘+ Wat3’ relates to the type 3 of FlexX water
Based on the analysis of the docking quality parameters for the reference docking experiments, the performance of the programs improves in the following order, independently of explicit presence of water molecules in the binding site: FlexX < MOE < eHiTs < Autodock 3 (Fig. 3a, b, c, d, e, respectively; Supplementary Material Tables 1–5; Tables 5 and 6). That suggests that Autodock 3 is highly reliable for docking short GAGs to proteins both with and without explicitly added water molecules in the binding site. The other three programs used here are very much complex-dependent (Supplementary Tables 1–5), and their results should not be taken for granted alone without cross-checking with the data obtained by other docking approaches. In summary, in the reference docking experiments all four tested docking programs perform significantly better with than without explicit water molecules in the protein binding site.
Table 5.
Autodock 3 performance with de novo solvent placement
| PDB ID | W− top posea RMSD (Å) | W+ top poseb RMSD (Å) | W− best pose ranka (RMSD (Å)) | W+ best pose rankb (RMSD (Å)) | W− correct posesa in top 50 | W+ correct posesb in top 50 | W− correct posesa in top 10 | W+ correct posesb in top 10 |
|---|---|---|---|---|---|---|---|---|
| 1DBO | 1.93 | 1.42 | 42 (1.27) | 11 (1.06) | 15 | 27 | 0 | 6 |
| 1OJN | 2.90 | 1.37 | 8 (1.11) | 2 (1.13) | 6 | 9 | 4 | 6 |
| 1RWH | 1.99 | 1.13 | 7 (0.97) | 2 (0.87) | 8 | 13 | 7 | 9 |
| 1G5N | 4.41 | 4.28 | 29 (1.48) | 27 (1.82) | 5 | 5 | 1 | 1 |
| 1T8U | 2.10 | 3.57 | 2 (1.84) | 3 (2.27) | 4 | 2 | 3 | 1 |
| 3E7J | 1.79 | 1.20 | 1 (1.79) | 1 (1.20) | 5 | 3 | 2 | 3 |
| 2HYU | 2.26 | 3.60 | 16 (1.91) | 4 (2.29) | 8 | 7 | 2 | 3 |
| 2BRS | 2.77 | 3.01 | 34 (1.13) | 15 (1.45) | 8 | 16 | 0 | 8 |
| 1BFB | 3.35 | 3.58 | 14 (1.88) | 6 (2.31) | 5 | 3 | 4 | 1 |
| 3IN9_1 | 3.54 | 2.75 | 32 (0.87) | 34 (0.87) | 20 | 16 | 0 | 2 |
| 3IN9_2 | 1.18 | 1.22 | 22 (0.99) | 13 (0.85) | 32 | 43 | 9 | 10 |
| Mean | 2.57 ± 0.93 | 2.47 ± 1.21 | 19 (1.39 ± 0.41) | 11 (1.47 ± 0.60) | 10.5 | 13.1 | 2.9 | 4.5 |
aDocking runs without explicit solvent
bDocking runs with explicit solvent
Table 6.
eHiTs performance with de novo solvent placement
| PDB ID | W− top posea RMSD (Å) | W+ top poseb RMSD (Å) | W− best pose ranka (RMSD (Å)) | W+ best pose rankb (RMSD (Å)) | W− correct posesa in top 50 | W+ correct posesb in top 50 | W− correct posesa in top 10 | W+ correct posesb in top 10 |
|---|---|---|---|---|---|---|---|---|
| 1DBO | 3.14 | 4.58 | 13 (1.67) | 4 (1.46) | 3 | 4 | 2 | 2 |
| 1OJN | 5.79 | 6.83 | 41 (1.32) | 46 (1.51) | 4 | 8 | 3 | 5 |
| 1RWH | 9.35 | 1.99 | 19 (2.56) | 42 (1.68) | 2 | 4 | 0 | 1 |
| 1G5N | 10.97 | 5.71 | 38 (5.68) | 43 (4.53) | 0 | 0 | 0 | 0 |
| 1T8U | 11.25 | 3.27 | 50 (4.91) | 37 (2.92) | 0 | 6 | 0 | 2 |
| 3E7J | 11.47 | 1.65 | 18 (2.02) | 8 (0.89) | 1 | 9 | 0 | 8 |
| 2HYU | 11.00 | 4.99 | 41 (6.62) | 49 (3.31) | 0 | 3 | 0 | 1 |
| 2BRS | 7.74 | 5.42 | 4 (2.07) | 28 (2.05) | 5 | 4 | 3 | 1 |
| 1BFB | 11.53 | 7.15 | 50 (5.57) | 30 (5.55) | 0 | 0 | 0 | 0 |
| 3IN9_1 | 6.43 | 5.57 | 44 (4.37) | 48 (4.22) | 0 | 0 | 0 | 0 |
| 3IN9_2 | 5.47 | 7.59 | 32 (2.98) | 27 (4.15) | 1 | 0 | 0 | 0 |
| Mean | 8.55 ± 2.98 | 4.97 ± 1.98 | 32 (3.61 ± 1.87) | 33 (2.93 ± 1.53) | 1.5 | 3.5 | 0.7 | 1.8 |
aDocking runs without explicit solvent
bDocking runs with explicit solvent
For the de novo docking calculations, ligand and solvent were removed from the initial crystal structures, no crystallographic data on solvent positioning was used, and the structure of the non-occupied binding site was minimized (see Methods section). In this case, only GRID-predicted waters (a total of 117) in combination with the “solvent exclusion” Csp3 probe (a total of 20 overlapping waters were discarded) were used for de novo solvent placement (Table 4). We carried out our calculations with Autodock 3 and eHiTs, as these two methods were performing best in our reference docking experiments (see above). With explicit solvent, Autodock 3 performs slightly better than without solvent for the top binding pose and for the number of correctly predicted binding poses in the 50 top solutions, and it performs significantly better for ranks of the best pose and for the number of correctly predicted binding poses in the 10 top solutions. However, with explicit solvent Autodock 3 shows a slight increase in RMSD of the top pose (Table 5). Energies of the top solutions for the docking without solvent are in general more favourable than for the corresponding solutions for the docking with solvent, which indicates that the Autodock 3 scoring function considers water-mediated interactions to be weaker than direct ones. For eHiTs all docking quality parameters significantly improve when explicit water molecules are used (Table 6).
Table 4.
De novo water molecules placement
| PDB ID | De novo watersa | Excluded watersb | Overlap with ligandc | RMSD bck/all (Å)d |
|---|---|---|---|---|
| 1DBO | 2 | 0 | 0 | 0.027/0.224 |
| 1OJN | 11 | 0 | 0 | 0.026/0.238 |
| 1RWH | 15 | 2 | 1 (1.5 Å) | 0.024/0.129 |
| 1G5N | 8 | 3 | 1 (1.6 Å) | 0.187/0.261 |
| 1T8U | 8 | 2 | 0 | 0.034/0.192 |
| 3E7J | 8 | 2 | 0 | 0.049/0.169 |
| 2HYU | 9 | 3 | 1 (1.1 Å) | 0.166/0.306 |
| 2BRS | 7 | 1 | 1 (0.9 Å) | 0.093/0.337 |
| 1BFB | 13 | 4 | 2 (0.7, 1.5 Å) | 0.113/0.317 |
| 3IN9_1 | 7 | 2 | 0 | 0.041/0.252 |
| 3IN9_2 | 9 | 1 | 0 | 0.071/0.237 |
aTotal number of GRID-generated water molecules used in de novo docking experiments
bNumber of water molecules excluded by use of Csp3 atomic probe
cNumber of water molecules overlapping with ligand in crystal structure in the docking experiments (distance to ligand)
dRMSD is calculated for backbone and all atoms for receptor binding site residues after minimization in comparison to the initial crystal structure of each complex
Analyzing all these data, it is very important to be aware that the comparison of the docking results with and without explicit solvent in the binding site, as it is done in this study, is not strictly equivalent to the comparison of the systems in vacuo with and without a fixed number of explicitly added water molecules. If fact, most of the modern docking programs, including the programs we used, already implicitly take into account effects of solvation in a certain way. Therefore, the analysis provided here carries conceptual and qualitative rather than methodological and quantitative meaning. We show that the accurate placement of explicit solvent into the systems, which already take into account hydration implicitly, could still potentially improve docking results. In this context, our observations are in agreement with the work of Wong et al., which demonstrated that inclusion of explicit water molecules in implicit solvent model for MM-PBSA free energy calculations leads to a better agreement with experimental data [49].
Considering the challenges of accurate water molecules positioning prior to docking, and based on our observations, we believe that docking highly charged molecules, GAGs in particular, should include a on the fly sampling step accounting for solvent. At each sampling step, water molecules should be added into energetically favourable positions in the binding site instead of fixing them prior to docking. A similar sampling procedure using a Monte Carlo approach has been proposed for protein–protein docking by van Dijk et al. [31]. Although this kind of sampling is prone to increase the computational expenses, it would offer good possibilities to improve docking performance.
Docking GAGs-disaccharides to IL-8 monomer
We run docking experiments for six heparin/heparan sulfate disaccharides using monomeric IL-8 as a receptor to compare our results with available binding experimental data [46] quantitatively. We used Autodock 3, as it performed best for docking GAGs in our tests. IL-8 heparin binding site is significantly bigger than the size of the used disaccharides, which makes it a good system to draw a conclusion about the computational abilities of the docking approach to observe the specificity of predicted binding poses of disaccharides. According to site-directed mutagenesis, the heparin binding site of IL-8 is comprised of positively charged residues localized on the C-terminal α-helix and in the proximal loop (H23, K25, R65, K69, K72, R73) [47]. Previous computational studies also indicate the importance of these residues for heparin binding [14, 46]. Our results show that there is no preference towards any specific binding pose independently of presence of water molecules in the binding site, and the bound poses, though structurally very different (Fig. 4), do not differ in scoring significantly. Clustering of the obtained docking poses also does not indicate any trend for binding specificity. According to our docking results, the average cluster size for the biggest three clusters is eigth members for 150 top poses when clustering is done at the level of 2 Å RMSD. Only for three out of six disaccharides one of the highly ranked clusters is within the biggest three clusters, which means poor clustering of the docking solutions in general (Table 7). We also calculated docking energy values for the top scoring pose and average energy of the ensemble of retained 150 top poses, which was calculated as weighed mean with the weights proportional to the probabilities of the energetical states of docking solutions, and compared them with the thermodynamic data for these six heparin/heparan sulfate disaccharides obtained by isothermal fluorescent titration [46]. While experimental data demonstrate high sulfate-position dependent specificity of disaccharides recognition by IL-8, we do find neither this nor any correlation between experimental data and the calculated docking energies. This shows the challenges that Autodock 3 has to distinguish the specificity of GAGs disaccharides binding to the IL-8 heparin binding site by only scoring and ranking the solutions.
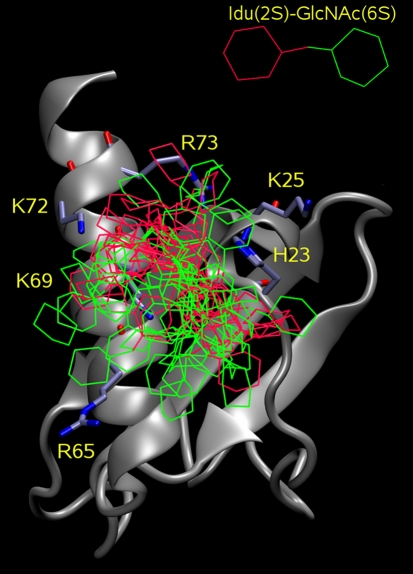
Fig. 4.
Results for the docking of Idu(2S)-GlcNAc(6S) to IL-8 with Autodock 3: 50 top docking solutions. The residues of heparin binding site are labeled and shown in licorice, the pyranose rings of disaccharides are in lines: red—Ido(2S) and green—GlcNAc(6S)
Table 7.
Clustering 150 docking solutions for disaccharides in IL-8 heparin binding site (RMSD = 2 Å)
| Disaccharides | CLUSTERS_WATa (−) | CLUSTERS_WATb (+) |
|---|---|---|
| Idu(2S)-GlcNAc(6S) | 13 (7), 27 (6), 43 (5) | 28 (10), 18 (8), 11 (5) |
| Idu-GlcNS(6S) | 10 (14), 12 (10), 37 (10) | 13 (30), 10 (17), 8 (12) |
| Idu-GlcNS | 64 (9), 42 (7), 1 (6) | 38 (10), 9 (8), 27 (7) |
| Idu-GlcNAc | 1 (8), 3 (7), 71 (6) | 2 (7), 19 (7), 27 (7) |
| Idu(2S)-GlcNS | 2 (7), 16 (5), 51 (5) | 14 (8), 38 (6), 2 (5) |
| Idu-GlcN(6S) | 10 (22), 17 (9), 34 (6) | 10 (24), 32 (13), 7 (9) |
aRanks of three biggest clusters (number of solutions in cluster) when docking with no explicit water molecules
bRanks of three biggest clusters (number of solutions in cluster) when docking with explicit water molecules
To define how much the results for docking GAGs disaccharides to IL-8 are influenced by electrostatic interactions, we carried out additional docking calculations with other GAGs-disaccharides: hyaluronic acid derivatives (GlcUA-GlcNAc, GlcUA-GlcNAc(6S), GlcUA(2S)-GlcNAc(4,6S), GlcUA(3S)-GlcNAc(4,6S)) and chondroitin sulfates (GlcUA-GalNAc(4S), GlcUA-GalNAc(6S)). For these disaccharides and the above-mentioned heparin/heparan sulfate disaccharides, we found a strong correlation between the docking energies and the charge of the disaccharides, which is higher for docking with explicitly added water molecules (adjusted correlated coefficient R 2 is 0.71 and 0.99, respectively). This may be attributed to a strong electrostatic impact for binding, which could explain the limitation of Autodock 3 ability, as well as docking approaches in general, to find specific solutions for GAGs disaccharides in a relatively large binding site comprising of many positively charged residues. In case of such systems, high electrostatic impact guides docking to yield unspecific binding modes within the top scoring poses.
Conclusion
Due to the key role of GAGs in intercellular communication processes, a good understanding of the rules governing their molecular recognition is crucial for exploiting their full potential to be used in rational engineering. Particularly, an important aspect in these lines is the role of solvent in mediating GAG-protein interactions. In this work we analyze the abundance of water molecules in GAG-protein interfaces and investigate the challenges of adding explicit water molecules in GAG-protein docking experiments. We find that GAG-protein interfaces are more hydrated than protein–protein interfaces, and we observe that, from a dynamic point of view, half of interactions in GAG-protein interfaces are water-mediated. We carry out a reference docking study with Autodock 3, eHiTs, MOE and FlexX for a dataset of GAG-protein complexes to investigate how solvent positioning in the binding site affects docking performance. We use the GRID program to de novo predict positions of water molecules in the binding site and to calculate possible areas of solvent displacement upon ligand binding. By using this GRID-based procedure of de novo solvent placement we achieve slight improvements in docking performance in comparison to docking results obtained without explicit solvent. Among the used docking methods, we observe that Autodock 3 performs best. In the analysis of the docking results of GAG-disaccharides and IL-8 in terms of energies of the best scoring poses, we notice that Autodock 3 yields very unspecific results for this system, which do not correlate with thermodynamic experimental data and are strongly biased towards electrostatic interactions.
Our study underlines the importance of water molecules in GAG-protein recognition, and it suggests the need for novel docking approaches for GAG-protein systems that should take into account proper localization and energetic properties of interfacial solvent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work has been funded by the German Research Council SFB-TRR 67. We would like to thank Ralf Gey for technical assistance, Orr Ravitz (SimBioSys Inc.) for technical support with eHiTs and Kristina Grabowski (Chemical Computing Group) for technical support with MOE.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Sergey A. Samsonov, Phone: +49-351-46340071, FAX: +49-351-46340087, Email: sergeys@biotec.tu-dresden.de
M. Teresa Pisabarro, Email: mayte@biotec.tu-dresden.de.
References
- 1.Jackson R, Busch S, Cardin A. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Phys Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 2.Angulo J, Ojeda R, de Paz J, Lucas R, Nieto P, Lozano R, Redondo-Horcajo M, Giménez-Gallego G, Martín-Lomas M (2004) The activation of fibroblast growth factors (FGFs) by Glycosaminoglycans: influence of the sulfation pattern on the biological activity of FGF-1. Chem Bio Chem 5:55–61 [DOI] [PubMed]
- 3.Faham S, Hileman R, Fromm J, Linhardt R, Rees D. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 4.Hintze V, Moeller S, Schnabelrauch M, Bierbaum S, Viola M, Worch H, Scharnweber D. Modifications of hyaluronan influence the interaction with human bone morphogenetic protein-4 (hBMP-4) Biomacromolecules. 2009;10:3290–3297. doi: 10.1021/bm9008827. [DOI] [PubMed] [Google Scholar]
- 5.Dementiev A, Petitou M, Herbert J, Gettins P. The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity. Nat struct Mole Biol. 2004;11:863–867. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- 6.Salek-Ardakani S, Arrand J, Shaw D, Mackett M. Heparin and heparan sulfate bind interleukin-10 and modulate its activity. Blood. 2000;96:1879–1888. [PubMed] [Google Scholar]
- 7.Yoon SI, Logsdon N, Sheikh F, Donnelly R, Walter M. Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J Biol Chem. 2006;281:35088–35096. doi: 10.1074/jbc.M606791200. [DOI] [PubMed] [Google Scholar]
- 8.Lamoureux F, Picarda G, Garrigue-Antar L, Baud’huin M, Trichet V, Vidal A, Miot-Noirault E, Pitard B, Heymann D, Redini F. Glycosaminoglycans as potential regulators of osteoprotegerin therapeutic activity in osteosarcoma. Cancer Res. 2009;69:526–536. doi: 10.1158/0008-5472.CAN-08-2648. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi N, Coombe D, Mancera R. Platelet endothelial cell adhesion molecule 1 (PECAM-1) and its interactions with glycosaminoglycans: 1. Molecular modeling studies. Biochemistry. 2008;47:4851–4862. doi: 10.1021/bi702455e. [DOI] [PubMed] [Google Scholar]
- 10.Sartipy P, Johansen B, Camejo G, Rosengren B, Bondjers G, Hurt-Camejo E. Binding of human phospholipase A2 type II to proteoglycans. Differential effect of glycosaminoglycans on enzyme activity. J Biol Chem. 1996;271:26307–26314. doi: 10.1074/jbc.271.42.26307. [DOI] [PubMed] [Google Scholar]
- 11.Dieckmann C, Renner R, Milkova L, Simon J. Regenerative medicine in dermatology: biomaterials, tissue engineering, stem cells, gene transfer and beyond. Exp Dermatol. 2010;19:697–706. doi: 10.1111/j.1600-0625.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 12.Woods R, Tessier M. Computational glycoscience: characterizing the spatial and temporal properties of glycans and glycan-protein complexes. Curr Opin Struct Biol. 2010;20:575–583. doi: 10.1016/j.sbi.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taroni C, Jones S, Thornton J. Analysis and prediction of carbohydrate binding sites. Prot Eng. 2000;13:89–98. doi: 10.1093/protein/13.2.89. [DOI] [PubMed] [Google Scholar]
- 14.Bitomsky W, Wade R. Docking of Glycosaminoglycans to heparin-binding proteins: validation for aFGF, bFGF, and antithrombin and application to IL-8. J Am Chem Soc. 1999;121:3004–3013. doi: 10.1021/ja983319g. [DOI] [Google Scholar]
- 15.Agostino M, Jene C, Boyle T, Ramsland P, Yuriev E. Molecular docking of carbohydrate ligands to antibodies: structural validation against crystal structures. J Chem Info Model. 2009;49:2749–2760. doi: 10.1021/ci900388a. [DOI] [PubMed] [Google Scholar]
- 16.Guerrini M, Guglieri S, Casu B, Torri G, Mourier P, Boudier C, Viskov C. Antithrombin-binding octasaccharides and role of extensions of the active pentasaccharide sequence in the specificity and strength of interaction. Evidence for very high affinity induced by an unusual glucuronic acid residue. J Biol Chem. 2008;283:26662–26675. doi: 10.1074/jbc.M801102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerzmann A, Neumann D, Kohlbacher O. SLICK—scoring and energy functions for protein-carbohydrate interactions. J Chem Inf Model. 2006;46:1635–1642. doi: 10.1021/ci050422y. [DOI] [PubMed] [Google Scholar]
- 18.Kerzmann A, Fuhrmann J, Kohlbacher O, Neumann D. BALLDock/SLICK: a new method for protein-carbohydrate docking. J Chem Inf Model. 2008;48:1616–1625. doi: 10.1021/ci800103u. [DOI] [PubMed] [Google Scholar]
- 19.Forster M, Mulloy B. Computational approaches to the identification of heparin-binding sites on the surfaces of proteins. Biochem Soc Trans. 2006;34:431–434. doi: 10.1042/BST0340431. [DOI] [PubMed] [Google Scholar]
- 20.Mulloy B. The specificity of interactions between proteins and sulfated polysaccharides. Anais da Academia Brasileira de Ciências. 2005;77:651–664. doi: 10.1590/S0001-37652005000400007. [DOI] [PubMed] [Google Scholar]
- 21.Baron R, Setny P, Andrew McCammon J. Water in cavity—ligand recognition. J Am Chem Soc. 2010;132:12091–12097. doi: 10.1021/ja1050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirschner K, Yongye A, Tschampel S, González-Outeiriño J, Daniels C, Foley L, Woods R. GLYCAM06: a generalizable biomolecular force field. Carbohydr J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Brady J. Anisotropic solvent structuring in aqueous sugar solutions. J Am Chem Soc. 1996;118:12276–12286. doi: 10.1021/ja962108d. [DOI] [Google Scholar]
- 24.Pagnotta S, McLain S, Soper A, Bruni F, Ricci M. Water and trehalose: how much do they interact with each other? J Phys Chem B. 2010;114:4904–4908. doi: 10.1021/jp911940h. [DOI] [PubMed] [Google Scholar]
- 25.Almond A, Sheehan J, Brass A. Molecular dynamics simulations of the two disaccharides of hyaluronan in aqueous solution. Glycobiology. 1997;7:597–604. doi: 10.1093/glycob/7.5.597. [DOI] [PubMed] [Google Scholar]
- 26.Almond A, Brass A, Sheehan J. Deducing polymeric structure from aqueous molecular dynamics simulations of oligosaccharides: predictions from simulations of hyaluronan tetrasaccharides compared with hydrodynamic and X-ray fibre diffraction data. J Mole Biol. 1998;284:1425–1437. doi: 10.1006/jmbi.1998.2245. [DOI] [PubMed] [Google Scholar]
- 27.Almond A, Sheehan J. Glycosaminoglycan conformation: do aqueous molecular dynamics simulations agree with x-ray fiber diffraction? Glycobiology. 2000;10:329–338. doi: 10.1093/glycob/10.3.329. [DOI] [PubMed] [Google Scholar]
- 28.Almond A, Sheehan J. Predicting the molecular shape of polysaccharides from dynamic interactions with water. Glycobiology. 2003;13:255–264. doi: 10.1093/glycob/cwg031. [DOI] [PubMed] [Google Scholar]
- 29.Roberts B, Mancera R. Ligand-protein docking with water molecules. J Chem Inf Model. 2008;48:397–408. doi: 10.1021/ci700285e. [DOI] [PubMed] [Google Scholar]
- 30.Thilagavathi R, Mancera R. Ligand-protein cross-docking with water molecules. J Chem Inf Model. 2010;50:415–421. doi: 10.1021/ci900345h. [DOI] [PubMed] [Google Scholar]
- 31.van Dijk A, Bonvin A. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22:2340–2347. doi: 10.1093/bioinformatics/btl395. [DOI] [PubMed] [Google Scholar]
- 32.Teyra J, Doms A, Schroeder M, Pisabarro M. SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces. BMC Bioinf. 2006;7:104. doi: 10.1186/1471-2105-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teyra J, Pisabarro M. Characterization of interfacial solvent in protein complexes and contribution of wet spots to the interface description. Proteins Struct Funct Bioinf. 2007;67:1087–1095. doi: 10.1002/prot.21394. [DOI] [PubMed] [Google Scholar]
- 34.Samsonov S, Teyra J, Pisabarro TM. A molecular dynamics approach to study the importance of solvent in protein interactions. Proteins Struct Funct Bioinf. 2008;73:515–525. doi: 10.1002/prot.22076. [DOI] [PubMed] [Google Scholar]
- 35.Samsonov S, Teyra J, Anders G, Pisabarro T. Analysis of the impact of solvent on contacts prediction in proteins. BMC Struct Biol. 2009;9:22. doi: 10.1186/1472-6807-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8 molecular dynamics package. http://ambermd.org
- 37.Duan Y, Wu C, Chowdhury S, Lee M, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 38.Huige C, Altona C. Force field parameters for sulfates and sulfamates based on ab initio calculations: Extensions of AMBER and CHARMm fields. J Comput Chem. 1995;16:56–79. doi: 10.1002/jcc.540160106. [DOI] [Google Scholar]
- 39.Sattelle B, Almond A. Less is more when simulating unsulfated glycosaminoglycan 3D-structure: Comparison of GLYCAM06/TIP3P, PM3-CARB1/TIP3P, and SCC-DFTB-D/TIP3P predictions with experiment. J Comput Chem. 2010;31:2932–2947. doi: 10.1002/jcc.21589. [DOI] [PubMed] [Google Scholar]
- 40.Lafont V, Schaefer M, Stote R, Altschuh D, Dejaegere A. Protein-protein recognition and interaction hot spots in an antigen-antibody complex: free energy decomposition identifies “efficient amino acids”. Proteins Struct Funct Bioinf. 2007;67:418–434. doi: 10.1002/prot.21259. [DOI] [PubMed] [Google Scholar]
- 41.Chemical Computing Group Inc. MOE v2005.06
- 42.Goodford P. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 43.Morris G, Goodsell D, Halliday R, Huey R, Hart W, Belew R, Olson A. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1999;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 44.Zsoldos Z, Reid D, Simon A, Sadjad S, Johnson P. eHiTs: A new fast, exhaustive flexible ligand docking system. J Mole GraphModel. 2007;26:198–212. doi: 10.1016/j.jmgm.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mole Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 46.Krieger E, Geretti E, Brandner B, Goger B, Wells T, Kungl A. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: support from isothermal fluorescence titrations. Proteins Struct Funct Bioinf. 2004;54:768–775. doi: 10.1002/prot.10590. [DOI] [PubMed] [Google Scholar]
- 47.Kuschert G, Hoogewerf A, Proudfoot A, Chung C, Cooke R, Hubbard R, Wells T, Sanderson P. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- 48.R-package. R Development Core Team (2006) R: a language and environment for statistical computing. Vienna, Austria. http://www.r-project.org
- 49.Wong S, Amaro R, McCammon A. MM-PBSA captures key role of intercalating water molecules at a protein-protein interface. J Chem Theory Comput. 2009;5:422–429. doi: 10.1021/ct8003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.