Abstract
Nucleotide excision repair (NER) and ataxia telangiectasia mutated (ATM)/ATR (ATM- and RAD3-related) DNA damage checkpoints are among the major pathways that affect the chemotherapeutic efficiency of the anticancer drug cisplatin. Xeroderma pigmentosum group A (XPA) protein plays a crucial role in NER including both global genome repair (GG-NER) and transcription-coupled repair (TC-NER) subpathways, and has been a potential target for improving cisplatin therapeutic effects. We report here that XPA translocates from the cytosol into the nucleus after DNA damage induced by UV irradiation and cisplatin, a mimetic of UV damage, in human cells with or without p53 deficiency. However, the damage-induced response of XPA nuclear import was significantly slower in p53-deficient cells than in p53-proficient cells. We also found that while XPA is imported into the nucleus upon cisplatin or UV damage in an ATR-dependent manner in p53-proficient A549 lung cancer cells, the ATR checkpoint pathway has no effect on the XPA nuclear import in p53-deficient H1299 lung cancer cells. Similarly, the XPA nuclear translocation is not regulated by ATM checkpoint or by p38MAPK/MK2 either. Our findings suggest that NER is independent on the major DNA damage checkpoint pathways in H1299 (p53-/-) cells and that DNA damage responses are mechanistically different between p53-proficient and p53-deficient cells. Our results also highlight the possibility of selectively targeting XPA nuclear import as a way to sensitize cisplatin anticancer activity, but targeting ATR/ATM-dependent checkpoints may not be helpful in killing p53-deficient cancer cells.
Keywords: DNA damage checkpoints; p53; Nucleotide excision repair (NER); ataxia telangiectasia,; Xeroderma pigmentosum group A
Introduction
Chemotherapy is a critical clinical intervention for cancer patients. Cisplatin is one of the three most commonly used chemotherapeutic drugs [1]. Cisplatin induces DNA intra- and interstrand diadducts and DNA-protein crosslinks [2], which are the main cause of its cytotoxicity and hence its anti-cancer therapeutic effects. DNA repair of cisplatin-induced DNA damage is a major factor in modulating the therapeutic efficacy of cisplatin [2, 3]. In humans, bulky DNA lesions produced by ultraviolet (UV) irradiation or by UV-mimetic agents such as cisplatin can only be removed by nucleotide excision repair (NER) [4, 5]. Therefore, the status of NER is an important factor in the success of chemotherapy using cisplatin [2, 6].
The DNA repair protein Xeroderma pigmentosum group A (XPA) is an indispensable factor for NER including both subpathways: transcriptioncoupled NER (TCR-NER) and global genome NER (GGR-NER). XPA is believed to verify the damage sites following initial recognition of a lesion, stabilize repair intermediates, and be involved in recruiting other NER factors [7–12]. To our knowledge, XPA has been reported to only function in NER pathways, and consistently has been reported to be the major factor that limits the repair of cisplatin-induced DNA damage [13–15]. Regulating XPA at transcriptional or post-transcriptional level would affect the NER activity and repair of UV- or cisplatin-induced DNA lesions [4, 13–16].
The DNA damage checkpoints survey the structural integrity of the genome and coordinate multiple cellular pathways to ensure efficient removal of DNA damage. The ATM- and ATR-dependent checkpoint pathways are two central components of the DNA damage response machineries in human cells. These pathways are comprised of a series of DNA damage sensors, signal mediators and transducers, and downstream effecter molecules [5, 17, 18]. Among the downstream effectors of the ATR checkpoint are the three major checkpoint proteins Chk1, p53 and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein kinase-2 (MK2). These proteins can be directly activated by ATR following UV irradiation [19–21] although p38/MK2 also can be activated independent of ATR [20].
It is believed that the checkpoint pathways play an important role in regulating the NER processes [17, 22, 23]. However, the underlying mechanisms remain elusive. Our previous studies indicated that the ATR checkpoint is required for regulation of DNA damage-induced XPA nuclear import and phosphorylation (24, 25). When the XPA nuclear translocation process is inhibited by disrupting the ATR-XPA interaction, DNA repair efficiency is significantly reduced [24]. Our recent data (unpublished observation) also suggests that UV-induced XPA nuclear import is regulated by ATR through a p53 signaling pathway. Given that UV irradiation-and cisplatin-generating DNA lesions can only be repaired by NER in humans, and p53-deficient cancer cells rely on the ATR/p38/MK2 pathway for survival of DNA damages rather than the ATR/p53 signaling pathway [20, 25], it is of great interest to determine whether the DNA damage-induced XPA nuclear import occurs in p53 deficient cancer cells and whether the import is regulated by different checkpoint pathways in p53-proficient and deficient cancer cells.
Materials and methods
Cell culture, drugs, and antibodies
Cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were grown at 37°C in 5% CO2. UVC irradiation was performed using a 254 nm lamp at a flounce of 0.83 J/m2/sec. For time course analysis cells were incubated in culture conditions for the indicated amounts of time. Cisplatin (Sigma Chemical Co.) was dissolved in 0.9% NaCl to make a 3 mM stock solution just before use. A final concentration of 30 uM was obtained by making a 1/100 dilution of the stock solution into culture medium. UCN-01 and SB203580 were purchased from Sigma Chemical Co. and Calbiochem Chemical Co., respectively. Both UCN-01 and SB203580 were dissolved in DMSO to make stock solutions of 1 mM and 10 mM which were diluted into cell culture medium to 250 nM and 10 µM, respectively. For Western blotting, primary rabbit polyclonal antibody against XPA, mouse monoclonal antibody against PARP, rabbit polyclonal antibody against p53, mouse monoclonal antibody against Chk1, and goat anti-MK2 polyclonal antibodies were purchased from Santa Cruz Biotechnology Co. A FITC-conjugated primary mouse anti-actin antibody was obtained from Sigma Chemical Co.
RNAi
siRNA duplexes were synthesized by GenePharma Co. using the following sequences: MK2 siRNA: sense strand UGACCAUCACCGAGUUUAUdTdT and antisense strand AUAAACUCGGUGAUGGUCAdTdT; Chk1 siRNA: sense strand ACAGUAUUUCGGUAUAAUATT and antisense strand UAUUAUACCGAAAUACUGUTG; ATR siRNA: sense strand CCUCCGUGAUGUUGCUUGATT, and antisense strand UCAAGCAACAUCACGGAGGTT; ATM siRNA: sense strand CAUACUACUCAAAGACAUUTT, and antisense strand AAUGUCUUUGAGUAGUAUGTT.
The siRNA transfection reagent was purchased from Polyplus Transfection and the transfections were carried out by following manufacturer's instructions. Briefly, cells were grown to 30-40% confluency and washed with PBS and antibiotic-free medium. siRNA duplexes were added to a small volume of FBS and antibioticfree medium and incubated with transfection reagent for 10 min. This mixture then was added to cells in FBS- and antibiotic-free medium at a final siRNA concentration between 5-10 nM. After 5-7 hour incubation, FBS and antibiotic were added into the transfection medium and incubation continued. Experiments accessing the levels of protein expression or the effects of UV irradiation were initiated 48 or 72 hrs after transfection. For time course experiments after UV irradiation, siRNA-containing medium was temporarily removed during the UV irradiation.
Subcellular fractionation and Western blotting
Subcellular fractionation was performed using the Proteo JETTM cytoplasmic and nuclear protein extraction kit (Fermentas) by following the procedures suggested by the manufacturer. Briefly, 10 volumes of cell lysis buffer with protease inhibitors were added to 1 volume of packed cells. After a brief vortexing and incubation on ice for 10 min, cytoplasm was separated from nuclei by centrifugation at 500 × g for 7 min at 4 ºC. Isolated nuclei were washed once or twice with 500 µL of the nuclei washing buffer and then collected by centrifugation. The collected nuclear pellets were re-suspended in ice-cold nuclear storage buffer, and 1/10 volume of the nuclear lysis reagent was added. The nuclei were lysed with inversions for 15 min at 4°C. Nuclear lysates were collected by centrifugation at 20,000 × g for 12 min at 4 ºC. The lysates then were mixed with 2x SDS loading buffer, boiled for 10 minutes and loaded into SDS-PAGE for Western blot analysis. In all of the fractionation experiments, protein levels of β-actin and PARP were assessed as cytoplasmic and nuclear protein loading controls, respectively.
Results
XPA translocates into the nucleus from the cytosol upon cisplatin- and UV-DNA damage
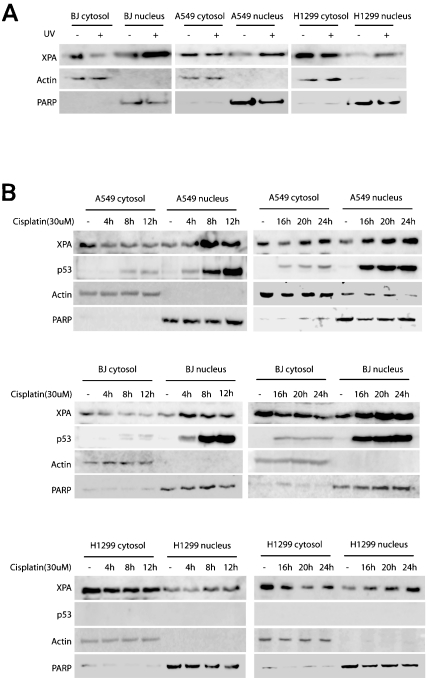
Consistent with previous observations [24, 26], the current data demonstrate that UV irradiation induced nuclear import of XPA in the normal foreskin primary cell line BJ, the human lung adenocarcinoma cell line A549, and a p53-deficient non-small cell lung carcinoma cell line H1299 (Figure 1A). XPA is an indispensable factor of NER. To determine whether the nuclear import of XPA is part of the cellular response to cisplatin-induced DNA damage, BJ, A549 (wildtype p53) and H1299 (p53-/-) cells were incubated with cisplatin for the indicated time periods (Figure 1B). The nuclear import of XPA started no later than 4 hrs after cisplatin addition in BJ and A549 cells. The nuclear translocation of XPA in H1299 cells, however, occurred later than in BJ and A549 cells as significant nuclear import of XPA was observed only after 8 hrs of incubation with cisplatin. With 20 hrs of exposure to cisplatin cytoplasmic XPA protein started to recover to pre-exposure levels in A549 cells, even though the DNA-damage induced nuclear accumulation of XPA continued to the 24 hour time point. Different from A549 cells, the increase of cytoplasmic XPA protein in H1299 cells was delayed to the 24 hour time point with cisplatin, although a similar increase of the nuclear XPA was observed. In agreement with the previous reports [27, 28], cisplatin induced over-expression and nuclear accumulation of p53 in A549 and BJ cells (Figure 1B).
Figure 1.
UV irradiation- and cisplatin-induced DNA damage promotes nuclear import of XPA. A. The normal foreskin cells BJ, human lung adenocarcinoma cells A549, and p53-deficient non-small cell lung carcinoma cells H1299 were subjected to 20 J/m2 UV-C irradiation followed by a 2-hr recovery. Fractionation and Western blotting were performed to assess the subcellular localization of XPA. B. The chemotherapeutic agent cisplatin induced cytoplasmic-tonuclear translocation of XPA. BJ, A549 and H1299 were incubated with DMSO (mocktreatment) or 30 uM cisplatin for the indicated time points. Fractionation and Western blotting were performed to assess the expression and intracellular localization of XPA. PARP and β-actin were probed as nuclear and cytoplasmic protein loading controls, respectively.
Neither Chk1 nor MK2 is required for damageinduced XPA nuclear import
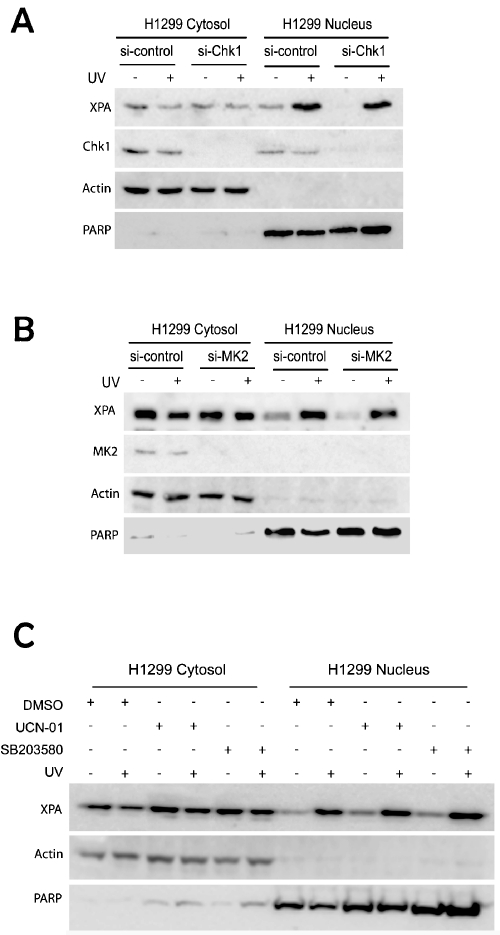
Our recent study demonstrated that UV-induced XPA nuclear import is dependent on the ATR/p53 signaling pathway in the p53-proficient cancer cells. However, the data in Figure 1 showed no defects of XPA nuclear import in p53-deficient cancer cells H1299. Since both MK2 and Chk1 kinases can transmit damage signals from ATR independent of p53 to arrest cell cycle progression, it is of interest to determine whether Chk1 and/or MK2 are required for the UV-induced nuclear import of XPA in H1299 cells. As shown in Figure 2A, no difference in XPA nuclear translocation was observed between the control siRNA and the Chk1 silencing cells, indicating that Chk1-mediated signaling is not required for the nuclear import of XPA. Similar result was also obtained for cells with MK2 knockdown by siRNA (Figure 2B). These results were further confirmed using two selective kinase inhibitors, UCN-01 and SB203580. UCN-01 inhibits both Chk1 and MK2 activity while SB203580 mainly targets the p38/MK2 pathways. These inhibitors were incubated with H1299 cells to stop the signal transductions mediated by Chk1 and/or MK2. Consistent with the siRNA knockdown results, inhibition of the two kinases did not change the UV-induced XPA nuclear import (Figure 2C).
Figure 2.
DNA damage-induced XPA nuclear import is not affected by checkpoint proteins Chk1 and MK2 in the p53-deficient cancer cells. A. H1299 cells were transfected with control siRNA or Chk1 siRNA. After 48 hrs, transfected cells were mock or 20 J/m2 UV-C treated and further incubated for 2 hr for recovery. The subcellular location of XPA was then assessed by fractionation and Western blotting. B. Control siRNA- or MK2 siRNA-transfected H1299 cells were mock-treated or irradiated with 20 J/m2 UV-C followed by a 2 hr recovery. Fractionation and Western blotting were employed to analyze the subcellular localization of XPA and the expression of MK2. C. H1299 cells were pretreated with DMSO, 250 nM UCN-01 or 10 uM SB203580 for 1 hr. Cells then were mock-treated or irradiated with 20 J/m2 UV-C followed by a 2 hr recovery in the presence of the inhibitors. The fractionated samples were analyzed by Western blotting.
ATR and/or ATM are not required for damageinduced XPA nuclear import
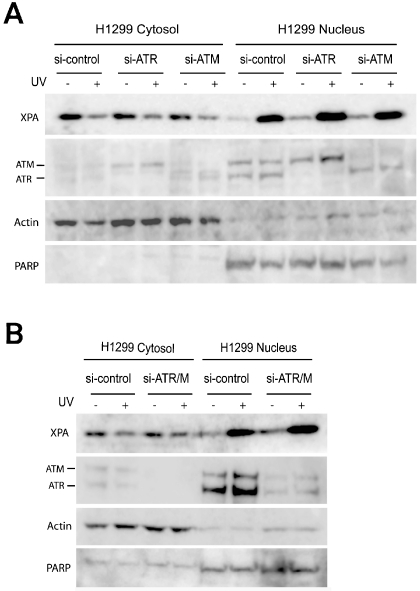
The observation that Chk1 and MK2 were not required for damage-induced XPA nuclear import in H1299 (p53-/-) cells implies that ATR may not be involved in regulation of nucleotide excision repair in p53 deficient H1299 cells. To test the hypothesis, ATR and ATM were depleted in H1299 cells. As shown in Figure 3, ATR and ATM were depleted by siRNA transfections individually (Figure 3A), or simultaneously (Figure 3B). However, the UV-induced cytosol-to-nucleus translocation of XPA was not affected, as indicated by the equal reduction of XPA in cytosol and increasing XPA in the nucleus between the control siRNA- and ATR/ATM siRNA-transfected cells. The result indicates that neither ATR nor ATM was required for the damage-induced XPA nuclear import.
Figure 3.
DNA damageinduced XPA nuclear import does not dependent on ATR and/or ATM in p53 deficient H1299 cancer cells. A. Cells transfected with control siRNA, ATR siRNA, and ATM siRNA were treated with UV (20 J/m2), followed by subcellular fractionation. The samples were then analyzed by Western blotting. B. ATR and ATM were simultaneously depleted by siRNA transfections, followed by UV irradiation of the cells (20 J/m2). Subcellular fractions were then subjected to Western blot analysis.
Discussion
DNA damage checkpoints and DNA repair are two major types of cellular DNA damage response pathways and a close coordination between these two pathways is believed to be crucial for maintaining the genome integrity and stability. Our recent studies indicated that the ATRdependent checkpoint pathway is required for regulation of the DNA damage-induced XPA translocation from cytoplasm to nucleus (24, 25). Interestingly, the results from this study indicate that although this ATR dependence occurs in p53-proficient normal and cancer cells treated with cisplatin or UV, it does not apply in p53-deficient lung cancer cells such as H1299 cells. This was further confirmed by the independence of XPA nuclear import on Chk1 and p38/MK2, two major downstream substrates of ATR checkpoint signaling. It should be noted that XPA is a crucial factor of NER and that XPA-deficient cells exhibit the highest UV sensitivity among other NER factors [29]. In addition, XPA nuclear import appears essential for the activity of NER [24, 30]. Thus, our findings suggest that NER is independent of ATR-dependent checkpoint in the p53-deficient cancer cells. XPA nuclear import, or likely NER, also was found to be independent of the ATM checkpoint in the p53-deficient cancer cells. These observations imply that all major known DNA damage checkpoints have no effect on NER in p53-deficient cancer cells. In spite of this, the fact that XPA nuclear import is a DNA damage-induced event indicates that it is a regulated event in cells. We propose that an alternative checkpoint pathway may be responsible for the regulation. Taken together, our results suggest that the cellular DNA damage responses to cisplatin treatments are different in p53 proficient and deficient cells.
One of the major challenges in treating cancer patients with cisplatin is the drugs side-effects: it introduces DNA damage to cancer cells while also causing damage to normal cells [1]. One solution for this problem is to identify mechanistic differences of DNA damage responses between normal and cancer cells. The current study attempts to define the unique mechanism of DNA damage responses in p53-deficient lung cancer cells as p53 is the most commonly mutated gene in human cancers, particularly the lung cancer. Thus, three cell lines including the human normal primary cell line BJ, the p53- proficient lung cancer cell line A549, and the p53-deficient lung cancer cell line H1299 were treated with cisplatin or UV-C irradiation. A pronounced cytoplasm-to-nucleus translocation of XPA was observed in each of these cell lines (Figure 1), which is consistent with our previous observations on other types of cells treated with UV. However, unlike in p53-proficient cells the damage-induced XPA import is not dependent on ATR or other major known checkpoints in the p53-deficient H1299 lung cancer cells (Figure 2 and Figure 3). These observations reveal a mechanistic difference of DNA damage responses between p53-proficient and p53-deficient lung cancer cells. This difference highlights a possibility of increasing drug sensitivity in p53-deficient cancer cells without sensitization of normal cells. This could possibly be achieved by targeting XPA nuclear import directly; however, repair of damaged DNA in the patients normal cells would also be affected. A better, more specific target would be to disrupt the alternative regulatory pathway for XPA nuclear import used in p53-deficient cancer cells and, thus, inhibit their DNA repair. This would allow the NER of cisplatin-damaged DNA to proceed in the patients normal cells. Obviously, this requires further investigation to identify the novel regulation mechanism responsible for the damage-induced XPA import in the p53-deficient cancer cells.
Acknowledgements
This work was supported by National Institutes of Health grants CA86927 and GM083307 (to Y.Z.).
References
- [1].Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- [2].Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- [3].Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinumbased chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- [4].Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665–1667. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- [5].Sancar A, Lindsey-Boltz LA, UnsalKacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- [6].Chaney SG, Sancar A. DNA repair: enzymatic mechanisms and relevance to drug response. J Natl Cancer Inst. 1996;88:1346–1360. doi: 10.1093/jnci/88.19.1346. [DOI] [PubMed] [Google Scholar]
- [7].Yang Z, Roginskaya M, Colis LC, Basu AK, Shell SM, Liu Y, Musich PR, Harris CM, Harris TM, Zou Y. Specific and Efficient Binding of Xeroderma Pigmentosum Complementation Group A to Double-Strand/Single-Strand DNA Junctions with 3'- and/or 5'-ssDNA Branches. Biochemistry. 2006;45:15921–15930. doi: 10.1021/bi061626q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- [9].Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- [10].Liu Y, Liu Y, Yang Z, Utzat C, Wang G, Basu AK, Zou Y. Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage. Biochemistry. 2005;44:7361–7368. doi: 10.1021/bi047598y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guzder SN, Sommers CH, Prakash L, Prakash S. Complex formation with damage recognition protein Rad14 is essential for Sac-charomyces cerevisiae Rad1-Rad10 nuclease to perform its function in nucleotide excision repair in vivo. Mol Cell Biol. 2006;26:1135–1141. doi: 10.1128/MCB.26.3.1135-1141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shell SM, Zou Y. Other proteins interacting with XP proteins. Adv Exp Med Biol. 2008;637:103–112. doi: 10.1007/978-0-387-09599-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wu X, Fan W, Xu S, Zhou Y. Sensitization to the cytotoxicity of cisplatin by transfection with nucleotide excision repair gene xeroderma pigmentosun group A antisense RNA in human lung adenocarcinoma cells. Clin Cancer Res. 20039:5874–5879. [PubMed] [Google Scholar]
- [15].Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer. 2005;4:18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kang TH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- [18].Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- [19].Helt CE, Cliby WA, Keng PC, Bambara RA, O'Reilly MA. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J Biol Chem. 2005;280:1186–1192. doi: 10.1074/jbc.M410873200. [DOI] [PubMed] [Google Scholar]
- [20].Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tibbetts RS, Brumbaugh KM, Williams JM, Sark-aria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Auclair Y, Rouget R, Affar el B, Drobetsky EA. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc Natl Acad Sci U S A. 2008;105:17896–17901. doi: 10.1073/pnas.0801585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Auclair Y, Rouget R, Drobetsky EA. ATR kinase as master regulator of nucleotide excision repair during S phase of the cell cycle. Cell Cycle. 2009;8:1865–1871. doi: 10.4161/cc.8.12.8800. [DOI] [PubMed] [Google Scholar]
- [24].Shell SM, Li Z, Shkriabai N, Kvaratskhelia M, Brosey C, Serrano MA, Chazin WJ, Musich PR, Zou Y. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J Biol Chem. 2009;284:24213–24222. doi: 10.1074/jbc.M109.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MA, Wang X, Linding R, Ong SE, Weaver D, Carr SA, Yaffe MB. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol Cell40:34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu X, Shell SM, Liu Y, Zou Y. ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation. Oncogene. 2007;26:757–764. doi: 10.1038/sj.onc.1209828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- [28].Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- [30].Knudsen NO, Andersen SD, Lutzen A, Nielsen FC, Rasmussen LJ. Nuclear translocation contributes to regulation of DNA excision repair activities. DNA Repair (Amst) 2009;8:682–689. doi: 10.1016/j.dnarep.2009.03.005. [DOI] [PubMed] [Google Scholar]