Abstract
During meiotic prophase homologous chromosomes find each other and pair. Then they synapse, as the linear protein core (axial element or lateral element) of each homologous chromosome is joined together by a transverse central element, forming the tripartite synaptonemal complex (SC). Ten uncloned Zea mays mutants in our collection were surveyed by transmission electron microscopy by making silver-stained spreads of SCs to identify mutants with non-homologous synapsis or improper synapsis. To analyse the mutants further, zyp1, the maize orthologue of the Arabidopsis central element component ZYP1 was cloned and an antibody was made against it. Using antibodies against ZYP1 and the lateral element components AFD1 and ASY1, it was found that most mutants form normal SCs but are defective in pairing. The large number of non-homologous synapsis mutants defective in pairing illustrates that synapsis and pairing can be uncoupled. Of the ten mutants studied, only dsy2 undergoes normal homologous chromosome recognition needed for homologous pairing. The dsy2 mutation fails to maintain the SC. ZYP1 elongation is blocked at zygotene, and only dots of ZYP1 are seen at prophase I. Another mutant, mei*N2415 showed incomplete but homologous synapsis and ASY1 and AFD1 have a normal distribution. Although installation of ZYP1 is initiated at zygotene, its progression is slowed down and not completed by pachytene in some cells and ZYP1 is not retained on pachytene chromosomes. The mutants described here are now available through the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/).
Keywords: Desynaptic, homologous chromosomes, maize, meiosis, meiotic mutants, pairing, recombination, synapsis, synaptonemal complex
Introduction
Meiosis is a specialized cell division that is required to halve the chromosome complement in cells and is a feature of all organisms with a sexual life cycle. The most significant event in meiotic prophase is the pairing of homologous chromosomes. Pairing, the alignment of homologous chromosomes, is initiated at the leptotene–zygotene transition. It is followed in zygotene by synapsis, which is defined as the intimate association of homologues as joined together by the synaptonemal complex (SC) (Page and Hawley, 2004; Hamant et al., 2006). Although meiotic recombination involves the recognition of DNA structure at the level of atomic resolution for the alignment of identical or near identical sequences of bases, homologous chromosome pairing and synapsis can work at a macromolecular level measured in microns.
Early in prophase I the characteristic leptotene chromosome forms as a linear protein axis or axial element (AE) is assembled between two sister chromatids. Cohesin complexes, required for sister chromatid cohesion, are an essential component of the AE. As homologous chromosomes start to pair, their AEs become aligned in parallel and the central element (CE) is assembled between the AEs, to form tripartite SCs (reviewed in Zickler and Kleckner, 1999; Page and Hawley, 2004). In maize, the SC first appears in zygotene and is initiated preferentially at sites near the telomeres and also at several internal sites (Burnham et al., 1972; Zickler and Kleckner, 1999). The chromosomes remain synapsed throughout pachytene as recombination is completed. In many organisms, the SC is required for the recombination event to mature into a chiasma that holds homologues together after the SC falls apart in diplotene. Finally, during diakinesis the chromosomes condense, thicken, and detach from the nuclear envelope.
To study the spatial kinetics of synapsis in maize, 3D Structured Illumination Microscopy was used, a method of ultra-high resolution light microscopy that overcomes the 200 nm limit of resolution of conventional light microscopy, and reaches a lateral resolution of at least 100 nm. The two AEs of synapsed SCs are clearly resolved with a 194 nm spacing and they coil around each other as a left handed helix of variable pitch. AFD1, an AE component, showed a bilaterally symmetrical pattern on the paired axes (Wang et al., 2009). At late zygotene/early pachytene, most unsynapsed regions in nuclei are associated with interlocks, suggesting that the resolution of interlocks between intertwined non-homologous chromosomes may be a rate-limiting step to complete synapsis. By end of pachytene, all interlocks are resolved in wild-type meiocytes.
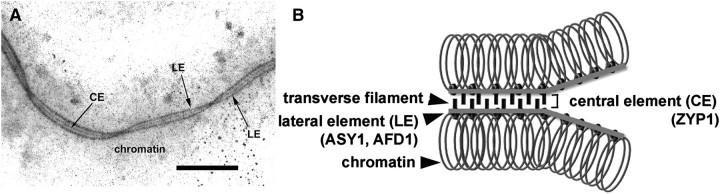
The ultrastructure of the SC in maize and most other organisms is similar: it contains two electron-dense lateral elements (LEs) and one central element consisting of transverse filaments. Each lateral element, called the axial element before synapsis, is associated with the core of each homologous chromosome (Fig. 1). The composition and function of the SC at a molecular level is not well understood, although the studies on mutants deficient in synapsis and the use of antibodies made against purified SCs have allowed the identification of many proteins in the SC, including ASY1/HOP1 and AFD1/REC8 as AE/LE components (Hollingsworth et al., 1990; Watanabe and Nurse, 1999; Armstrong et al., 2002; Cai et al., 2003; Golubovskaya et al., 2006) and the coiled coil protein ZIP1 as a CE component. ZIP1 orthologues have been identified in several species (Meuwissen et al., 1992; Sym et al., 1993; Page and Hawley, 2001; Colaiacovo et al., 2003; Higgins et al., 2005; Wang et al., 2010). Although their primary amino acid sequences share limited similarity, all ZIP1 proteins possess an extended coiled coil-rich segment located in the centre of the protein, flanked by largely globular domains (Zickler and Kleckner, 1999; Page and Hawley, 2004).
Fig. 1.
Image of silver-stained maize synaptonemal complex showing the position of the central elements (CE) and the lateral elements (LE) in (A). Bar=1 μm. (B) A diagram of the tripartite structure of synaptonemal complex showing locations of LE and CE proteins.
The afd1 gene (absence of first division 1) in maize encodes an alpha kleisin, REC8-like protein that is a component of the cohesin complex and is responsible for regulating sister chromatid cohesion during meiosis (Golubovskaya and Mashnenkov, 1975; Golubovskaya et al., 2006). AFD1 is also required for AE elongation. Immunofluorescence of AFD1 demonstrated that it is recruited to chromosome axes at leptotene, and appears as numerous foci along the AE/LE in zygotene and pachytene. Homologous pairing, synapsis, and proper distribution of RAD51 depended on full AE elongation, providing the basis for a model in which AFD1 helps regulating homologous pairing, recombination and synapsis via its role in AE elongation and the subsequent distribution of the recombination machinery.
Maize, a diploid (2n=20) monocot grass of the Poaceae, is one of the few organisms with a large sequenced genome in which meiotic prophase events are amenable to analysis by a combination of cytological, genetic, molecular, and biochemical techniques. Using maize, the link between recombination and the cytological observation of crossing-over was first demonstrated by HB Creighton and B McClintock (1931). Maize was used to identify the leptotene–zygotene transition (prezygotene) as a cytologically-distinct stage in meiosis (Dawe et al., 1994), and to analyse the kinetics of chromosome pairing, bouquet formation, AE and SC formation, and recombination among other meiotic features (Dawe et al., 1994; Bass et al., 1997, 2000; Franklin and Cande, 1999; Carlton et al., 2003; Franklin et al., 2003; Pawlowski et al., 2003, 2004, 2009; Golubovskaya et al., 2006; Li et al., 2007; Wang et al., 2009). Using forward genetics screens based on sterility, over 60 mutants representing at least 35 genes have been identified in maize mostly by Inna Golubovskaya and associates (Golubovskaya et al., 2003; Cande et al., 2009). The majority of mutants in this collection fail to form or maintain bivalents, usually due to defects in homologous pairing, synapsis or recombination. Table 1 lists 15 mutants in our collection with abnormal synapsis. To characterize synapsis in these mutants further, the maize zyp1 gene was cloned and a ZYP1 antibody was generated. Using antibodies against ZYP1 and AFD1 and other methods such as transmission electron microscopy (TEM) of silver-stained SCs, the synaptic phenotypes of most of these mutants were determined. The criteria used to classify the phenotypes of mutants with problems in synapsis, and the behavior of the SC in several mutants, including one, dsy2, that is defective in the loading of ZYP1 onto paired chromosomes, are described here.
Materials and methods
Plant material
The inbred lines of maize (B73 or A344) were grown for about 6 weeks under greenhouse conditions and harvested throughout the year. Maize meiotic mutants were grown along with their wild-type siblings. The origin of all mutants is listed in the references associated with Table 1 and mutants from Dr Golubovskaya's personal collection are now available through the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/).
Cloning of Zea mays zyp1
Total RNA from A344 young tassel was isolated by Trizol (Invitrogen). Primers RW84 and RW85, which are specific to putative Zm zyp1 EST sequences, were used to amplify the predicted coding regions of zyp1 by RT-PCR. The amplified fragment was cloned and sequenced. The sequence was then used to design gene-specific primers. RACE (Rapid Amplification of cDNA Ends) was carried out with 3′ and 5′ RACE systems (Invitrogen) using gene-specific primers RW104, RW105, and RW109. RACE PCR products were cloned and sequenced. The maize zyp1 coding sequence was deposited in GenBank (accession number HQ116413).
Primers
RT-PCR and RACE-PCR primers used to amplify maize zyp1 were: RW84 (5′-GGAAACCTAGCTAGCAGTGAAAGTGAAAAG), RW85 (5′-CCACCGTTGTGCCATGTTCCTCCTTA), RW104 (5′-AACTGTTCTTTTCACTTTCACTGCTA), RW105 (5′-AAGCATGATTCTGAGAGGTATTTG), and RW109 (5′-ATTTTCTCCTCTTGGGCCATTTCATA) (see Supplementary Fig. S2 at JXB online for primer positions).
Antibody production and Western blot
To generate anti-ZYP1 antibody, a partial zyp1 cDNA corresponding to amino acids 15–345 of the ZYP1 protein was cloned into the pGEX plasmid in translational fusion with GST (see Supplementary Fig. S2 at JXB online). The protein was expressed in E. coli BL21. Upon induction using IPTG, the GST-ZYP1 fusion protein aggregated as insoluble inclusion bodies. Two gentle, non-ionic detergents (sarkosyl and Triton X-100) were used to break up and solubilize the inclusion bodies (Frangioni and Neel, 1993). The GST-ZYP1 fusion protein was purified with GST purification kit (GE Healthcare life sciences) and the GST tag was then cleaved using PreScission protease. The resulting protein was used to produce a polyclonal antibody in Guinea Pig (Covance). For Western blot analysis, 30 mg protein samples were separated by 6% SDS-PAGE and then transferred onto a polyvinylidene fluoride membrane (Millipore). Hybridization was performed using polyclonal primary antibody against ZYP1 protein (1:1000). Donkey anti-guinea pig antibody conjugated with horseradish peroxidase (1:5000) was used to detect the proteins. Protein bands were visualized by enhanced chemiluminescence substrate.
Cytology
For the survey, the families segregating for a particular meiotic gene were used. To discriminate mutant versus wild-type siblings, young tassels from 15 plants in each family were fixed in Farmer's fixative (3:1 ratio of 95% ethanol to glacial acetic acid) for 1–2 h. Immature anthers were stained with 2% acetocarmine, squashed, and observed with a light microscope to detect mis-segregrating chromosomes at diakinesis-metaphase I (Golubovskaya et al., 1993). For FISH, immunostaining, and immunoFISH, anthers with the appropriate developmental stage from the same floret and adjacent florets were fixed at room temperature in 4% formaldehyde in Buffer A (15 mM PIPES-NaOH (pH 6.8), 80 mM KCl, 20 mM NaCl, 2 mM EDTA, 0.5 mM EGTA, 0.2 mM spermine, 0.5 mM spermidine, 1 mM DTT, 0.32 M sorbitol) for 45 min as described in Golubovskaya et al. (2002). Meiocytes were embedded in polyacrylamide and handled for indirect immunofluorescence as described in Golubovskaya et al. (2006). Newly polymerized acrylamide pads attached to a coverslip were washed with 1× PBS and cells were permeabilized for 1 h in 1× PBS, 1% Triton X-100, and 1 mM EDTA, and then blocked for 2 h in 1× PBS, 3% BSA, 1 mM EDTA, and 0.1% Tween 20. Pads were incubated overnight in a humid chamber with a rat anti-AFD1 antibody (1:50) (Golubovskaya et al., 2006), guinea pig anti-ZYP1antibody, or a rabbit anti-AtASY1 antibody (1:200), kindly provided by FC Franklin (University of Birmingham, UK) (Armstrong et al., 2002). After washes, pads were incubated with Cy3-conjugated F(ab′)2 donkey anti-rat IgG (1:50) and Alexa 488-conjugated F(ab′)2 donkey anti-rabbit IgG (1:100) or Alexa 488-conjugated donkey anti-guinea pig IgG (1:100) secondary antibodies for 2 h. Chromosomes were stained with 10 μg ml−1 DAPI. FISH was performed as described in Golubovskaya et al. (2002).
Staging criteria were as described previously (Dawe et al., 1994; Bass et al., 1997; Golubovskaya et al., 2002; Wang et al., 2009). The protocol described in Golubovskaya et al. (2002) was used to take images of maize meiocytes. Images were acquired on a Delta Vision (Applied Precision) imaging station: an Olympus IX70 inverted microscope with ×100, 1.35 NA oil-immersion lens and a photometric (Roper Scientific) CCD. All images were taken with a Z step size of 0.2 μm, saved as 3-D stacks, and subjected to constrained iterative deconvolution. Three-dimensional data analysis and two-dimensional image creation were performed using the DeltaVision/SoftWoRx software package (Applied Precision). Two-dimensional images were converted to TIFF and opened in Photoshop on a Macintosh computer.
TEM of chromosome spreads of male meiocyte nuclei was used to study the extent of synapsis. The spreads were treated with silver nitrate, which stains the AEs present at leptotene and early zygotene that later become the lateral elements of the SC, after the SC is formed between paired chromosomes at pachytene. SC spreads were prepared and analysed as described in Golubovskaya et al. (2002).
Results
In our mutant collection, 15 uncloned mutants were identified that appear to have problems undergoing synapsis (Table 1). When acetocarmine stained squashes of mutant meiocytes were analysed by light microscopy at diakinesis–metaphase I, all 15 mutants had reduced but a variable number of synapsed chromosomes, i.e. bivalents (see also Golubovskaya et al., 2003). However, the molecular bases of their synaptic problems are unknown. A variety of cytological techniques was used here to describe the synaptic phenotypes of ten of the mutants further, as a first step in determining why they fail to undergo normal synapsis. This includes using TEM of silver-stained prophase chromosome spreads, FISH using telomere probes, and immunostaining with antibodies raised against proteins (ASY1, AFD1, and ZYP1) that participate in SC formation to identify improper synapsis. Based on this analysis, two classes of mutants can be defined. First, some mutants may fail to form or maintain a normal synaptonemal complex or the SC forms slowly or not at all. This phenotype will be referred to here as improper synapsis. Secondly, in some mutants the SC is formed between non-homologous chromosomes and in the most obvious examples, chromosomes fold back upon themselves or switch partners during SC formation. In these mutants the structure of the SC is normal but pairing of homologous chromosomes is defective. This phenotype is referred to as non-homologous synapsis. Although asynaptic and desynaptic mutants, i.e. mutants incapable of forming or maintaining an SC, have an improper synapsis phenotype, most of the previously named asynaptic and desynaptic mutants of maize (see Table 1) are incorrectly specified since they form normal SCs. When these mutants were originally named, they were often classified as asynaptic or desynaptic based on having univalents at diakinesis. However, with the exceptions of dsy2 and mei*N2415, these mutants have a non-homologous synapsis phenotype.
SCs in wild-type and mutant cells
Silver-stained spreads of synapsing chromosomes as visualized by TEM are an important tool for monitoring SC behaviour in mutant meiocytes as compared with the wild type (Fig. 2). Although chromatin morphology and the overall 3D organization of the nucleus is not retained in these preparations, many aspects of normal and abnormal synapsis can be distinguished including the ability to form the tripartite SC, the extent of homologous and/or non-homologous synapsis, and the formation and resolution of interlocks (Golubovskaya et al., 2002).
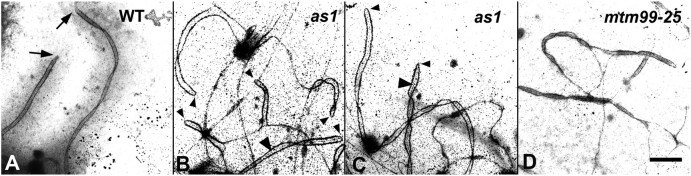
Fig. 2.
Transmission electron microscopy of silver-stained spreads of synaptonemal complexes (SC) from wild-type A344 inbred and as1 and mtm-99-25 mutants. Bar=1 μm. (A) Fragments of two SCs and associated chromatin at the pachytene stage of wild-type showing complete synapsis of homologous chromosomes. Note that LEs of each SC are not connected at the telomere ends (arrows). (B, C) Short and long foldbacks, and hairpin-like structures in the middle of chromosomes in as1 showing non-homologous synapsis. Compare the ends of foldbacks (arrowheads) and the normal telomere ends (arrows in A). Note that the central element (CE) has assembled between the LEs of foldbacks (large arrowheads). (D) An early diplotene in mtm99-25. Both pairing partner switches and foldbacks are present in the same chromosome and a CE has formed between non-homologous chromosomes. The SC is beginning to disassemble; unsynapsed AEs disappear while synapsed LEs are more stable.
In regular meiosis at pachytene, all chromosomes are completely synapsed and the tripartite SC structures are established (Figs 1A, 2A). As shown in Fig. 2B–D, meiotic mutants show several major types of irregular chromosome synapsis. Completely unsynapsed chromosomes can be seen as univalents (data not shown). Meiocytes with incomplete synapsis contain unsynapsed regions which are often found at the ends of chromosomes in mutants. When unsynapsed regions occur interstitially, the unsynapsed axes look like bubbles (Fig. 4A–H). Non-homologous chromosome synapsis has been observed in a majority of the mutants listed in Table 1. Two types of non-homologous chromosome synapsis can be distinguished: an exchange of synaptic partners when more than two chromosomes are involved in non-homologous synapsis, and foldbacks, when a single chromosome folds and synapses with itself forming a pin-shaped loop within or at the end of a chromosome. The exchange of synaptic partners with another chromosome can occur once or multiple times and can involve several different SCs (Figs 2D). The foldbacks can be short or long; sometimes a whole chromosome is completely folded onto itself (Figs 2B, C, 4O–Q). Foldbacks and partner exchanges may coexist in the same nucleus. Moreover, a single chromosome can be involved in both types of non-homologous synapsis (Fig. 2D).
Fig. 4.
Axial element and central element behaviour in mutants. (A–D) In as1 at pachytene, DAPI-stained chromosomes (blue in merged image) look fully synapsed at higher magnification. AFD1 (red) is installed along the entire chromosome including an unsynapsed region that forms a bubble (arrowhead), and ZYP1 (green) is installed between synapsed chromosomes except at the bubble. The merged images of ZYP1 and AFD1 are yellow because of colocalization of two signals. (E–H) In mei*N2415, synapsis is retarded at zygotene. Immunostaining with ASY1 (green) and AFD1 (red) and DAPI staining of chromatin (blue) shows incomplete synapsis of homologous chromosomes. Arrowheads point to two unsynapsed bubbles seen between short regions where chromosomes are paired and synapsed. (I–L) Early zygotene nucleus of mtm00-10 immunostained with antibodies against ASY1 (green) and AFD1 (red) and with DAPI-stained chromatin (blue). The chromosome axes are longer in mtm00-10 than in the wild type. A small foldback (arrow) and a synapsed chromosome region (small arrowhead) are stained with AFD1 but not with ASY1. In the merged image (L), unsynapsed chromosome threads are stained with both antibodies, and their staining is discontinuous and does not overlap along the chromosome axis (large arrowhead). The synapsed regions are intense red due to visualization of AFD1 staining only (small arrowhead). (M, N) Irregular synapsis in mtm00-10 zygotene nucleus. The image of immunostaining (M) with antibodies against ASY1 (green), AFD1 (red), and DAPI-stained chromatin (blue) shows a foldback (arrow) and non-homologous synapsis (small arrowhead) associated with an interlock (large arrowhead). (N) A cartoon of the foldback and interlock. (O–Q) In mtm99-25, a foldback of a pachytene chromosome is shown at high magnification with DAPI staining for chromatin (O, blue in Q) and immunostaining with an antibody against ZYP1 (P, green in Q). ZYP1 is installed between the two axes of the folded chromosome. Bar=1 μm. (R) In mei*N2415, a part of a zygotene nucleus immunostained with antibodies against ASY1 (green) and AFD1 (red), and DAPI-stained chromatin (blue). The unsynapsed chromosome (arrow) stained only with ASY1 (green), is twisted and forms a few bubbles. It appears to be entangled with a synapsed chromosome (stained with AFD, red) forming an interlock. Except for the bar in (Q), other bars=5 μm.
Unexpectedly, some meiotic mutants showed a preference for partner exchanges while other mutants show more foldbacks. dsy1-1 showed a preference for partner exchanges (Timofejeva and Golubovskaya, 1991; Golubovskaya et al., 1997). The mtm99-25 mutants (Figs 2D, 4O–Q) exhibited random synapsis: homologous synapsis co-existed with all categories of non-homologous synapsis including foldbacks in the same nuclei. Irregular synapsis began early in zygotene and persisted throughout late prophase. However, asynaptic1 (as1) and mtm00-09 prophase chromosomes preferentially form foldbacks, as illustrated in spreads prepared from as1 (Fig. 2B and C). In mtm00-09 (data not shown), whole chromosome foldbacks are frequent.
Analysis of molecular components of synaptonemal complex
The identification of genes encoding components of the SC by forward and reverse genetics has permitted molecular analysis of synapsis in both wild-type and meiotic mutants. The 3D microscopy of prophase nuclei (or meiocytes) immunostained with antibodies raised against the AE proteins ASY1, AFD1, and the CE protein ZYP1 is important for establishing the kinetics of SC formation (Golubovskaya et al., 2006; Wang et al., 2009). ASY1 and AFD1 appear in the nucleus as early as leptotene (see Supplementary Fig. S1 at JXB online). Chromosomes are seen as distinctive thin threads at this stage, and both proteins are recruited to the chromosome cores (see Supplementary Fig. S1A–D at JXB online). ASY1 staining is brighter than AFD1 at leptotene. At zygotene, when the bouquet is formed and chromosomes start to synapse, both proteins are visible on unsynapsed regions of chromosomes and have an equally bright staining (data not shown). At pachytene, when all chromosomes are synapsed, the AFD1 immunostaining is much brighter than ASY1 (see Supplementary Fig. S1I–L at JXB online). It has previously been suggested that the ASY1 epitope is masked by the assembly of the CE since the differences in intensity of staining are lost if fixation did not adequately preserve the structure of condensed chromatin (Golubovskaya et al., 2006). This difference in staining between AFD1 and ASY1 is useful for detecting unsynapsed versus synapsed regions of chromosomes (Wang et al., 2009).
The full sequence of the maize zyp1 gene was obtained using a reverse genetics approach. In Arabidopsis, ZYP1a/ZYP1b encode closely related proteins with structural similarity to the transverse filament protein of the SC and are thought to be yeast Zip1 homologues. The two Arabidopsis genes are functionally redundant and have an extended coiled-coil region flanked by globular domains at the N and C termini (Higgins et al., 2005). Using the sequence of Arabidopsis ZYP1a/ZYP1b genes, a BLAST search was conducted to find potential homologues in the maize genome. Two maize EST sequences, CD437886 and DY401328, sharing similarity to the coiled-coil domain of AtZYP1a/ZYP1b genes and the rice gene ZEP1 (Wang et al., 2010) were found in GenBank (http://www.ncbi.nlm.nih.gov/). The maize zyp1 gene transcript is 3061 bp long and encodes a 100.4 kDa protein of 867 amino acids (see Supplementary Fig. S2 at JXB online). The coiled-coil regions share ∼68% similarity with Arabidopsis ZYP1a/ZYP1b genes. Although Arabidopsis has two ZYP1 genes, only one zyp1 gene was found in the maize genome (http://www.maizegdb.org/). The N-terminus region (aa 15–345), the domain responsible for parallel homodimer formation, was chosen to be expressed in E. coli and used to develop a polyclonal antibody. The antibody was tested by Western blot analysis and showed specificity to ZYP1 protein. It detected a ∼100 kDa protein in the extract prepared from anthers, and the band is absent in extracts prepared from somatic tissues (see Supplementary Fig. S2 at JXB online).
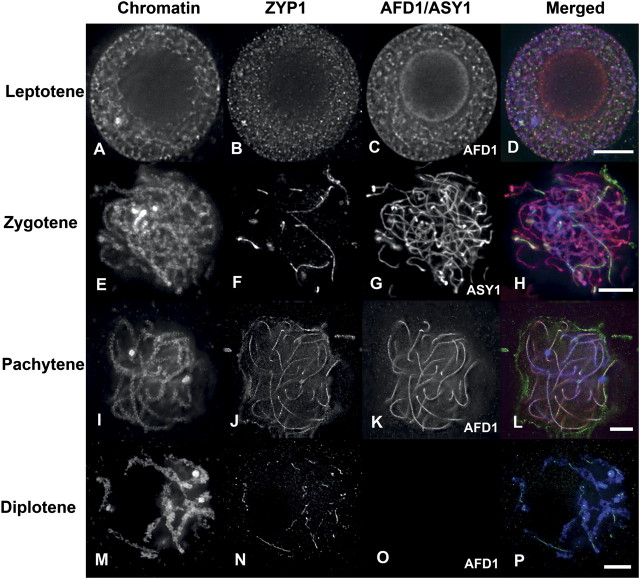
Immunolocalization experiments on maize meiocytes showed that ZYP1 appears at leptotene as diffuse staining the nucleus (Fig. 3A–D). During zygotene, the ZYP1 loads onto synapsed regions and forms linear signals between homologous chromosomes (Fig. 3E–H). At early zygotene, ZYP1 is loaded on at the subtelomeric regions associated with the telomere bouquet and only a few short stretches of staining can be observed in the interstitial regions of chromosomes. At pachytene, ZYP1 forms a continuous line between homologous chromosomes (Fig. 3I–L). At late pachytene, ZYP1 becomes fragmented and begins to disperse from the chromosomes (Fig. 3M–P). This staining pattern indicated that ZYP1 is likely to be a central element component of the SC.
Fig. 3.
ZYP1 protein behaviour in wild-type A344. DAPI staining (blue in merged) image for chromatin and immunostaining with antibodies against ASY1 or AFD1 (red) and ZYP1 (green) are shown for the prophase stages. The merged image appears yellow where red and green signals overlap (see L). In leptotene (A–D), both proteins are present in the nucleus; AFD1(C) is beginning to polymerize on chromosomes whereas ZYP1 (B) occurs as small dots throughout the nucleus. In zygotene (E–H), ZYP1 (F) is beginning to load onto synapsed chromosomes at regions in close juxtaposition. In pachytene (I–L), synapsis is complete and ZYP1 (J) is installed between all homologous chromosomes. In diplotene (M–P), the SC has disassembled and ZYP1 is maintained only near chiasmata. Bar=5 μm.
Distribution of SC components in most mutants is normal
Meiotic mutants with problems in synapsis have been surveyed using double ASY1-AFD1 immunostaining with the goal of discovering mutants that fail to distribute these proteins normally during synapsis (Table 1). As described elsewhere, the afd1-1 mutant did not show any staining for the AFD1 protein and this was accompanied by poor staining and irregular assembly of ASY1 and ZYP1 proteins (Golubovskaya et al., 2006). However, all other mutants examined so far do not show any irregularity in the timing or appearance of these AE proteins, and their pattern of localization on prophase chromosomes was similar to that observed in the wild type. In mtm99-25, for example, foldbacks were found with ZYP1 assembled inside the chromosome loops (Fig. 4O–Q). as1 was selected to demonstrate the behaviour of SC proteins during non-homologous synapsis (Fig. 4A–D; see Supplementary Fig. S3 at JXB online). Although non-homologous synapsis occurs during prophase in as1 meiocytes (Fig. 2B, C), neither ASY1, AFD1, nor ZYP1 showed any irregularities in timing of appearance and location on chromosomes from early leptotene through pachytene.
Visualization of intermediates in homologous synapsis in mutants
In general, mutants with problems in synapsis progress through meiotic prophase more slowly than wild-type cells, and this has allowed the visualization of intermediates in synapsis that are rarely observed in wild-type meiocytes. The progression of synapsis in the mei*N2415 mutant is more retarded than in other mutants. In mei*N2415 meiocytes, some chromosomes fail to start or accomplish synapsis and install CEs, and, consequently, univalents appear at zygotene, and persist until MI (data not shown). At zygotene, many chromosomes formed spectacular multiple desynaptic bubbles (Fig. 4E, H, R; see Supplementary Fig. S4 at JXB online). These bubbles persist until late pachytene when most chromosomes finally accomplish synapsis and deposit ZYP1. A desynapatic bubble was only observed once in a wild-type meiocyte during zygotene (Fig. 3E–H). Interestingly, the distribution of desynaptic bubbles along chromosomes demonstrates that, as synapsis is initiated, juxtaposition of homologous chromosomes is not limited to subtelomeric regions, but also occurs in interstitial regions. This is also supported by observations of partial deposition of ZYP1 at interstitial regions on pachytene chromosomes in mei*N2415 (Fig. 4R; see Supplementary Fig. S4 at JXB online), even though homologous chromosomes show proper juxtaposition along their length.
Another mutant with extremely slow prophase I progression, mtm00-10, differs in detail from mei*N2415. In mtm00-10 mutant, non-homologous synapsis as well as other irregularities start early in zygotene. In addition, the chromosome axes in this mutant at leptotene–zygotene are longer than chromosomes axes in the wild type or other mutants. The distribution of axial element components along these extended axes confirm that AFD1 and ASY1 form foci along the axes but the foci are not colocalized (Fig. 4I–L). Moreover, the extended chromosome morphology permitted visualization of foldbacks and interlocks, which previously were only observed using high resolution microscopy methods such as TEM (Fig. 2) and 3D structured illumination microscopy (Wang et al., 2009). Foldbacks are a prominent feature of abnormal synapsis in mtm00-10 and interlocks associated with foldbacks were observed in mtm00-10 nuclei (Fig. 4M, N).
desynaptic2 (dsy2) fails to assemble ZYP1
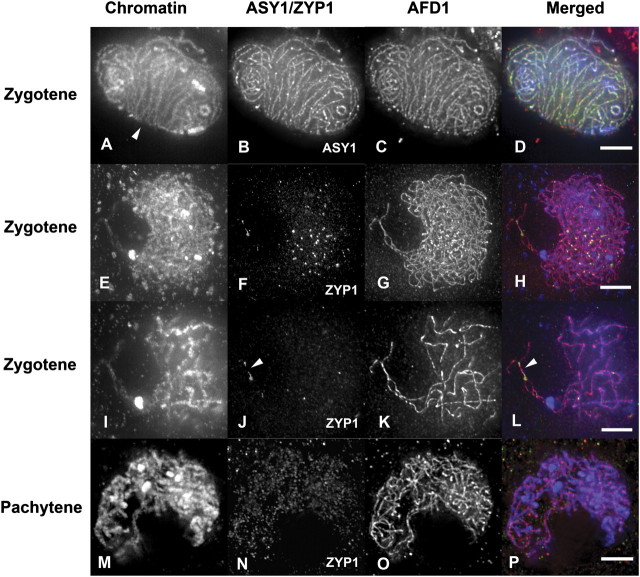
A TEM study of silver-stained chromosome spreads from dsy2 cells revealed desynaptic bubbles on closely aligned homologous chromosomes, however, there was no indication of non-homologous synapsis (Franklin et al., 2003). Double immunostaining with antibodies against ASY1 and AFD1 revealed that both proteins were loaded onto chromosome cores normally in dsy2 meiocytes, suggesting that the formation of the LE is unaffected by the dsy2 mutation (Fig. 5). Bouquet formation is also normal in the mutant (Fig. 5A–D). However, the central element is not assembled between homologues even when they are in close juxtaposition. ZYP1 staining was only observed on a few short regions of closely aligned homologues during zygotene. However, at pachytene, ZYP1 staining is no longer observed (Fig. 5, compare 5F and 5N). It seems that ZYP1 is able to load at certain sites on pairing chromosomes that are in close alignment; however, deposition cannot be extended along the chromosome axes nor maintained between chromosomes to the end of pachytene. Since heterochromatic knobs are paired (Fig. 5K, L), the homology search and pairing processes appear to be normal. These observations suggest that dsy2 plants carry a mutation that affects the loading of ZYP1 onto paired chromosome axes.
Fig. 5.
ZYP1 is not loaded onto paired chromosomes in desynaptic 2 (dsy2). Immunostaining of dsy2 meiocytes with antibodies against ASY1 or ZYP1 (green in merged) and AFD1 (red), and DAPI (blue) for chromatin shows pairing of homologous chromosomes but abnormal synapsis and improper loading of ZYP1. (A–D) Immunostaining with ASY1 (green) and AFD1 (red) shows that both proteins are loaded normally on the chromosome axes in zygotene, and a bouquet has formed (arrowhead in A). (E–L) Immunostaining with ZYP1 (green) and AFD1 (red) shows that ZYP1 foci are scattered throughout the zygotene nucleus. No long filaments were observed. In (I–L) a projection based on 10 optical sections (rather than 70 as in E–H) is shown at higher magnification. Several ZYP1 foci (J) are seen on homologous chromosomes in close juxtaposition (arrowhead), but ZYP1 fails to extend along chromosomes (L). (M–P) ZYP1 staining (green) has disappeared from the pachytene nucleus but AFD1 staining (red) is still present. Bar=5 μm.
Discussion
The non-homologous synapsis phenotype
The synaptonemal complex is an evolutionarily conserved structure formed in almost all organisms that undergo meiosis. There are multiple reasons why it may fail to form and function properly in mutants. As has been shown previously for several maize mutants, failure to pair homologous chromosomes (e.g. phs1), to undergo homologous recombination (e.g. rad51 double mutants), to form axial elements (e.g. afd1), or a bouquet (e.g. pam1), all lead to abnormal synapsis phenotypes (Golubovskaya et al., 2002, 2006; Pawlowski et al., 2004; Li et al., 2007). As a first step in analysing our collection of maize meiotic mutants, an attempt was made to classify genes according to the events they control. The failure to undergo normal synapsis is a prominent feature of many of these meiotic mutants. The underlying molecular lesions leading to these phenotypes are not known.
Most of our abnormal synapsis mutants show an inability to pair properly. These include as1, dsy1, mtm00-09, mtm00-10, mtm99-14, mtm99-25, mtm99-30 and dsyCS (Table 1). These mutants illustrate that pairing and synapsis can be uncoupled. Provided chromosomes are in close juxtaposition and the chromosomes have normal AEs, a central element can form between the two LEs even if they are not on homologues forming a tripartite SC. In haploid maize (Ting, 1971) and other plants as well as in some interspecific hybrids, when homologous partners are not available, non-homologous synapsis occurs (Jenkins, 1985). These results imply that there must be mechanisms to promote SC formation between homologues. However, if homologous synapsis is delayed or cannot occur, chromosomes are able to synapse non-homologously (Pawlowski and Cande, 2005).
Table 1.
Meiotic mutants with synaptic defects leading to univalents at diakinesis
| Name of mutant | Reference | Alleles | Phenotype |
| asynaptic 1 (as1) | (Beadle, 1930) | as1 | N.H.a |
| desynaptic 1 (dsy1), 2 alleles | (Golubovskaya and Mashnenkov, 1976; Timofejeva and Golubovskaya, 1991; Bass et al., 2003) | dsy1-1, dsy1-9101 | N.H. |
| desynaptic2 (dsy2) | (Golubovskaya, 1989; Franklin et al., 2003) | dsy2 | Improper synapsis (desynaptic) |
| mei*N2415 | MG. Neuffer's mutant collection, obtained from Maize Genetics Cooperation Stock Center | ms*N2415 former name | Improper synapsis (slow) |
| mtm99-14b | (Golubovskaya et al., 2003) | mtm99-14 | N.H. |
| mtm99-25 | (Golubovskaya et al., 2003) | mtm99-25 | N.H. |
| mtm00-10 | mtm00-10 | N.H. | |
| mtm99-30 | mtm99-30 | N.H. | |
| mtm00-09 | (Golubovskaya et al., 2003) | mtm00-09 | N.H. |
| desynapticCS | (Staiger and Cande, 1990; Golubovskaya et al., 2003) | dsyCS | N.H. |
| desynaptic 9303 | (Golubovskaya et al., 2003) | dsy*9303 | Not known |
| desynaptic 9305 | dsy*9305 | Not known | |
| desynaptic 9904 | dsy9904a | Not known | |
| desynaptic 9905 | dsy9905a | Not known | |
| desynaptic 9906a | dsy9906a | Not known |
N.H. represents nonhomologous synapsis.
mtm represents maize targeted mutagenesis.
When acteo-carimine stained squashes of meiocytes were analyzed by light microscopy at diakinesis –metaphase I, all nuclei had reduced but variable numbers of synapsed chromosomes, i.e. bivalents, when compared to wild type meiocytes (see also (Golubovskaya et al., 2003).
Pairing is known to require the installation of the recombination machinery in most organisms. The phs1 and double rad51 mutants fail to complete homologous recombination and, consequently, show non-homologous synapsis, with extensive pairing partner switches. This occurs after a delay to initiate homologous synapsis. RAD51 is not loaded onto zygotene chromosomes in phs1. When meiocytes of the phs1 mutant are in zygotene and early pachytene as defined by stage-specific chromosome morphology (Dawe et al., 1994; Golubovskaya et al., 2006), the chromosomes are completely unsynapsed. In late pachytene the chromosomes synapse quickly but non-homologously and by light microscopy synapsis appears to be complete. However, monitoring homologous synapsis by FISH with a probe for the 5S rDNA locus on chromosome 2L showed that almost all 5S rDNA foci remain unpaired at both stages: at zygotene, when chromosomes are unsynapsed and at pachytene when chromosomes are completely synapsed (Pawlowski et al., 2004). The behaviour of non-homologously synapsed chromosomes in the double rad51 mutant is similar, but recombination can occur between non-homologous chromosomes leading to non-homologous chiasma formation and multivalents at metaphase I (Li et al., 2007). The prophase chromosomes in pam 1, also show random synapsis, including foldbacks, and extensive interlocks as they pair. However, this mutant is unique in that it affects the movement of telomeres on the nuclear envelope retarding the formation of the telomere bouquet (Golubovskaya et al., 2002). Its impact on pairing and synapsis may be indirect by preventing the proper alignment of homologous chromosomes as they pair.
Although most of the irregular synapsis mutants examined have problems in pairing there are some unexplained complications. Several show a bias for foldbacks versus pairing partner switches and as1 and mtm00-09 fall into this group. Chromosomes of as1 mutants were involved in both large and small foldbacks, but not in exchanging partners; and in mtm00-09 whole prophase chromosome foldbacks occurred frequently. The numerous foldbacks in as1 could explain the puzzling observations of discontinuous gaps in SC structure in longitudinally sectioned pachytene chromosomes as observed using TEM (Maguire and Riess, 1991). Random chromosome synapsis characterizes both dsy1 and mtm99-25, and it occurs at early zygotene and persists throughout pachytene. However, the mtm-99-25 meiocytes have more severe irregularities; the same chromosome can exchange their partners for synapsis, and simultaneously form foldbacks (Fig. 2D).
What could explain a bias for foldbacks? During homologous recombination, repair of double strand breaks is usually made using the homologue not the sister as a template (Zickler and Kleckner, 1999). Perhaps in the mutants that have a bias towards foldbacks there is more repair using the sister as a template and this promotes SC initiation at kinks in the chromosome leading to self synapsis, i.e. foldbacks. Alternatively, the molecular lesion leading to foldbacks could be due to an altered chromosome morphology that favours sister chromatid interactions at the expense of homologous chromosome interactions. The phenotypes of mtm99-25 and mtm00-10 support this idea. Both mutants have altered chromosome morphology. In mtm00-10 zygotene chromosomes have elongated axes (Fig 4I–L), and in mtm99-25, heterochromatin is less condensed at zygotene and pachytene than in the wild type (see Supplementary Fig. S5 at JXB onine).
In a few mutants synapsis appears to be normal except for a slowing of the kinetics of SC formation. In these mutants, for some chromosomes, SC formation is completed during pachytene, synapsis is normal, and chiasma formation is unaffected. But for other chromosomes, synapsis is incomplete and leads to the formation of desynaptic bubbles which could block the formation of chiasmata, or synapsis is not initiated leading to univalents at metaphase I. mei*N2415 falls into this class. Since this is an EMS mutation, it is likely that this is a point mutation not a null and chromosome behaviour may reflect a mild phenotype affecting either regulation of CE formation or the kinetics of CE assembly. The mtm00-10 mutation has a more complicated phenotype, consistent with having a non-homologous synapsis phenotype. The kinetics of synapsis is much slower than normal in this mutant but non-homologous synapsis especially foldbacks occur early in zygotene. As mentioned above, chromosome axes are longer than normal. These mutants have been used for identifying intermediate stages in homologous synapsis such as the formation of desynaptic bubbles. A meiotic progression phenotype leading to arrest or delay of synapsis is characteristic of genes affecting recombination events in yeast, i.e. the zmm mutants (Börner et al., 2004). The mutant mei*2415 is likely to fall into the category because at metaphase I a delay in chiasma resolution, as well as a delay in homologous synapsis and an extended prophase I was observed.
With two exceptions, the mutant phenotypes observed are not due to a failure to form proper AEs or CEs. By using markers for CE and LE components it was determined that AEs and CEs form with normal timing and location in most of the mutations in our collection. It has previously been shown that afd1, a REC8 orthologue (Watanabe and Nurse, 1999) is essential for the maintenance of the AEs of the SC as well as in the regulation of sister chromatid cohesion (Golubovskaya and Mashnenkov, 1975; Golubovskaya et al., 2006; Wang et al., 2009). In this survey of mutants, dsy2 has been identified as another mutant essential for SC formation. It is a true synaptic mutant and the primary function of DSY2 may be to regulate the assembly of CE components, including ZYP1, on to paired chromosomes that show proper juxtaposition.
ZYP1 loading is blocked in the dsy2 mutant
The maize zyp1 gene was cloned based on its homology similarity to the At ZYP1a/ZYP1b genes and its amino acid sequence shares ∼40% identity to At ZYP1a/ZYP1b proteins. The maize, rice, and Arabidopsis ZYP1 proteins all have a similar domain organization in common with yeast Zip1, including two coiled-coil regions and globular domains at the N and C termini (Higgins et al., 2005; Wang et al., 2010). In addition, the distribution of maize ZYP1 protein during zygotene and pachytene is consistent with localization to the central element of the SC. Although no maize zyp1 mutants with strong phenotypes have been obtained, the maize zyp1 gene cloned in this study is likely to be a central element component of the maize SC.
Immunostaining with ZYP1 antibodies demonstrated that cells of dsy2 mutants are defective in ZYP1 installation on chromosomes. Bouquet formation and homologous chromosome recognition, as shown by the pairing of homologous knobs is normal in the mutant. Although CE installation starts at zygotene, the time when paired chromosomes begin synapsis, CE deposition fails to be maintained and only a few limited sites of ZYP1 assembly are found. ZYP1 is not maintained on chromosomes and disappears at pachytene, while in wild-type and in mutants with non-homologous synapsis, ZYP1 stains the entire axis of the synapsed chromosome. Previous studies showed that neither the switching of chromosome partners nor foldbacks was observed in the dsy2 mutant. RAD51 immunostaining is normal during leptotene, whereas the total number of RAD51 foci per nucleus during zygotene is reduced (∼125 versus ∼500) and elongated RAD51 structures are present on synapsing chromosomes (Franklin et al., 2003). Lacking normal SC formation to stabilize homologue interactions, RAD51 function may be affected, resulting in a significant reduction in the number of chiasmata and bivalent chromosomes at diakinesis. It is predicted that DSY2 regulates CE assembly. Experiments are in progress to identify dsy2 by map-based cloning.
Conclusion
To our surprise, analysis of the synaptic phenotype of mutants showing reduced numbers of bivalents has led to the identification of a large class of mutants that are defective in pairing, as opposed to SC formation. These mutants illustrate that synapsis and pairing can be uncoupled and suggest that there must be mechanisms to promote SC formation between homologues. However, non-homologous synapsis may be a default state in meiocytes since it can occur if homologous synapsis is delayed. One mutant, dsy2, is defective in the formation of the CE of the SC and may regulate ZYP1 assembly on paired chromosomes.
Supplementary data
Supplementary data is available at JXB online.
Supplementary Fig. S1. LE/AE behaviour of wild-type A344 line.
Supplementary Fig. S2. Amino acid sequence of ZYP1 and Western blot analysis.
Supplementary Fig. S3. Axial and central element behaviour in asynaptic1.
Supplementary Fig. S4. Retarded synapsis in mei*N2415.
Supplementary Fig. S5. Heterochromatin in mtm99-25 is less condensed.
Acknowledgments
We thank Chris Franklin for providing the ASY1 antibody, Joshua Wong for help in cloning zyp1, and Lisa Harper for critical reading of the manuscript. This research was supported by a grant from the National Institutes of Health GM 048547 to WZ Cande.
Glossary
Abbreviations
- SC
synaptonemal complex
- AE
axial element
- CE
central element
- LE
lateral element
- TEM
transmission electron microscopy
References
- Armstrong SJ, Caryl AP, Jones GH, Franklin FC. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. Journal of Cell Science. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- Bass HW, Bordoli SJ, Foss EM. The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L.) cause distinct telomere-misplacement phenotypes during meiotic prophase. Journal of Experimental Botany. 2003;54:39–46. doi: 10.1093/jxb/erg032. [DOI] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. Journal of Cell Biology. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Seda JW, Agard DA, Cande WZ. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. Journal of Cell Science. 2000;113:1033–1042. doi: 10.1242/jcs.113.6.1033. [DOI] [PubMed] [Google Scholar]
- Beadle GW. Genetic and cytological studies of a Mendelian asynaptic in Zea mays. Cornell Agriculture Experimental Station Memorandum. 1930;129:1–23. [Google Scholar]
- Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Burnham CR, Stout JT, Weinheimer WH, Kowles RV, Philips PL. Chromosome pairing in maize. Genetics. 1972;71:111–126. doi: 10.1093/genetics/71.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Dong F, Edelmann RE, Makaroff CA. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. Journal of Cell Science. 2003;116:2999–3007. doi: 10.1242/jcs.00601. [DOI] [PubMed] [Google Scholar]
- Cande WZ, Golubovskaya I, Wang CJR, Harper L. Meiotic genes and meiosis in maize. In: Bennetzen J, Hake S, editors. The maize handbook. New York: Springer; 2009. [Google Scholar]
- Carlton PM, Cowan CR, Cande WZ. Directed motion of telomeres in the formation of the meiotic bouquet revealed by time course and simulation analysis. Molecular Biology of the Cell. 2003;14:2832–2843. doi: 10.1091/mbc.E02-11-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Developmental Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Creighton HB, McClintock B. A correlation of cytological and genetical crossing-over in Zea mays. Proceedings of the National Academy of Sciences, USA. 1931;17:492–497. doi: 10.1073/pnas.17.8.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RK, Sedat JW, Agard DA, Cande WZ. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 1994;76:901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Analytical Biochemistry. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Franklin AE, Cande WZ. Nuclear organization and chromosome segregation. The Plant Cell. 1999;11:523–534. doi: 10.1105/tpc.11.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, Golubovskaya IN, Bass HW, Cande WZ. Improper chromosome synapsis is associated with elongated RAD51 structures in the maize desynaptic2 mutant. Chromosoma. 2003;112:17–25. doi: 10.1007/s00412-003-0242-8. [DOI] [PubMed] [Google Scholar]
- Golubovskaya I, Grebennikova ZK, Avalkina NA, Sheridan WF. The role of the ameiotic1 gene in the initiation of meiosis and in subsequent meiotic events in maize. Genetics. 1993;135:1151–1166. doi: 10.1093/genetics/135.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya IN. Meiosis in maize: mei genes and conception of genetic control of meiosis. Advanced Genetics. 1989;26:149–192. [Google Scholar]
- Golubovskaya IN, Grebennikova ZK, Auger DL, Sheridan WF. The maize desynaptic1 mutation disrupts meiotic chromosome synapsis. Developmental Genetics. 1997;21:146–159. [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ. Alleles of afd1 dissect REC8 functions during meiotic prophase I. Journal of Cell Science. 2006;119:3306–3315. doi: 10.1242/jcs.03054. [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Harper LC, Pawlowski WP, Schichnes D, Cande WZ. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.) Genetics. 2002;162:1979–1993. doi: 10.1093/genetics/162.4.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya IN, Mashnenkov AS. Genetic control of meiosis. I. Meiotic mutation in corn (Zea mays L.) afd, causing the elimination of the first meiotic division. Genetika (Russ) 1975;11:810–816. [Google Scholar]
- Golubovskaya IN, Mashnenkov AS. Genetic control of meiosis: II A desynaptic mutant in maize induced by N-nitroso-N-methylurea. Genetika (Russ) 1976;12:7–14. [Google Scholar]
- Golubovskaya IN, Sheridan WF, Harper LC, Cande WZ. Novel meiotic mutants of maize identified from Mu transposon and EMS mutant screens. Maize Genetics Cooperation Newsletter. 2003;77:10–13. [Google Scholar]
- Hamant O, Ma H, Cande WZ. Genetics of meiotic prophase I in plants. Annual Review of Plant Biology. 2006;57:267–302. doi: 10.1146/annurev.arplant.57.032905.105255. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes and Development. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Jenkins G. Synaptonemal complex formation in hybrids of Lolium temulentum× Lolium perenne (L.). II. Triploid. Chromosoma. 1985;92:387–390. [Google Scholar]
- Li J, Harper LC, Golubovskaya I, Wang CR, Weber D, Meeley RB, McElver J, Bowen B, Cande WZ, Schnable PS. Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics. 2007;176:1469–1482. doi: 10.1534/genetics.106.062604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP, Riess RW. Synaptonemal complex behavior in asynaptic maize. Genome. 1991;34:163–168. doi: 10.1139/g91-025. [DOI] [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO Journal. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes and Development. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annual Review of Cell and Developmental Biology. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Cande WZ. Coordinating the events of the meiotic prophase. Trends in Cell Biology. 2005;15:674–681. doi: 10.1016/j.tcb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Cande WZ. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. The Plant Cell. 2003;15:1807–1816. doi: 10.1105/tpc.012898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Timofejeva L, Meeley RB, Sheridan WF, Cande WZ. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science. 2004;303:89–92. doi: 10.1126/science.1091110. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Wang CJ, Golubovskaya IN, Szymaniak JM, Shi L, Hamant O, Zhu T, Harper L, Sheridan WF, Cande WZ. Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proceedings of the National Academy of Sciences, USA. 2009;106:3603–3608. doi: 10.1073/pnas.0810115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Cande WZ. Microtubule distribution in dv, a maize meiotic mutant defective in the prophase to metaphase transition. Developmental Biology. 1990;138:231–242. doi: 10.1016/0012-1606(90)90193-m. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Timofejeva LP, Golubovskaya IN. A new type of desynaptic gene in maize revealed by the microspreading method of synaptonemal complexes. Cytologia (Russ) 1991;33:3–8. [Google Scholar]
- Ting YC. Further studies on the synaptonemal complex of haploid maize. Genetics. 1971;68:67. [Google Scholar]
- Wang CR, Carlton PM, Golubovskaya IN, Cande WZ. Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics. 2009;183:905–915. doi: 10.1534/genetics.109.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. The Plant Cell. 2010;22:417–430. doi: 10.1105/tpc.109.070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annual Review of Genetics. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.