Abstract
General stress proteins, Gls24 and GlsB, were previously shown to be involved in bile salts resistance of Enterococcus faecalis and in virulence. Here, we identified 2 gene clusters in Enterococcus faecium each encoding a homolog of Gls24 (Gls33 and Gls20; designated on the basis of their predicted sizes) and of GlsB (GlsB and GlsB1). The sequences of the gls33 and gls20 gene clusters from available genomes indicate distinct lineages, with those of hospital-associated CC17 isolates differing from non-CC17 by ∼7% and ∼3.5%, respectively. Deletion of an individual locus did not have a significant effect on virulence in a mouse peritonitis model, whereas a double-deletion mutant was highly attenuated (P < .004) versus wild-type. However, mutants lacking either gls33-glsB, gls20-glsB1, or both all exhibited increased sensitivity to bile salts. These results suggest that gls-encoded loci may be important for adaptation to the intestinal environment, in addition to being important for virulence functions.
Enterococci have emerged as important opportunistic pathogens over the past 3 decades and now rank second among pathogens isolated from health care-associated infections in humans [1, 2]. Recent surveillance data have revealed a significant change in the ratio of E. faecium to E. faecalis isolated from nosocomial infections; as recently as the 1990s, E. faecium represented <10% of total enterococcal infections, but now it accounts for more than one-third of enterococcal clinical isolates identified to the species level in the United States and in some European hospitals [2–4]. Concurrently, clinical E. faecium isolates are much more resistant to multiple antibacterial agents, particularly ampicillin and vancomycin [5–7], than are E. faecalis isolates, making treatment of infections caused by this organism problematic. Furthermore, E. faecium isolates from nosocomial sources more often carry virulence-associated genes, such as an intact acm gene (encoding a collagen-binding adhesin) [8, 9], hyl (a putative hyaluronidase) [10–12], and esp (enterococcal surface protein) [13, 14], and most of the 14 fms genes encoding E. faecium surface proteins of the MSCRAMM (microbial surface components recognizing adhesive matrix molecules) family [15–17], including pili [15,18–20].
Bacteria are known to encounter various stress conditions in their natural environments and in the host when causing infections. Although these processes and their determinants remain uncharacterized for E. faecium, several factors involved in stress response in other gram-positive bacteria are thought to be involved in bacterial adaptation to hostile environments in the host and have been associated with virulence [21–25]. Previous studies with E. faecalis have shown that growth in the absence of glucose elicits the production of 42 glucose starvation proteins [26]. Because the expression of one of these proteins, Gls24, was shown to be induced under other stress conditions also, such as the presence of bile salts and cadmium chloride, it was designated as a general stress (Gls) protein. Insertional inactivation of the gls24 gene [27, 28] led to a reduced survival rate in the presence of bile salts. Subsequently, our mutation and complementation data identified glsB, located immediately downstream of gls24, as also being involved in bile salts tolerance of E. faecalis strain OG1RF. Disruption of gls24 in E. faecalis OG1RF resulted in significant attenuation in a mouse peritonitis model [27] and a rat endocarditis model [29], whereas no effect was observed with glsB in either model, indicating that gls24, but not glsB, is important for E. faecalis virulence. Our data also demonstrated protection by anti-Gls24 immune serum against E. faecalis infection in the mouse peritonitis model, suggesting that Gls24 may be useful as a target for immunotherapy [27].
In this study, we used in silico analysis to search for E. faecalis gls24 homologs in E. faecium and found 2 closely located gene clusters, each containing a homolog of gls24 and of glsB. The 2 gls24-glsB-like gene pairs were deleted independently and in combination to evaluate their role in bile salts response and in a mouse peritonitis model.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions
Escherichia coli, E. faecalis, and E. faecium strains and plasmids used are listed in Table 1. Plasmid constructs were given pTEX numbers, and their host strains were assigned respective TX numbers. E. coli strains were grown routinely in Luria-Bertani media (Difco Laboratories) and enterococci in either brain heart infusion (BHI) or Todd-Hewitt (TH) broth or agar (Difco Laboratories) at 37°C. The following antibiotic concentrations were used with enterococci: chloramphenicol, 10 μg/mL; erythromycin, 15 μg/mL, gentamicin, 125 μg/mL. With E. coli, the concentrations used were chloramphenicol, 10 μg/mL; erythromycin, 200 μg/mL; and gentamicin, 25 μg/mL.
Table 1.
Bacterial Strains and Plasmids Used in This Study
| Strains/plasmids | Relevant characteristics | Reference or source |
| Strains | ||
| Enterococcus faecium | ||
| TX16 (DO) | Sequenced strain; endocarditis isolate, Eryr, Gms | [30] |
| TX1330 | Sequenced strain; faecal isolate from a healthy volunteer, Erys, Gms | [31] |
| TX6060 | TX1330Δgls33-glsB::erm(B), single deletion mutant; Eryr | This study |
| TX6066 | TX1330Δgls20-glsB1, single deletion mutant | This study |
| TX6067 | TX1330Δgls33-glsB::erm Δgls20-glsB1, double deletion mutant, Eryr | This study |
| TX6064 | TX6060 with pAT392:: gls33-glsB, complementation construct, Eryr, Gmr | This study |
| TX6071 | TX6066 with pAT392::gls20-glsB1, complementation construct, Gmr | This study |
| TX6063 | TX6060 with pAT392, Eryr, Gmr | This study |
| TX6070 | TX6066 with pAT392, Gmr | This study |
| Escherichia coli | ||
| DH5α | E. coli cloning host | Stratagene |
| TX6059 | DH5α (pTEX6059) | This study |
| TX6065 | DH5α (pTEX6065) | This study |
| TX6065a | DH5α (pTEX6065a) | This study |
| TX6072 | DH5α with pAT392::gls33-glsB | This study |
| TX6073 | DH5α with pAT392::gls20-glsB1 | This study |
| Plasmids | ||
| pMSP3535 | Shuttle plasmid used as a source of erm(B) to create pTEX5504ts | [32] |
| pTEX5501ts | Temperature-sensitive suicide plasmid, used for deletion mutagenesis of E. faecium, Chlr, Gmr | [33] |
| pTEX5504ts | Derived from pTEX5501ts, used for deletion mutagenesis of E. faecium, Eryr, Gmr | This study |
| pAT392 | Vector used for complementation, Gmr | [34] |
| pTEX6059 | Plasmid for gls33-glsB deletion; flanking regions of the gls33-glsB on either side of erm(B) cloned into pTEX5504ts; Eryr, Gmr | This study |
| pTEX6065 | Plasmid for gls20-glsB1 deletion; flanking regions of gls20-glsB1 amplified using primers gls20-B1del1F, gls20-B1del1a R and gls20-B1del2F, gls20-B1del2R and cloned into pTEX5501ts; Chlr, Gmr, | This study |
| pTEX6065a | Plasmid for gls20-glsB1 deletion; flanking regions of the gls20-glsB1 amplified using primers gls20-B1del1F, gls20-B1del1b R and gls20-B1del2F, gls20-B1del2R and cloned into pTEX5501ts; Chlr, Gmr | This study |
| pTEX6072 | Plasmid for complementation; fragment containing gls33-glsB (1250 bp) cloned to pAT392 | This study |
| pTEX6073 | Plasmid for complementation; fragment containing gls20-glsB1 (853 bp) cloned to pAT392 | This study |
NOTE. Superscript “s” designates sensitivity, “r” designates resistance. Chl, chloramphenicol; Ery, erythromycin; Gm, gentamicin.
Standard Molecular Techniques
DNA from E. faecium isolates was prepared following the hexadecyltrimethyl ammonium bromide method [35]. Agarose plugs containing genomic DNA were digested with SmaI, and pulsed-field gel electrophoresis (PFGE) was performed as described previously [36], but with ramped pulse times beginning with 2 s and ending with 28 s, at 200 V for 23 h.
Construction of pTEX5504ts Vector
To construct plasmid pTEX5504ts, a polymerase chain reaction (PCR)-amplified erm(B)-containing fragment (using primers erm F and erm R; Supplementary Table 1) from the E. faecalis shuttle vector pMSP3535 [32] was digested with ApaI and HindIII and then ligated into similarly digested pTEX5501ts [33].
Construction of Deletion Mutants
E. faecium deletion mutants were constructed by allelic replacement, as described previously [33]. Briefly, upstream and downstream fragments (both ∼1 kb) flanking gls33 and glsB were amplified from E. faecium TX1330 genomic DNA using primers designed from the sequenced strain TX16 (for primers, see Supplementary Table 1) and ligated into pTEX5504ts; the construct was then transformed into E. coli DH5α to obtain TX6059 (Table 1) and confirmed by sequencing. After electroporation into E. faecium TX1330 (wild-type [WT]), single-crossover integrants (TX1330::pTEX6059; see Table 1) were selected on BHI plates that contained gentamicin and erythromycin. One single cross-over integrant grown for 12 serial passages at 42°C was serially diluted, plated, and grown at 37°C on nonselective media. These plates were then replica plated onto erythromycin and gentamicin plates to identify colonies that had lost the integrated plasmid by double crossover recombination. This deletion mutant, designated as TX6060 [TX1330Δgls33-glsB::erm(B)], was confirmed by sequencing, hybridization, and PFGE.
The same protocol was used to construct a clean gls20-glsB1 deletion, except that another temperature-sensitive vector derivative, pTEX5501ts [33], was used. The upstream and downstream fragments flanking gls20-glsB1 were cloned in this vector, resulting in pTEX6065.The resulting deletion mutant strain was designated as TX6066 (TX1330Δgls20-glsB1).
Attempts to create a gls20-glsB1 deletion in the gls33-glsB deletion mutant (TX6060), using the above construct (pTEX6065, above and in Table 1), were unsuccessful, so another reverse primer (gls20-B1del1b R, Supplementary Table 1) was designed for amplification of the upstream flanking region of gls20-glsB1 and to construct pTEX6065a (Table 1). As a result, the double deletion mutant, designated as TX6067 (TX1330Δgls33-glsB::erm Δgls20-glsB1), includes a 68-bp 5′ segment of the gls20 coding sequence.
Complementation Constructs
To complement the TX1330Δgls33-glsB::erm and TX1330Δgls20-glsB1 mutants in trans, a 1250-bp fragment containing the gls33-glsB open reading frames plus the gls33 ribosomal binding site (see Supplementary Table 1 for primers) and a 853-bp fragment containing the gls20-glsB1 open reading frames plus the gls20 ribosomal binding site (Supplementary Table 1) were cloned under control of the P2 promoter of the shuttle vector, pAT392 [34]. The resulting constructs were designated as TX6064 (TX6060 with pAT392::gls33-glsB) and TX6071 (TX6066 with pAT392::gls20-glsB1).
Growth Curves
Growth curves in BHI were determined as described previously [33].
Reverse Transcriptase (RT)-PCR
Isolation of total RNA from E. faecium TX1330, TX6060, TX6066, and TX6067, grown to mid-exponential phase in BHI, and RT-PCR (see Supplementary Table 1 for primers) were performed as described previously [15]. A 585-bp fragment of the gyrase gene (gyrA) of TX1330 was amplified as an internal control, and reactions without reverse transcriptase were used as additional controls to confirm the absence of DNA contamination in the total RNA preparation.
Antibodies
Anti-Gls24 antibodies raised in rabbits against E. faecalis OG1RF recGls24 [27] were affinity purified using recGls24 as the binding ligand, as described previously [9].
Protein Extraction and Western Blot
Surface protein extracts from E. faecium were prepared using 0.2% Zwittergent 3–12 (Calbiochem) as described previously [27]. Protein extracts were electrophoresed by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were incubated with affinity-purified anti-Gls24-specific antibodies followed by horseradish peroxidase-conjugated goat anti-rabbit antibodies. The blots were then developed with Supersignal West Pico chemiluminescent substrate (Pierce).
Bile Salts Resistance Assay
Resistance to bile salts was determined as described previously [27]. In brief, after 24 h growth in BHI at 37°C, E. faecium WT and mutants were centrifuged and resuspended in the same volume of fresh BHI. From these, 20 μL was then inoculated into 1 mL of fresh BHI containing 0.3% bile salts. After a 30 min incubation at 37°C, E. faecium cells were serially diluted and plated. The percentage of survival was calculated using a previously described equation [27].
Mouse Peritonitis Model
Four inocula (ranging from 2.6 × 108 to 3.8 × 109) of E. faecium TX1330 (WT), gls mutants, and complemented strains, grown in BHI broth premixed with 50% sterile rat fecal extract, were individually injected intraperitoneally into outbred ICR mice (Harlan Sprague Dawley) using 10 mice per inoculum, and mice were observed for 5 days, following our previously published method [37]. Preapproved guidelines of the Animal Welfare Committee of the University of Texas Health Science Center at Houston were followed throughout the animal experiments.
Statistical Analyses
For the bile salts assay, survival rates of WT versus gls mutants were compared using 1-way ANOVA (analysis of variance) with Bonferroni correction for multiple comparisons. The unpaired t test was used for complementation constructs versus the respective vector controls. For the peritonitis model, comparison of survival curves at similar inocula was performed using a log-rank test.
RESULTS AND DISCUSSION
Paralogous gls Loci in E. faecium
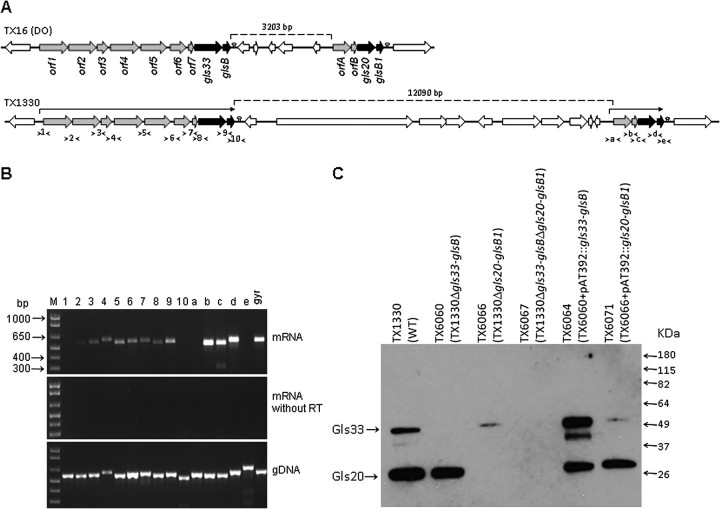
Because factors involved in the adaptation of E. faecium to stress environments have not been described, we searched the genomes of E. faecium strains TX16 (DO; endocarditis-derived) and TX1330 (healthy human volunteer feces-derived) for homologs of gls genes of E. faecalis that assist in resistance to bile salts [27, 28]. In both genomes, we identified 2 genes encoding close homologs of E. faecalis gls24 (88%–90% similarity with Gls24 of E. faecalis V583 and 87% similarity between the 2 E. faecium paralogs). One is encoded as part of a 9-gene cluster and the second as part of a 4-gene cluster, separated by ∼3.2 (TX16) or 12.1 (TX1330) kb regions (Figure 1A; see below). Each of these Gls24 homologs was followed by an 80 aa ORF exhibiting >90% similarity with GlsB of E. faecalis V583 (ca. 98% similarity between the 2 paralogs). The gls24 homolog found in the 9-gene cluster has an additional 130 aa at the N-terminus, compared with the E. faecalis homolog and the other E. faecium paralog, and was designated as Gls33 based on its predicted molecular mass of 33 kDa. The Gls24 homolog of the 4-gene cluster was named as gls20 (predicted protein 20 kDa) and the 2 GlsB homologs as GlsB and GlsB1, respectively. Of note, although E. faecalis OG1RF has a single gls24 locus, E. faecalis V583 has an additional gls-like locus in its pathogenicity island [38]. The percent similarities among different ORFs of each gls cluster of E. faecium and also versus E. faecalis are shown in Supplementary Table 2.
Figure 1.
Organization of the 2 gls loci of Enterococcus faecium and their transcriptional and protein expression analysis. A, Schematic representation of these loci in E. faecium TX16 and TX1330. The annotations were predicted from bioinformatics analysis for orf1 to orf7 and orfA to orfB, which are shown in Supplementary Table 2. Predicted transcriptional terminators downstream of glsB and glsB1 are indicated with lollypops. Arrows above each of the 2 gls clusters of TX1330 indicate messenger RNA (mRNA) co-transcripts. Location of each intergenic primer pair for reverse transcriptase polymerase chain reaction (RT-PCR) is shown with arrow heads (see panel B below). B, RT-PCR analysis of the 2 gls clusters of TX1330. Top gel, RT-PCR with DNase-treated total RNA (30 ng) isolated from mid-exponential cells; middle gel, control reaction of the same RNA preparation amplified without reverse transcriptase; bottom gel, control reaction amplified with genomic TX1330 DNA. An intragenic region of the gyrase gene was used as an additional control. Numbering and lettering of the lanes corresponds to the intergenic primer pairs shown in panel A. M, molecular weight marker. C, Western blot analysis of Zwittergent whole-cell extracts from E. faecium TX1330, its 3 gls deletion mutants, and complementation derivatives. The membrane was probed with affinity-purified anti-Gls24 (E. faecalis) antibodies [26], which cross-react with both Gls33 and Gls20. Locations of bands corresponding to Gls33 and Gls20 are indicated with arrows.
Transcriptional Analysis of the gls33 and gls20 Loci Confirmed That These Genes Are Co-Transcribed as 9- and 4-Gene Operons, Respectively
As shown in Figure 1A, in both TX16 and TX1330, gls33 and glsB are preceded by 7 ORFs oriented in the same direction and separated from each other by -8 (overlapping) to 79 bp (orf1 to orf7), whereas gls20 and glsB1 are preceded by 2 similarly oriented ORFs (orfA and orfB, separated by 12 bp). Annotations for the genes encoding ORF1 to ORF7 and ORFA to ORFB are listed in Supplementary Table 2. Because of short or overlapping intergenic regions and the absence of identifiable rho-independent transcriptional terminator-like sequences, we predicted that orf1 to glsB and orfA to glsB1 may be co-transcribed.
RT-PCR of messenger RNA (mRNA) isolated from TX1330, using primers spanning the intergenic regions of the 2 gene clusters, indicated that the genes from orf1 to glsB and from orfA to glsB1 are expressed by TX1330 cells during mid-exponential growth and that the genes in each cluster are expressed as a single mRNA transcript (Figure 1B). No PCR products were detected with primer pairs designed to amplify upstream regions of orf1 and orfA, which begin before their predicted promoter sequences and extend into their 5′ intragenic regions, indicating that the genes further upstream are not part of these operons. A strong hair-pin loop 133 bp downstream of glsB (free energy -22.9 kcal/mol) and another 43 bp downstream of glsB1 (-19.9 kcal/mol) were identified as transcriptional terminators for these operons. Consistent with these predictions, no PCR product was detected with primer pairs that include the 3′ end of glsB or glsB1 and extend beyond the predicted transcriptional terminators. Taken together, our analyses indicate that both gene clusters, from orf1 to glsB and orfA to glsB1, are transcribed as single polycistronic mRNA and, thus, are organized as operons. However, the gene compositions of both of these operons are only partially similar to the 6-gene gls operon present in E. faecalis [27].
Organization of Both gls Loci Is Highly Conserved among Sequenced E. faecium Isolates of Clinical and Nonclinical Origin
Our most recent searches with other currently available E. faecium draft genomes (by the Broad Institute, MA, and available at NCBI [39]) also identified orf1,2,3,4,5,6,7-gls33-glsB and orfA,B-gls20-glsB1 operons in 8 of the 9 genomes; the missing orf1-glsB locus in one of the genomes may be attributable to the incomplete status of this genome assembly (currently 220 contigs). The sequences of these clusters from the 11 strains (including TX16 and TX1330) were found to represent 2 distinct groups. Group A consists of 6 strains (all from clinical origin and belonging to the hospital-associated genogroup CC17 [40]) with >99% identity to each other over the entire orf1-gls33-glsB cluster but only ∼93% identity with the 5 group B isolates (3 derived from community human feces and 2 from human blood, all non-CC17 and belonging to unrelated STs that differ by more than 5 alleles). Similarly, the orfA-gls20-glsB1 cluster showed 100% identify within Group A isolates but ∼96.5% identity with the B isolates. Interestingly, of the 8 E. faecium draft genomes where both gls loci are located in the same contig, the distance between the 2 loci (glsB and orfA) is ∼3.2 kb in all 4 group A isolates (including TX16) and ∼12.1 kb in the 4 group B isolates (including TX1330). More than 90% of the ∼3.2 kb spacer region of TX16 is also present in the longer spacer of TX1330, which is predicted to carry 5 additional ORFs. Thus, our analysis suggests that there are 2 common variants of this region in E. faecium, in terms of sequence length and gene composition.
Characterization of gls33-glsB and gls20-glsB1 Deletion Mutants
We initially constructed an insertion mutant of gls33 by single cross-over mutagenesis in TX1330 and tested it in a mouse peritonitis model. Although our results showed delayed mortality of the gls33 insertion mutant, compared with WT TX1330, this difference did not reach statistical significance (data not shown). To circumvent disadvantages associated with single cross-over mutants (eg, instability of the mutant or possible expression of residual gls33), we next generated single and double deletion mutants of the gls loci in TX1330 (Supplementary Figure 1A, panel A). Comparison of growth curves of TX1330 (WT), TX6060 (TX1330Δgls33-glsB::erm(B), TX6066 (TX1330Δgls20-glsB1), and TX6067 [TX1330Δgls33-glsB::erm(B) Δgls20-glsB1] showed almost identical growth rates (OD and CFU) in BHI (data not shown), indicating that these genes are not essential for in vitro growth of E. faecium.
i. mRNA Studies.
Analysis of the genes located immediately upstream of gls33 (orf6-orf7) and gls20 (orfA-orfB) in these 3 mutants by RT-PCR showed that the gene deletions had no effect on expression of the upstream genes (versus WT), indicating that gls33-glsB and gls20-glsB1 are not involved in transcriptional autoregulation of these operons (Supplementary Figure 1B). Furthermore, the transcription of the remaining gls gene pair in each of the 2 single deletion mutants showed no difference from the WT (Supplementary Figure 1B), suggesting that the 2 gls loci do not cross-regulate each other's expression, at least not in BHI.
ii. Protein Studies.
Because Gls24 of E. faecalis is similar to Gls33 and Gls20 (see above) and, thus, antibodies against these proteins may cross-react, we used an affinity-purified polyclonal anti-Gls24 antibody in a Western blot analysis of surface protein extracts from WT TX1330 and the 3 deletion mutants. We detected 2 bands in the TX1330 extract (Figure 1C) corresponding to Gls33 and Gls20, although they migrated at ∼50 kDa and ∼28 kDa in SDS-PAGE, respectively (our previous study with Gls24 of E. faecalis also showed migration at ∼28 kDa; this may be due to the pI values of these proteins). In addition, the larger band was absent in the gls33-glsB deletion (TX6060) and the lower band in the gls20-glsB1 deletion (TX6066), whereas both were absent in the double deletion mutant (TX6067). Hence, this confirms the lack of protein expression of Gls33 and Gls20 in surface extracts of their respective deletion mutants. Furthermore, complementation of the deleted genes in trans restored expression of Gls33 and Gls20 to WT levels in TX6064 (TX6060 with pAT392::gls33-glsB) and TX6071 (TX6066 with pAT392:: gls20-glsB1), respectively (Figure 1C). The extra band in TX6064 is likely to be a degradation product of the over-expressed protein. Because TX6064 is a complementation construct of a single deletion mutant lacking only Gls33, it is expected to have the other band, which is Gls20. A similar band pattern was observed in >3 experiments.
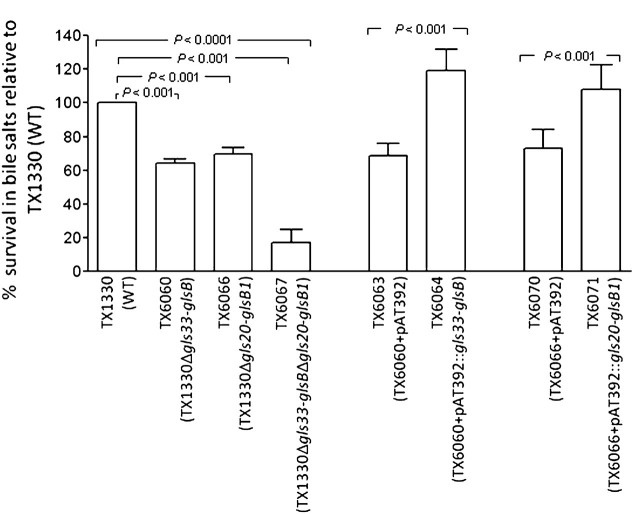
Contribution of the Two gls Loci to Survival in the Presence of Bile Salts
To study the role of the gls33-glsB and gls20-glsB1 gene pairs in resistance to bile salts, the different mutants and complementation derivatives were examined for viability upon exposure to 0.3 % bile salts, a concentration that is physiologically relevant [41]. All 3 gls deletion mutants were more sensitive to bile salts than TX1330 (P < .001; ANOVA). The mean percentages of survival ± SD of TX6060, TX6066, and TX6067 were 64% ± 2%, 70% ± 4%, and 17% ± 8%, respectively, compared with WT (Figure 2). The reduction by the double deletion mutant versus each of the single deletion mutant was also significant (P < .001). As shown in Figure 2, complementation of the 2 single deletions, TX6060 and TX6066, with gls33-glsB (TX6064) and gls20-glsB1 (TX6071), respectively, resulted in restoration of tolerance to bile salts to levels ≥WT (119 ± 12 by TX6064, P < .001 versus the vector alone control TX6063 and versus TX6060; 108 ± 15 by TX6071, P < .001 versus the vector alone control TX6070 and versus TX6066). These results are in agreement with the restored protein expression observed by Western blots (see above). Thus, our data indicate that both gls loci are required for full tolerance of bile salts at concentrations likely to be encountered by enterococci in the upper human intestinal tract, under the conditions tested.
Figure 2.
Comparison of three gls deletion mutants and complementation constructs versus wild-type (WT) and vector alone controls for survival after an exposure to bile salts. The mean and SD for combined results are shown from ≥5 independent experiments. Survival rates of WT versus the 3 gls deletion mutants were compared by analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons, and complementation constructs versus the vector only controls by the unpaired t test.
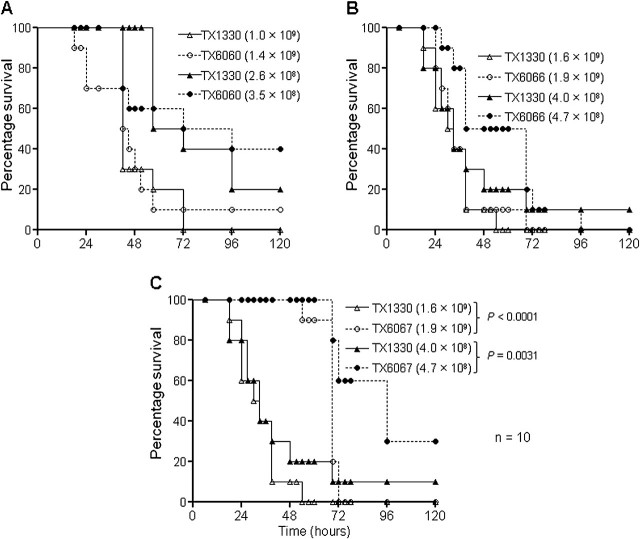
Importance of the gls33-glsB and gls20-glsB1 Loci for E. faecium Virulence
To test the effects of different gls deletions in a mouse peritonitis model, mice were infected with a range of CFUs. As shown in Figure 3A and B, neither the gls33-glsB (TX6060) nor the gls20-glsB1 deletion (TX6066) mutants showed attenuation versus the WT in either total death or time to death over a period of 5 days. Similarly, no difference was observed with complementation constructs TX6064 and TX6071 versus vector alone controls TX6063 and TX6070, respectively (data not shown). Of note, we also found that ∼95% of TX6064 and TX6071 colonies isolated from peritoneal fluid of mice had lost the complementing plasmid in vivo. Using a range of inocula (4.0 ×108 to 3.8 × 109 CFU), the double deletion mutant, TX6067, was significantly (P < .004) attenuated versus WT in this model (Figure 3C), indicating that, although both gls33-glsB and gls20-glsB1 are important for virulence in this model, the individual genes are able to compensate for each other's absence. Because of the observed instability of the complementing plasmid noted above, we did not attempt further to complement the double deletion mutant, because other vectors have also shown instability.
Figure 3.
Effect of the gls deletions on Enterococcus faecium virulence in a mouse peritonitis model. Kaplan-Meier survival curves after injecting 2 different inocula of (A) wild-type (WT) and TX6060 (TX1330Δgls33-glsB), (B) WT and TX6066 (TX1330Δgls20-glsB1), and (C) WT and TX6067 (TX1330Δgls33-glsBΔgls20-glsB1). A log-rank test was used for comparisons of survival rates. Survival rates of each single deletion mutant at 2 inocula shown above were nonsignificant versus WT, with P values .1 to .7.
In summary, we have identified 2 closely located conserved paralogs, gls33-glsB and gls20-glsB1, encoding Gls-like proteins in E. faecium. RT-PCR analysis demonstrated that both loci are expressed as larger operons, with the gls33-glsB operon encompassing 7 additional upstream genes and the gls20-glsB1 operon encompassing 2 additional upstream genes. Analysis of regions containing gls genes from available sequenced genomes identified that (1) the 2 gls clusters are close to each other in all strains, (2) the gls region occurs as 2 common variants with the 2 operons separated by either a 3.2 or a 12.1 kb region in different strains, and (3) the 2 variants show ∼7% sequence divergence between their gls33-glsB operons and ∼3.5% between their gls20-glsB1operons. Importantly, 1 of the variants was exclusively found in the hospital-associated CC17 genogroup, suggesting the possibility that evolution of this locus preceded CC17 emergence. Deletion of each gls-like locus resulted in decreased tolerance to bile salts and, in our assay, a double mutant lacking both loci showed additional decreased tolerance. Furthermore, in the peritonitis model, single gene pair deletions were not significantly attenuated, whereas deletion of both gls33-glsB and gls20-glsB resulted in significantly prolonged survival, compared with WT. Thus, gls33-glsB and gls20-glsB1 both play a role in bile salts tolerance and in experimental peritonitis, although one appears to be able to substitute, at least partially, the role of the other in the latter.
Supplementary Data
Supplementary data are available at http://jid.oxfordjournals.org/ online.
Funding
National Institutes of Health, Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases (grant R01 AI067861 to B.E.M.).
Supplementary Material
Acknowledgments
We thank Karen Jacques-Palaz for her technical assistance.
References
- 1.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st Century–a clinical super-challenge. N Engl J Med. 2009;360:439–43. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 2.Hidron AI, Edwards JR, Patel J, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Erratum in Infect Control Hosp Epidemiol. 2009;30:107. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 3.Top J, Willems R, van der Velden S, Asbroek M, Bonten M. Emergence of clonal complex 17 Enterococcus faecium in The Netherlands. J Clin Microbiol. 2008;46:214–9. doi: 10.1128/JCM.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werner G, Coque TM, Hammerum AM, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008:13. [PubMed] [Google Scholar]
- 5.Eliopoulos GM. Antimicrobial agents for treatment of serious infections caused by resistant Staphylococcus aureus and enterococci. Eur J Clin Microbiol Infect Dis. 2005;24:826–31. doi: 10.1007/s10096-005-0055-1. [DOI] [PubMed] [Google Scholar]
- 6.Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–21. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 7.Rice LB. Antimicrobial resistance in gram-positive bacteria. Am J Med. 2006;119:S11–9. doi: 10.1016/j.amjmed.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun. 2008;76:4120–8. doi: 10.1128/IAI.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun. 2008;76:4110–9. doi: 10.1128/IAI.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother. 2009;53:4240–6. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice LB, Carias L, Rudin S, et al. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187:508–12. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- 12.Rice LB, Lakticova V, Carias LL, Rudin S, Hutton R, Marshall SH. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J Infect Dis. 2009;199:342–9. doi: 10.1086/595986. [DOI] [PubMed] [Google Scholar]
- 13.Leendertse M, Heikens E, Wijnands LM, et al. Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J Infect Dis. 2009;200:1162–5. doi: 10.1086/605609. [DOI] [PubMed] [Google Scholar]
- 14.Willems RJ, Homan W, Top J, et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357:853–5. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- 15.Sillanpaa J, Nallapareddy SR, Prakash VP, et al. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology. 2008;154:3199–211. doi: 10.1099/mic.0.2008/017319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sillanpaa J, Prakash VP, Nallapareddy SR, Murray BE. Distribution of genes encoding MSCRAMMs and pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol. 2009;47:896–901. doi: 10.1128/JCM.02283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickx AP, van Wamel WJ, Posthuma G, Bonten MJ, Willems RJ. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189:8321–32. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sillanpaa J, Nallapareddy SR, Singh KV, et al. Characterization of the ebp pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence. 2010;1:236–46. doi: 10.4161/viru.1.4.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology. 2008;154:3212–23. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- 20.Hendrickx AP, van Luit-Asbroek M, Schapendonk CM, et al. SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium. Infect Immun. 2009;77:5097–106. doi: 10.1128/IAI.00275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abranches J, Martinez AR, Kajfasz JK, Chavez V, Garsin DA, Lemos JA. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–56. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Carbona S, Sauvageot N, Giard JC, et al. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol Microbiol. 2007;66:1148–63. doi: 10.1111/j.1365-2958.2007.05987.x. [DOI] [PubMed] [Google Scholar]
- 23.Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun. 2004;72:408–13. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janulczyk R, Ricci S, Bjorck L. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun. 2003;71:2656–64. doi: 10.1128/IAI.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotter PD, Emerson N, Gahan CG, Hill C. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol. 1999;181:6840–3. doi: 10.1128/jb.181.21.6840-6843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giard JC, Hartke A, Flahaut S, Boutibonnes P, Auffray Y. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res Microbiol. 1997;148:27–35. doi: 10.1016/S0923-2508(97)81897-9. [DOI] [PubMed] [Google Scholar]
- 27.Teng F, Nannini EC, Murray BE. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J Infect Dis. 2005;191:472–80. doi: 10.1086/427191. [DOI] [PubMed] [Google Scholar]
- 28.Giard JC, Rince A, Capiaux H, Auffray Y, Hartke A. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J Bacteriol. 2000;182:4512–20. doi: 10.1128/jb.182.16.4512-4520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nannini EC, Teng F, Singh KV, Murray BE. Decreased virulence of a gls24 mutant of Enterococcus faecalis OG1RF in an experimental endocarditis model. Infect Immun. 2005;73:7772–4. doi: 10.1128/IAI.73.11.7772-7774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arduino RC, Jacques-Palaz K, Murray BE, Rakita RM. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect Immun. 1994;62:5587–94. doi: 10.1128/iai.62.12.5587-5594.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coque TM, Patterson JE, Steckelberg JM, Murray BE. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J Infect Dis. 1995;171:1223–9. doi: 10.1093/infdis/171.5.1223. [DOI] [PubMed] [Google Scholar]
- 32.Bryan EM, Bae T, Kleerebezem M, Dunny GM. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44:183–90. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- 33.Nallapareddy SR, Singh KV, Murray BE. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006;72:334–45. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arthur M, Depardieu F, Snaith HA, Reynolds PE, Courvalin P. Contribution of VanY D, D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Current Protocols in Molecular Biology. Brooklyn, NY: Green Publishing Associates 1994; 2.4.1–2.4.2. [Google Scholar]
- 36.Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–63. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh KV, Qin X, Weinstock GM, Murray BE. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–20. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 38.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J Bacteriol. 2005;187:5709–18. doi: 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer KL, Carniol K, Manson JM, et al. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J Bacteriol. 2010;192:2469–70. doi: 10.1128/JB.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willems RJ, Top J, van Santen M, et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–8. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Deest BW, Fordtran JS, Morawski SG, Wilson JD. Bile salt and micellar fat concentration in proximal small bowel contents of ileectomy patients. J Clin Invest. 1968;47:1314–24. doi: 10.1172/JCI105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.