Abstract
Comparative gene mapping in mammals typically involves identification of segments of conserved synteny in diverse genomes. The development of maps that permit comparison of gene order within conserved synteny has not advanced beyond the mouse map that takes advantage of linkage analysis in interspecific backcrosses. Radiation hybrid (RH) mapping provides a powerful tool for determining order of genes in genomes for which gene-based linkage mapping is impractical. Comparative RH mapping of 24 orthologous genes in this study revealed internal structural rearrangements between human chromosome 17 (HSA17) and bovine chromosome 19 (BTA19), two chromosomes known previously to be conserved completely and exclusively at level of synteny. Only six of the 24 genes had been previously ordered on the human G3 RH map. The use of the G3 panel to map the other 18, however, produced parallel RH maps for comparison of gene order at a resolution of <5 Mb on the bovine linkage map and from 1 to 3 Mb in the human physical map.
Comparative gene mapping is a valuable tool for studying the evolutionary history of chromosomes and subsequently, for extrapolating information from one genome to another. The latter is extremely important to mammalian genetics in which a wealth of genomic information is being accumulated for humans and mice, whereas significant advances in both comparative medicine and animal-based agriculture await a better understanding of genomic organization in a variety of map-poor species. Even the map-rich human and mouse genomes will require increased comparative resolution to fully exploit gene-discovery strategies based on the merger of their respective maps (Carver and Stubbs 1997). Comparative mapping of bovine chromosomes relative to those of humans and mice has been advanced historically by the continual search for improved resolution of boundaries and genomic conservation. Somatic-cell hybrids segregating cattle chromosomes resulted in synteny maps of bovine homologs of previously mapped human genes (Womack and Moll 1986). This “cow on human” approach has revealed extensive conservation of synteny (Womack and Kata 1995) but provides no information about conservation of gene order. ZOO-FISH mapping in which human chromosome-specific painting probes were hybridized to cattle chromosomes (Hays 1995; Solinas-Toldo et al. 1995; Chowdhary et al. 1996) produced “human on cow” comparative maps, again without addressing gene order. Limited success in ordering cattle genes on their respective chromosomes has been achieved through linkage mapping, with both intraspecies (Barendse et al. 1997) and interspecies crosses (Gao and Womack 1997; Riggs et al. 1997; Yang and Womack 1997) and by in situ hybridization of probes to banded chromosomes. Although limited in both scope and power, these studies have demonstrated that rearrangement of gene order within segments of conserved synteny is very common and must be addressed for effective trans-species shuttling of information between the cow and the two map-rich mammals. Comparative mapping in other nonprimate species has led to similar conclusions.
Radiation hybrid (RH) mapping (Goss and Harris 1975) was recently rediscovered (Cox et al. 1990; Walter et al. 1994) as an effective approach to building ordered maps of sequence-tagged sites, regardless of allelic variation. Complete genomic coverage has been achieved in human RH maps constructed at different levels of resolution (Hudson et al. 1995, plus supplementary data from the Whitehead Institute/MIT Center for Genome Research, Human Genetic Mapping Project, Data Release, October 1996; Gyapay et al. 1996; Stewart et al. 1997) and more recently the approach has been extended to mapping in other species (McCarthy 1996), including cattle (Womack et al. 1997). A 5000-rad bovine whole-genome radiation hybrid (WG-RH) panel was constructed in our laboratory to provide a matrix for mapping the expanding pool of bovine-expressed sequence tags (ESTs), for integrating the existing bovine linkage maps that consist primarily of microsatellites, and first and foremost for constructing ordered comparative maps of the cattle genome relative to those of humans and mice.
We initiated comparative RH mapping in cattle to investigate rearrangements in gene order in human chromosome 17 (HSA17) and bovine chromosome 19 (BTA19), two chromosomes known to be conserved completely and exclusively at the level of synteny mapping (Solinas-Toldo et al. 1995; Yang and Womack 1995). A BTA19 RH map was constructed utilizing 17 coding sequences and 12 microsatellites selected for their spacing on published linkage maps (Yang et al. 1998). Most of the homologous human coding sequences had been mapped previously to HSA17, primarily by fluorescence in situ hybridization (FISH). Whereas comparative maps of FISH-mapped genes are sufficient for demonstrating the most dramatic evolutionary changes in chromosomes, subtle rearrangements cannot be defined. We found severe limitations in the “bovine RH/human FISH” comparative map because of overlap of gene assignments on the human cytogenetic map. In this study we demonstrate the power of parallel RH mapping, mapping orthlogous genes on RH maps of two genomes, to advance comparative mapping from the level of cytogenetic resolution to the megabase level of resolution provided by RH mapping.
RESULTS
Comparison of Gene Order by RH Mapping on BTA19 and HSA17
The WG-RH map of BTA19 has been expanded to include 36 markers, 24 of which are expressed genes (Fig. 1). Human orthologs of the 24 genes were mapped with the human G3 RH panel. The linear orders of orthologous loci on the G3 RH map of HSA17 and the BTA19 RH map are shown in Figure 2. Framework markers ordered at odds greater than 1000:1, including 14 of the 24 markers on BTA19 RH map and 21 of 24 on the HSA17 G3 map, are highlighted (blue).
Figure 1.
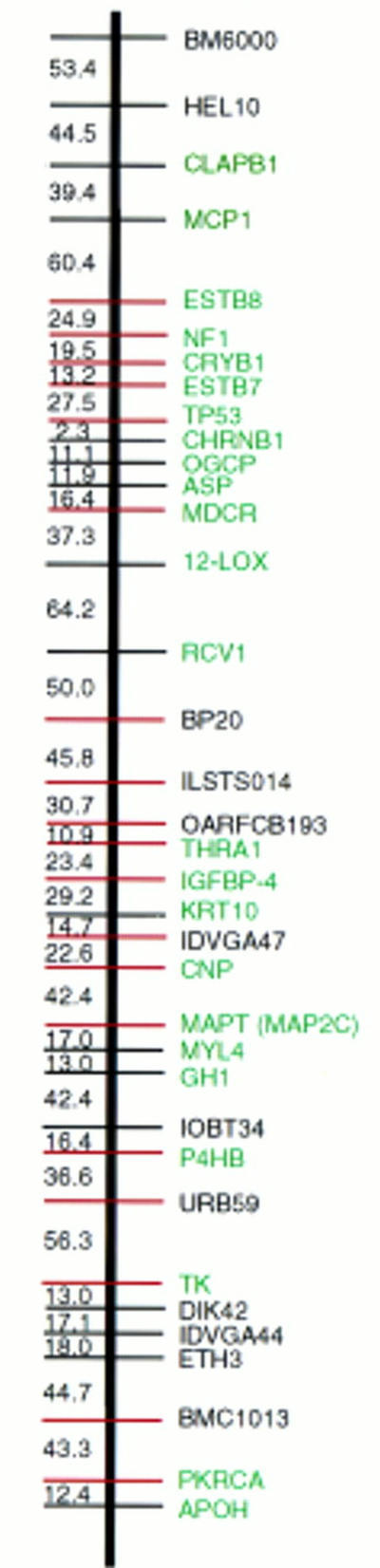
Comprehensive RH map of BTA chromosome 19. (Red) Framework markers ordered at odds >1000:1. (Green) Expressed genes and ESTs. Map units are cRays5000.
Figure 2.
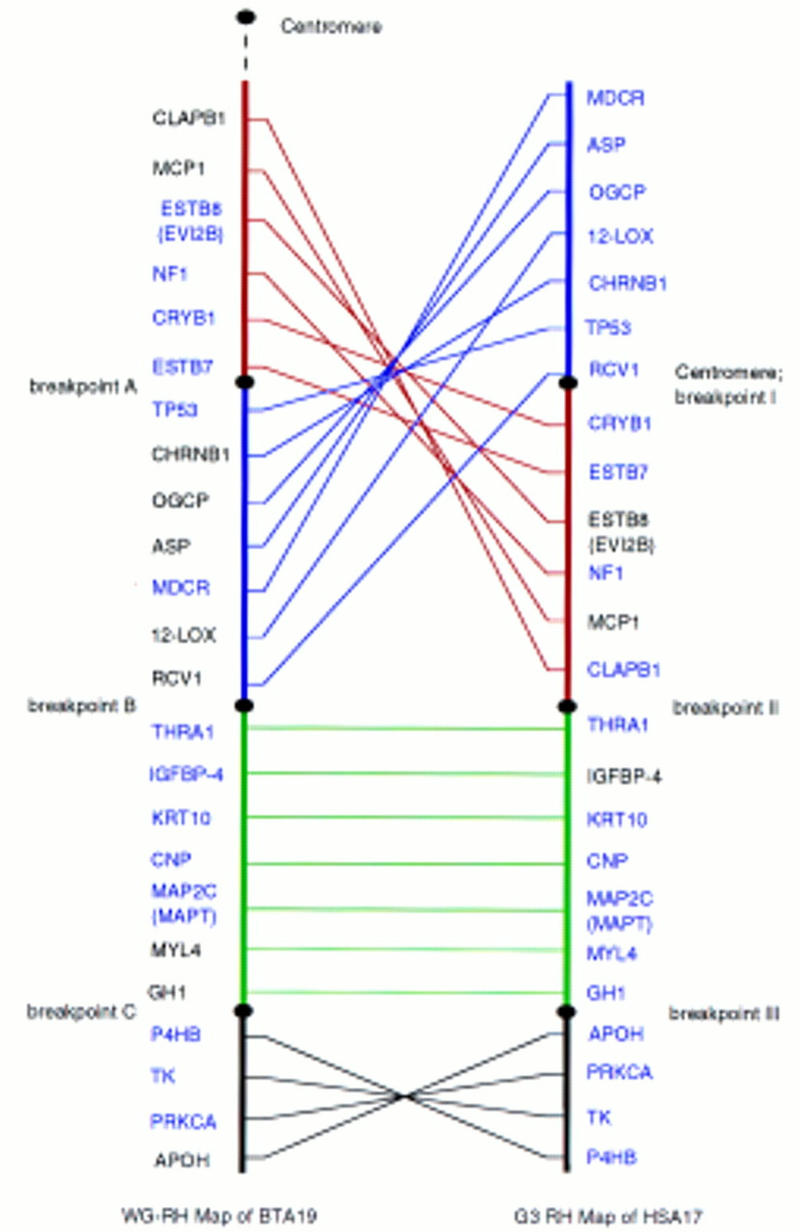
Parallel RH maps of BTA19 and HSA17. (Blue) Framework markers. The comparative RH map was divided into four regions with three breakpoints on each chromosome. Not only are the positions of region I (red) and region II (blue) inverted between human and bovine, but the gene orders within the two regions are also shuffled. (Green) Region III has the same gene order between human and cattle. The gene orders in region IV (black) between human and cattle are inverted.
Rearrangements in gene order are revealed for HSA17 and BTA19 and the parallel RH maps are divided into four regions based on these rearrangements (Fig. 2). Regions I (red) and II (blue) flank the human centromere and are the most proximal to the bovine centromere. The positions of region I and region II are inverted in the human and bovine linear maps and gene orders within these segments are also shuffled. Region III (green) is distal to regions I and II, with a totally conserved gene order in humans and cattle. Region IV (black) is the most distal and gene order in the two maps is inverted relative to GH1 and other markers in region III.
Previous studies have demonstrated that high retention frequencies around the selectable marker thymidine kinase (TK) could lead to the overestimate of the breakage probabilities (Lunetta et al. 1996). Not surprisingly, the TK marker with 100% retention frequency was not found linked to any marker on the G3 HSA17 map in this study. Its location was predicted to be within the gap between markers WI-7837 and SHGC-31110 near the distal end of HSA17 because of the high retention frequencies (∼90%) of the markers around the gap.
Identification of Rearrangement Breakpoints on HSA17 and BTA19
The four major regions of the comparative RH maps identified three major evolutionary breakpoints in the ancestral linage of HSA17 and BTA19. On HSA17, breakpoint I includes the area around the centromere. Breakpoint II is within a 100 cR10000 segment on the G3 map between the most distal gene of region II, CLAPB1, and the most proximal gene of region III, THRA1. Breakpoint III is localized between GH1 of region III and APOH of region IV, a distance of ∼61 cR10000. With the estimate of 1 cR10000 on the G3 map to equal to ∼29 kb (Stewart et al. 1997), the evolutionary breakpoints II and III on HSA17 can be narrowed down to physical distances of ∼3 Mb and <2 Mb, respectively.
BTA19 breakpoint A is included in the 30-cR5000 span between ESTB7 and TP53 on the established BTA19 RH map (Yang et al. 1998). Breakpoint B is in the 140-cR5000 span between RCV1 and THRA1 as determined by two-point analysis. Breakpoint C is localized to the to a 60-cR5000 span between GH1 and P4HB (Yang et al. 1998). On the BTA19 RH map, 1 cM equals ∼8 cR5000 (Yang et al. 1998), and a bovine cM is on the average ∼1 Mb (Barendse et al. 1997). Thus, the rearrangement breakpoints A, B, and C were identified within the estimated distances of ∼4, 18, and 8 Mb, respectively.
DISCUSSION
Parallel RH mapping was used for the first time as a comparative mapping tool to reveal internal structural rearrangements between HSA17 and BTA19, chromosomes totally conserved at the synteny level. Twenty-one of the 24 genes compared in this study were ordered on the HSA17 G3 map with odds greater than 1000:1. Fourteen of the 24 genes were ordered at 1000:1 odds on the BTA19 RH map. The other 10 were placed on the map to satisfy criteria of maximum likelihood and minimum breakage. The confidence of gene ordering on the bovine map will increase with the availability of additional RH panels and by the inclusion of more markers on this map.
Comparative mapping of animal genomes is now poised to move beyond the identification of segments of conserved syteny. Whereas high-resolution mapping is feasible with interspecific hybrid backcrosses in mice, ordering genes is problematic in most other animal species. RH panels are being constructed in a growing number of mammalian species and are theoretically possible for nonmammals as well. High-resolution human RH maps, derived from the G3, GB4, and other RH panels are available as reference maps. Although many genes useful for comparative mapping, such as the comparative-mapping anchor loci (Lyons et al. 1997), are not currently included on the G3 map, this study demonstrated that comparative markers can be added to the G3 map efficiently and with high levels of confidence. The systematic application of parallel RH mapping in mammalian species will lead to a new generation of comparative maps that focus on the order of genes within segments of conserved synteny. The significance of refined comparative maps will extend beyond our understanding of ancestral rearrangements in mammalian chromosome evolution. It will facilitate “comparative candidate positional cloning” whereby a genomic address for an economically important trait in a map-poor species can be translated to a conserved region of the human transcription map and its respective bin of potential candidate genes.
METHODS
Bovine RH Panel
A 5000-rad whole genome RH panel (Womack et al. 1997) was utilized in this study. A total of 100 hybrids were analyzed for the presence of markers as described below.
Gene Markers and Marker Typing
Twenty-four genes were mapped in both the bovine RH panel and the human G3 RH panel. All genotyping was done by PCR as described previously (Yang et al. 1998). The name, symbol, and primer information for the genes are summarized in Table 1.
Table 1.
Primer Information for Genes Used in This Study
 |
Ordering Genes on the BTA19 RH Map
Seventeen of the 24 genes used in this study had been RH mapped previously in cattle (Yang et al. 1998). The other seven genes, 12-LOX, APOH, CNP, IGFBP-4, MDCR, PRKCA, and RCV1 (Table 1; Fig. 1), were added to the bovine RH map in this study using the RHMAP 3.0 program package (Boehnke 1992; Lunetta et al. 1996).
Ordering Genes on the HSA17 G3 RH Map
Human homologs of six of the genes in this study, MYL4, NF1, OGCP, PRKCA, THRA1, and 12-LOX, had been ordered previously on the G3 map. To generate parallel comparative RH maps, we placed the other 18 genes on the map. PCR primers were retrieved from the public database Rhdb (http://www.ebi.ac.uk/RHdb), or designed using Macvector 4.1 software (Eastman Kodak, New Haven, CT) (Table 1). The 83 hybrids of the human G3 panel were purchased from Research Genetics (Huntsville, AL).
The two-point maximum likelihood analysis was first performed through the G3 map RH server of the Stanford Human Genome Center (SHGC) (http://www-shgc.stanford.edu/rhserver2/rhserver_form.html) and the 18 genes fell into three categories. The first category included ASP, CHRNB1, TP53, CRYB1, and CLAPB1, which were totally linked to framework markers on the G3 map. The second category included only the selectable marker, TK, which had a 100% retention frequency and was not linked to any framework marker on the map with lod > 3. The remaining 12 genes fell into the third category, which were linked to framework markers on HSA17 at lod > 6 but with some recombination. No further analysis was necessary for genes in the first two categories. For genes in the third group, however, the RHMAP 3.0 program package was used to further determine their locations relative to the linked-framework markers on the G3 map. For this purpose, the typing data of the 12 genes as well as all the framework markers linked to them at lod > 6 were included in the analysis.
Acknowledgments
We thank Elaine Owens, Janice Johnson, and Christy Fickey for excellent technical support. This project was supported by The Texas Agricultural Experiment Station and U.S. Department of Agriculture National Research Initiative grant no. 97-03984.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL JWOMACK@cvm.tamu.edu; FAX (409) 845-9972.
REFERENCES
- Barendse W, Vaiman D, Kemp SJ, Sugimoto Y, Armitage SM, Williams JL, Sun HS, Eggen A, Agaba M, Aleyasin SA, et al. A medium-density genetic linkage map of the bovine genome. Mamm Genome. 1997;8:21–28. doi: 10.1007/s003359900340. [DOI] [PubMed] [Google Scholar]
- Bishop MD, Kappes SM, Keele JW, Stone RT, Sunden SLF, Hawkins GA, Solinas-Toldo S, Fries R, Grosz MD, Yoo Y, Beattie CW. A genetic linkage map for cattle. Genetics. 1994;136:619–639. doi: 10.1093/genetics/136.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M. Multipoint analysis for radiation hybrid mapping. Ann Med. 1992;24:383–386. doi: 10.3109/07853899209147842. [DOI] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human–mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Fronicke L, Gustavsson I, Scherthan H. Comparative analysis of the cattle and human genomes: Detection of ZOO-FISH and gene mapping-based chromosomal homologies. Mamm Genome. 1996;7:297–302. doi: 10.1007/s003359900086. [DOI] [PubMed] [Google Scholar]
- Chumakov IM, Rigault P, Le Gall I, Bellanne-Chantelot C, Billault A, Guillou S, Solarue P, Guasconi G, Poullier E, Gros I, et al. A YAC contig map of the human genome. Nature. 1995;377:S175–S183. doi: 10.1038/377175a0. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Gilbert DJ. A genetic linkage map of the mouse: Current application and future prospects. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: A somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Gao Q, Womack JE. A genetic map of bovine chromosome 7 with an interspecific hybrid backcross panel. Mamm Genome. 1997;8:258–261. doi: 10.1007/s003359900405. [DOI] [PubMed] [Google Scholar]
- Goss SJ, Harris H. New method for mapping genes in human chromosomes. Nature. 1975;255:680–684. doi: 10.1038/255680a0. [DOI] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud’Homme JF, Dib C, Auffray C, et al. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- Hayes H. Chromosome painting with human chromosome-specific DNA libraries reveals the extent and distribution of conserved segments in bovine chromosomes. Cytogenet Cell Genet. 1995;71:168–174. doi: 10.1159/000134100. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Hu S-H, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick BW. Detection of a three-allele single strand conformation polymorphism (SSCP) in the fourth intron of the bovine growth hormone gene. Anim Genet. 1992;23:179–181. doi: 10.1111/j.1365-2052.1992.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Lunetta KL, Boehnke M, Lange K, Cox DR. Selected locus and multiple panel models from radiation hybrid mapping. Am J Hum Genet. 1996;59:717–725. [PMC free article] [PubMed] [Google Scholar]
- Lyons LA, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, O’Brien SJ. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nature Genetics. 1997;15:47–56. doi: 10.1038/ng0197-47. [DOI] [PubMed] [Google Scholar]
- McCarthy LA. Whole genome radiation hybrid mapping. Trends Genet. 1996;12:491–493. doi: 10.1016/s0168-9525(96)30110-8. [DOI] [PubMed] [Google Scholar]
- Moore SS, Byrne K, Berger KT. Characterization of 65 bovine microsatellites. Mamm Genome. 1994;5:84–90. doi: 10.1007/BF00292333. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Womack JE, Lyons LA, Moore KJ, Jenkins NA, Copeland NG. Anchored reference loci for comparative genome mapping in mammals. Nature Genet. 1993;3:103–112. doi: 10.1038/ng0293-103. [DOI] [PubMed] [Google Scholar]
- Riggs PK, Owens KE, Rexroad III CE, Amaral MEJ, Womack JE. Development and initial characterization of a Bos taurus × B. gaurus interspecific hybrid backcross panel. J Heredity. 1997;88:373–379. doi: 10.1093/oxfordjournals.jhered.a023121. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Boguski MS, Hudson TJ, Hui L, Ma J, Castle AB, Wu X, Silva J, Nusbaum HC, Birren BB, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- Solinas-Toldo S, Lengauer C, Fries R. Comparative genome map of man and cattle. Genomics. 1995;27:489–496. doi: 10.1006/geno.1995.1081. [DOI] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Walter MA, Spillett DJ, Tomas P, Weissenbach J, Goodfellow PN. A method for constructing radiation hybrid maps of whole genomes. Nature Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- Womack JE. Symposium: Bovine gene mapping: The goals and status of the bovine gene map. J Dairy Sci. 1993;76:1199–1203. doi: 10.3168/jds.S0022-0302(93)77449-4. [DOI] [PubMed] [Google Scholar]
- Womack JE, Kata SR. Bovine genome mapping: Evolutionary inference and the power of comparative genomics. Curr Opin Genet Dev. 1995;5:725–733. doi: 10.1016/0959-437x(95)80004-o. [DOI] [PubMed] [Google Scholar]
- Womack JE, Moll YD. A gene map of the cow: Conservation of linkage with mouse and man. J Hered. 1986;77:2–7. doi: 10.1093/oxfordjournals.jhered.a110160. [DOI] [PubMed] [Google Scholar]
- Womack JE, Johnson JS, Owens EK, Rexroad III CE, Schläpfer J, Yang Y. A whole genome radiation hybrid panel for bovine gene mapping. Mamm Genome. 1997;8:854–856. doi: 10.1007/s003359900593. [DOI] [PubMed] [Google Scholar]
- Yang Y-P, Womack JE. Human chromosome 17 comparative anchor loci are conserved on bovine chromosome 19. Genomics. 1995;27:293–297. doi: 10.1006/geno.1995.1045. [DOI] [PubMed] [Google Scholar]
- Yang Y, Womack JE. Construction of a bovine chromosome 19 map with an interspecies hybrid backcross. Mamm Genome. 1997;8:262–266. doi: 10.1007/s003359900406. [DOI] [PubMed] [Google Scholar]
- Yang Y-P, Rexroad III CE, Schläpfer J, Womack JE. An integrated radiation hybrid map of bovine chromosome 19 and ordered comparative mapping with human chromosome 17. Genomics. 1998;48:93–99. doi: 10.1006/geno.1997.5143. [DOI] [PubMed] [Google Scholar]


