Abstract
Astrocytes are the major cellular component of the central nervous system (CNS), and they play multiple roles in brain development, normal brain function, and CNS responses to pathogens and injury. The functional versatility of astrocytes is linked to their ability to respond to a wide array of biological stimuli through finely orchestrated changes in cellular gene expression. Dysregulation of gene expression programs, generally by chronic exposure to pathogenic stimuli, may lead to dysfunction of astrocytes and contribute to neuropathogenesis. Here, we review studies that employ functional genomics to characterize the effects of HIV-1 and viral pathogenic proteins on cellular gene expression in astrocytes in vitro. We also present the first microarray analysis of primary mouse astrocytes exposed to HIV-1 in culture. In spite of different experimental conditions and microarray platforms used, comparison of the astrocyte array data sets reveals several common gene-regulatory changes that may underlie responses of these cells to HIV-1 and its proteins. We also compared the transcriptional profiles of astrocytes with those obtained in analyses of brain tissues of patients with HIV-1 dementia and macaques infected with simian immunodeficiency virus (SIV). Notably, many of the gene characteristics of responses to HIV-1 in cultured astrocytes were also altered in HIV-1 or SIV-infected brains. Functional genomics, in conjunction with other approaches, may help clarify the role of astrocytes in HIV-1 neuropathogenesis.
Keywords: astrocytes, microarrays, gene expression profiling, HIV-1, HAD, brain, neurobiology, neurodegeneration, innate immunity
Introduction
Neuroglia are non-neuronal brain cells consisting of two broad cell categories, microglia and macroglia (Skoff and Knapp 1995; Streit 1995). Microglia originate in bone marrow and represent resident macrophages in the brain; they play major roles in innate immune responses and neuroinflammation (Streit 1995). Macroglia, similar to neurons, originate from neuroectoderm, and they include three major cell types, oligodendrocytes, radial glia, and astrocytes (Skoff and Knapp 1995). Oligodendrocytes are highly specialized glial cells responsible for myelinating axons. Radial glia serve as precursor cells during neurogenesis and, as Bergmann glia, modulate synaptic plasticity in adult brain (Skoff and Knapp 1995). Astrocytes are the most abundant, heterogeneous, and functionally versatile type of neuroglia (Schubert 1984; Skoff and Knapp 1995; Davies et al. 2000; Bachoo et al. 2004). Their roles range from structural and metabolic support in the brain, maintenance of brain homeostasis, responses to brain injury and pathogens, regulation of synaptic activity and neural signaling, to participation in brain development (reviewed in Benveniste et al. 1998; Bezzi and Volterra 2001; Danbolt 2001; Farina et al. 2007; Seth and Koul 2008; Silver and Steindler 2009).
This review focuses on the applications of functional genomics to the study of astrocyte biology in the context of HIV-1 infection and HIV-1-associated neurocognitive disorder (HAND). HAND (Antinori et al. 2007) and its most severe manifestation HIV-1 dementia (HAD) persist at a high rate in spite of the increasingly common use of antiretroviral therapies (McArthur 2004; Anthony et al. 2005; Ellis et al. 2007). Although the core pathophysiological defects of HAD are neuronal damage (Masliah et al. 1992; Everall et al. 1993; Navia and Price 1998), neurons rarely show evidence of HIV-1 infection (Wiley et al. 1986; Takahashi et al. 1996; Torres-Muñoz et al. 2001a). HIV-1 replicates in the brain in microglia and infiltrating macrophages (Gartner et al. 1986; Genis et al. 1992), and virus genome and proteins can also be found in a small and variable fraction of astrocytes in vivo, particularly in advanced brain disease (Saito et al. 1994; Tornatore et al. 1994; Ranki et al. 1995; Takahashi et al. 1996; An et al. 1999; Trillo-Pazos et al. 2003). The consensus view is that HIV-1 can be neuropathogenic through neurotoxic viral proteins and cellular toxins and inflammatory mediators secreted by HIV-1-infected macrophages and microglial cells, particularly in HIV-1 encephalitis (reviewed in Lipton and Gendelman 1995; Kolson and Pomerantz 1996; González-Scarano and Martín-García 2005). However, there is growing recognition that astrocytes also contribute to HIV-1-mediated neuropathogenesis, in several ways. First, astrogliosis, the presence of activated and hypertrophied astrocytes typically seen in HAD (Sharer et al. 1986; Budka 1991), may damage tissue by scarring. Second, as indicated above, HIV-1 can infect a small fraction of astrocytes in vivo. Productive infection of astrocytes with HIV-1 has significant effects on cell physiology in vitro (Kim et al. 2004; Cosenza-Nashat et al. 2006) and it associates with measurable neuropathology in a mouse model (Dou et al. 2006), suggesting that infected astrocytes, although infrequent, can have localized pathogenic effects. Third, astrocyte dysregulation resulting from exposure of the cells to HIV-1 particles, viral proteins, cytokines, and other mediators secreted by HIV-1-infected cells could have a broad impact on brain function (reviewed in Conant and Major 1998; Brack-Werner 1999; Wang et al. 2004). Studies in vitro indicate that many of these products significantly modulate astrocyte physiology, which in turn can alter essential interactions of astrocytes with other cells in the brain, particularly neurons. For example, exposure of cultured astrocytes to HIV-1, recombinant gp120, or viral transactivator Tat can induce several known mediators of neuropathogenesis, including inflammatory cytokines TNF-α and IL-1β, chemokines MCP-1 and IP-10, IL-6, and neurotoxin nitric oxide (Yeung et al. 1995; Conant et al. 1998; Lavi et al. 1998; Glabinski and Ransohoff 1999; Hesselgesser and Horuk 1999; Cota et al. 2000; Kutsch et al. 2000; McManus et al. 2000; Liu et al. 2002; El-Hage et al. 2005; Ronaldson and Bendayan 2006; Li et al. 2007). Binding of HIV-1 or exogenous gp120 to astrocytes can reprogram cellular gene expression (Su et al. 2002, 2003; Galey et al. 2003; Wang et al. 2004), while the critical astrocyte function of glutamate uptake can be inhibited by exposure of the cells to HIV-1, gp120, Tat, or TNF-α (Benos et al. 1994; Fine et al. 1996; Wang et al. 2003; Zhou et al. 2004). Defective glutamate transport by astrocytes can result in neuronal death by excitotoxicity (Danbolt 2001). In another potential neurotoxic mechanism, recombinant gp120 was shown to induce Ca++-dependent glutamate secretion by astrocytes and neuronal cell death in glial-neuronal co-cultures in a pathway involving signaling through CXCR4 and production of TNF-α (Holden et al. 1999; Bezzi et al. 2001). Because of their abundance and their many roles in the brain, it is clear that damage or dysregulation of astrocytes may significantly affect the normal physiological functioning of the nervous system.
Challenge of functional analysis of astrocytes
Astrocytes obtained from fetal or adult brain are morphologically and electrophysiologically diverse (Davies et al. 2000; Nakagawa and Schwartz 2004), and they show varied patterns of regional gene expression (Bachoo et al. 2004). It has been proposed that astrocytes stratify in vivo into subtypes or subpopulations distinguished by their anatomic location and brain region-specific functions (Bachoo et al. 2004). Molecular profiling of such astrocyte subgroups has only begun (Bachoo et al. 2004), and it may lead to a definition of astrocyte lineages such as those known for T cells. The lineages may vary in their responses to specific physiological and pathogenic stimuli. Laboratory studies of primary astrocytes generally use enriched GFAP-positive cell populations isolated from human fetal brain tissue or similarly prepared rodent fetal or neonatal astrocytes. These cultures represent cell pools originating from several anatomical regions of the brain, and they are morphologically and functionally heterogeneous. It is therefore difficult to ascertain whether responses of astrocytes to various stimuli in culture accurately reflect cellular changes under normal and pathological conditions in vivo. Some responses such as cell activation or secretion of chemokines (Conant et al. 1998; Cota et al. 2000; McManus et al. 2000; Ronaldson and Bendayan 2006) can be considered beneficial as part of normal neuroprotective function of the cells. Others such as inhibition of glutamate uptake or secretion of TNF-α (Benos et al. 1994; Holden et al. 1999; Bezzi et al. 2001; Wang et al. 2003) clearly have the potential to injure or impair neurons and contribute to the overall inflammatory environment in the brain. Another difficulty in unequivocal interpretation of astrocytic responses in culture is that some stimuli such as HIV-1 and gp120 can induce both neuroprotective (e.g., secretion of MCP-1) and neurotoxic astrocyte functions (e.g., glutamate uptake inhibition). Complicating matters further, as shown for HIV-1-infected macrophages (Lipton and Gendelman 1995; Kolson and Pomerantz 1996; González-Scarano and Martín-García 2005), unregulated stimulation of cells can cause excessive production of chemokines and cytokines, which rather than exerting their normal physiological functions can become neurotoxic and contribute to brain inflammation. These considerations underscore the challenges associated with functional evaluation of astrocytes in culture and extrapolation of the results to HIV-1 neuropathogenesis. Which of the HIV-1 effects on astrocytes in culture defines HIV-1-astrocyte interactions in vivo? Do stimuli such as gp120 or Tat induce different responses in different subpopulations of astrocytes? How can we distinguish normal from potentially pathogenic responses in the heterogeneous astrocyte populations in culture? Is it possible to identify molecular switches that can alter astrocyte physiology between normal and dysfunctional or harmful states?
One approach to attempt to analyze complex physiological responses of cells to external stimuli is by functional genomics, which permits objective and global determination of cellular gene expression profiles that are characteristic of normal and pathological conditions. Below, we describe applications of this approach to studies on astrocytes exposed to HIV-1 and HIV-1 proteins in vitro. The molecular profiles of cultured astrocytes are compared to those obtained from brain tissue from patients with HIV-1 dementia and macaques infected with simian immunodeficiency virus (SIV) to better understand the role of astrocytes in HIV-1 neuropathogenesis.
Gene expression profiling of cells and tissues: general considerations
Highly regulated programs of cellular gene expression underlie most physiological functions of a cell, including cellular responses to extracellular cues. Functional genomics, namely, a genome-wide transcriptional analysis using microarray technology, is a highly efficient method for analyzing complex gene networks altered in one pathological situation, and it has become a standard tool for molecular studies in many diseases (for reviews, see Dufva et al. 2002; D’Agata and Cavallaro 2004; Glanzer et al. 2004; Cobb et al. 2005; Hoheisel 2006). One of the greatest advantages of the functional genomics method is that it permits objective analysis of expressional changes of thousands of genes in a single experiment. This provides a global view of complex, interconnecting, and often coordinate genomic responses (a gene expression profile) in well-defined cell or tissue populations.
There are multiple microarray approaches using different RNA detection platforms, experimental protocols, and statistical analysis methods (for example, see Churchill 2002; Scharpf et al. 2003; Hardiman 2004; Korn et al. 2004). This technological diversity may pose a difficulty in comparing microarray data from different studies, even when using the same source material (Tan et al. 2003; Jarvinen et al. 2004; Parmigiani et al. 2004). To assure compatibility of microarray information across different laboratories, the functional genomics community developed binding recommendations for methodological standardization of microarray experiments. “Minimum Information about a Microarray Experiment” (MIAME) lists specific experimental requirements that are essential for reproducing the microarray study by another laboratory (Brazma et al. 2001). A more recent standard called “Microarray Quality Control Project” has been developed by the Food and Drug Administration (Chen et al. 2007). Microarray experiment output consists of scans of labeled cDNA or cRNA samples hybridized to oligonucleotide or cDNA probes on a microchip. To obtain gene expression measures, scan data are subjected to probe-level statistical analysis, including quality control for hybridization and normalization across the array (Irizarry et al. 2003). This is followed by calculation of average fold changes in gene expression between systems of interest and controls and statistical significance analysis per gene. Depending on the cut-off significance values assigned to the experiment, a list from hundreds to thousands of “significantly changed” genes is generated.
One of the challenges of functional genomics is reduction of the large amounts of gene expression data generated by a microarray experiment to functional information, namely, to implicate specific genes and biological pathways in the biological process of interest. Although even large datasets can be examined gene by gene, a more practical approach is to employ a growing battery of bioinformatics methods that allow organizing data according to predetermined criteria (Kapetanovic et al. 2004). In complex biological systems, changes in gene expression occur coordinately rather than as independent events. Expression of many genes depends on the expression of others within biological and regulatory pathways. Most of microarray “data mining” programs take advantage of these functional relationships. One of the most commonly used is Gene Ontology (GO) analysis that allows classifying transcripts in well-defined biological pathways (Pavlidis et al. 2004; Werner 2008); popular GO programs include EASE (Hosack et al. 2003) and MAPPFinder (Doniger et al. 2003). Other approaches include clustering analysis that groups genes using different algorithms presenting similar expression patterns (Tibshirani et al. 1999). A similarity in gene expression can indicate a coordinate biological event (Kerr et al. 2008). A more recent approach to identify significant but smaller changes in gene expression is the Gene Set Enrichment analysis, which follows behavior of clusters of functionally inter-related genes (gene sets) rather than individual genes (Kim and Volsky 2005; Nam and Kim 2008). Generally, a combination of bioinformatics methods is used for identifying candidate genes of interest for more detailed investigation. The biological significance of changes in expression of these genes should be validated in independent studies using Northern blot hybridization or quantitative RT-PCR to confirm altered expression of selected transcripts. The ultimate validation of microarray data rests on demonstrating altered expression of functions of proteins involved in biological processes that are suspected to contribute to the specific biological outcomes in the system subjected to gene expression profiling. An example of an informative microarray experiment conducted under MIAME specifications is described later in this work for analysis of mouse astrocytes exposed to HIV-1.
Functional genomics in the analysis of neurodegenerative diseases, including HAD
Functional genomics as a method of study of disease states has been applied with great success to identification of complex molecular pathways in carcinogenesis and more precise classification, detection, and prognosis of some cancers (Golub et al. 1999; Khan et al. 2001; Martin et al. 2001; Ramaswamy et al. 2003; Parmigiani et al. 2004). Application of functional genomics to diseases of the human nervous system has been slower, in part because these diseases involve multiple cell types rather than clonal expansion of a single-cell type, and the relevant tissues are generally not accessible during the lifetime of the patient (Glanzer et al. 2004). Nonetheless, large-scale gene expression profiling of tissues obtained at autopsy has revealed potential pathogenic pathways in Alzheimer’s disease (Blalock et al. 2004; Lukiw 2004), chronic schizophrenia (Hakak et al. 2001), multiple sclerosis (Lock et al. 2002), and viral encephalitis (Gebicke-Haerter 2005).
Functional genomics analyses of brain tissues from individuals with HIV CNS disease have begun to appear (Gelman et al. 2004; Masliah et al. 2004; Shapshak et al. 2004). Masliah et al. (2004) and Gelman et al. (2004) found a number of significant gene expression changes in the cortex of patients who died with HIV encephalitis (HIVE). Follow-up studies established a correlation between use of methamphetamine and up-regulation of interferon genes in patients who died with HIVE (Everall et al. 2005) and identified somatostatin as a potential marker of MDD in HIVE patients (Everall et al. 2006). Experimental SIV infections conducted in macaques identified several functional clusters that were up-regulated in encephalitic brain, including several histocompatibility antigens involved in antigen presentation and interferon-related genes (Roberts et al. 2003, 2004). The human and macaque microarray studies for which sufficient information was available for comparative analysis with studies in culture are listed in Table 1.
Table 1.
Gene profiling studies in HIV-1-infected astrocytes and HIV-1- or SIV-infected brains discussed in this review
| Citation | Cells | Microarray | Virus stimulus |
Analysis | Genes regulated/ total |
Conclusions |
|---|---|---|---|---|---|---|
| Astrocytes | ||||||
| Galey et al. 2003 | Primary human astrocytes |
NIA immuno and neuroarray |
HIV- gp120 |
HIV or gp120 vs non-HIV |
295/ 2,306 |
Differential effect of HIV-1 and gp120 in astrocytes. Gp120 has more profound effect but chemokine and cytokine induction occurs predomi- nantly by HIV infection |
| Kim et al. 2004 | Primary human astrocytes |
Affymetrix U133 A/B |
VSV- HIV |
VSV-HIV vs non- VSV-HIV |
734/ >44,000 |
Up-regulation of IFN antiviral responses, intercellular contacts, cell adhesion, and signaling. Down-regulation of cell cycle, DNA replication, and cell proliferation |
| Kramer-Hämmerle et al. 2005 | Astrocytoma— Nef expressing cells |
BD Bioscience Clontech |
Nef | Native Nef vs non- myristoylated Nef |
400/ 11,835 |
Up-regulation of small GTPase signaling, regulation of apoptosis, lipid metabolism. JAK/STAT and MAPK signaling pathways |
| Borjabad and Volsky 2009 (present work) |
Primary mouse astrocytes |
Affymetrix U74v2 | HIV | HIV vs non-HIV |
211/ 36,000 |
Up-regulation of immune response, inflammation, signal transduction, and chemotaxis. Down-regulation of signal transduction, development, and protein modification processes |
| Brain tissue | ||||||
| Sui et al. 2003 | Macaque—basal ganglia |
Clontech chemokine and cytokine array |
SHIV | SIVE vs non-SIVE |
50/277 | Changes in cytokine and its receptors expression during SHIV-encephalitis. Up-regulated genes participating in promoting monocyte infiltration, macrophage activation, and enhancement of virus replication. Down- regulation of neutrophic factors |
| Roberts et al. 2003 | Macaque— frontal lobe |
Affymetrix U95Av2 | SIV | SIVE vs ni | 98/ 12,625 |
Up-regulation in SIVE of genes implicated in monocyte entry to the brain, inflammation, IFN response, antigen presentation, production of neurotoxic effects, transcription factors, and others |
| Roberts et al. 2004 | Macaque— frontal lobe |
Affymetrix U95Av2 | SIV | SIV vs ni | 97/ 12,625 |
Up-regulation in acute SIV infection of genes involved in IFN and IL-6 pathways. Many of these genes also up-regulated in long-term infection and SIVE |
| Stephens et al. 2006 | Macaque— cortical brain |
Clontech Cytokine array |
SHIV | SHIV vs ni | 8 | Up-regulated genes, including Cripto-1 and genes implicated in inflammatory, neuroprotective, cognitive, and stress responses |
| Masliah et al. 2004 | Human—frontal cortex |
Affymetrix U95Av2 | HIV | HIVE vs non-HIVE |
133/ 12,625 |
Up-regulated pathways included neuroimmune and antiviral response, transcription, and signaling pathways. Down-regulated pathways included synaptic plasticity and transmission, cell cycle, signaling molecules, transcription factors, and cytoskeletal components |
It is clear that gene expression profiling of unfractionated brain tissue informs about the relative “average” RNA levels present in the tissue extract, regardless of any differences in expression in the individual cell types. Still, one can make predictions based on a relative abundance of cell types in the brain and changes in expression of cell-specific transcripts. With astrocytes constituting 50–60% of brain cell volume (Schubert 1984), it can be postulated that many of the differentially expressed genes in molecular profiles obtained from brain tissues of patients with HIV-1 neurocognitive disease represent physiological changes in astrocytes in vivo. It follows that comparisons of gene expression profiles of HAD brains with those from astrocytes subjected to pathogenic stimuli like HIV-1 in vitro may shed light on the role of astrocytes in HAD.
Gene expression profiling of human astrocytes challenged with HIV-1 or HIV-1 proteins
First attempts dedicated to identifying transcriptional responses of human astrocytes to HIV-1 used rapid subtraction hybridization (RaSH; Su et al. 2002, 2003). Fifteen transcripts were identified as being induced by the virus or viral gp120 envelope protein and termed astrocyte elevated genes (AEG); 10 genes were suppressed after HIV-1 infection or gp120 exposure and termed astrocyte suppressed genes or ASG (Su et al. 2002, 2003). Both AEG and ASG displayed temporal patterns of altered expression after HIV-1 infection of gp120 exposure; for example, expression of AEG-3 was highest 6 h after HIV-1 or gp120 exposure and declined thereafter, while expression of AEG-1 increased over time and peaked 7 days after the HIV-1 or gp120 stimulus (Su et al. 2003). These results demonstrated that human primary astrocytes respond to HIV-1 with extensive and temporal changes in cellular gene expression. The exposure to exogenous gp120 closely paralleled the effects of HIV-1 (Su et al. 2002, 2003), confirming previous observations indicating that gp120 binding itself, independent of infection, can exert profound global effects on astrocyte physiology (Benos et al. 1994; Wang et al. 2003).
RaSH and other subtraction hybridization methods are not high-flux procedures, and they have been designed primarily for discovery and cloning of novel differentially expressed genes (Boukerche et al. 2007). One of the genes discovered by RaSH in HIV-1-treated human astrocytes, AEG-1, was subsequently cloned and characterized as a mild tumor promoter and a factor controlling the expression of the astrocytic glutamate transporter EAAT2 (Kang et al. 2005).
Several studies undertook comprehensive gene expression profiling of astrocytes exposed to HIV-1 or viral proteins using high-flux microarray platforms for parallel detection of multiple differentially expressed genes (Galey et al. 2003; Kim et al. 2004; Kramer-Hämmerle et al. 2005). These reports are listed in Table 1. The table also includes the microarray analysis of primary mouse astrocytes exposed to HIV-1 described in this work (next section and Table 2 and Fig. 1). Finally, for comparison with astrocyte studies in vitro, Table 1 lists gene expression profiling studies with brain tissues from patients with HAD and SIV or SHIV-infected macaques. For all studies listed, we included information about the source of material used for microarray analysis, the microarray method used, the stimulus used in vitro or brain pathology for brain arrays, the total number of significantly changed (dysregulated) genes, and the main conclusions of the work. Each study in vitro found that HIV-1 or its proteins had profound effects upon astrocyte gene expression; it follows that in the HIV-1-infected brain, astrocytes subjected to HIV-1 exposure and infection are likely to suffer transcriptional changes that may contribute to the process of HAD.
Table 2.
Partial list of cellular significantly changed genes in mouse primary astrocytes exposed to HIV-1
| Up-regulated genes |
Down-regulated genes |
||||||
|---|---|---|---|---|---|---|---|
| Genet title | Gene symbol |
FC | t test | Gene title | Gene symbol |
FC | t test |
| Chemokines | Signal transduction | ||||||
| Chemokine (C–C motif) ligand 22 |
Ccl22 | 15.18 | 0.0002 | Regulator of G-protein signaling 2 |
Rgs2 | −4.69 | 0.0004 |
| Chemokine (C–X–C motif) ligand 1 |
Cxcl1 | 4.49 | 0.0001 | G-protein-coupled receptor 34 | Gpr34 | −3.20 | 0.0009 |
| Chemokine (C–C motif) ligand 17 |
Ccl17 | 4.16 | 0.0042 | RAS, guanyl releasing protein 3 |
Rasgrp3 | −2.98 | 0.0063 |
| Chemokine (C–X–C motif) ligand 7 |
Cxcl7 | 3.70 | 0.0142 | Chimerin (chimaerin) 2 | Chn2 | −2.22 | 0.0000 |
| Chemokine (C–X–C motif) ligand 11 |
Cxcl11 | 3.10 | 0.0020 | Epstein–Barr-virus-induced gene 2 |
Ebi2 | −2.15 | 0.0006 |
| Chemokine (C–X–C motif) ligand 5 |
Cxcl5 | 2.71 | 0.0017 | Purinergic receptor P2Y, G-protein-coupled 13 |
P2ry13 | −2.06 | 0.0010 |
| Chemokine (C–X–C motif) ligand 10 |
Cxcl10 | 2.67 | 0.0009 | Amyloid beta (A4) precursor protein binding A1 |
Apba1 | −2.05 | 0.0076 |
| Chemokine (C–C motif) ligand 4 |
Ccl4 | 2.51 | 0.0006 | Protein tyrosine phosphatase, receptor type, O |
Ptpro | −2.02 | 0.0125 |
| Chemokine (C–X–C motif) ligand 2 |
Cxcl2 | 2.39 | 0.0023 | RAB31, member RAS oncogene family |
Rab31 | −2.01 | 0.0309 |
| Chemokine (C–X3–C motif) ligand 1 |
Cx3cl1 | 2.38 | 0.0068 | Cell adhesion | |||
| Chemokine (C–C motif) ligand 3 |
Ccl3 | 2.22 | 0.0006 | Osteomodulin | Omd | −3.46 | 0.0033 |
| Chemokine (C–X–C motif) ligand 12 |
Cxcl12 | 2.15 | 0.0006 | CD34 antigen | Cd34 | −3.44 | 0.00001 |
| Interleukines | Procollagen, type XIV, alpha 1 | Col14a1 | −3.22 | 0.0011 | |||
| Interleukin 1 alpha | Il1a | 5.95 | 0.0002 | Prostaglandin-endoperoxide synthase 1 |
Ptgs1 | −3.02 | 0.0001 |
| Interleukin 6 | Il6 | 3.52 | 0.0001 | Nephronectin | Npnt | −2.67 | 0.0136 |
| Interleukin 1 beta | Il1b | 3.36 | 0.0001 | Metastasis suppressor 1 | Mtss1 | −2.38 | 0.0086 |
| Interleukin 12b | Il12b | 3.24 | 0.0012 | Phosphoglucomutase 5 | Pgm5 | −2.18 | 0.0117 |
| Interleukin 10 | Il10 | 2.36 | 0.0186 | Neural cell adhesion molecule 1 |
Ncam1 | −2.04 | 0.0319 |
| Inflammation | Sidekick homolog 2 (chicken) | Sdk2 | −2.02 | 0.0010 | |||
| Selectin, platelet | Selp | 2.87 | 0.0006 | Apoptosis | |||
| Acyloxyacyl hydrolase | Aoah | 2.86 | 0.0010 | Insulin-like growth factor 1 | Igf1 | −2.88 | 0.00004 |
| Selectin, platelet | Selp | 2.56 | 0.0004 | B-cell leukemia/lymphoma 2 | Bcl2 | −2.38 | 0.0079 |
| Tumor necrosis factor receptor superfamily, member 1b |
Tnfrsf1b | 2.03 | 0.0001 | Serum/glucocorticoid- regulated kinase |
Sgk | −2.01 | 0.0001 |
| Complement activation | Metabolism | ||||||
| Complement factor B | Cfb | 2.60 | 0.0016 | Hydroxyprostaglandin dehydrogenase 15 (NAD) |
Hpgd | −7.52 | 0.0001 |
| Complement component 1, r subcomponent |
C1r | 2.02 | 0.0030 | Phosphotriesterase related | Pter | −3.43 | 0.0005 |
| Other immune response | C1q-like 3 | C1ql3 | −2.26 | 0.0013 | |||
| Fc receptor, IgG, low affinity IIb |
Fcgr2b | 4.26 | 0.0003 | Arachidonate 5-lipoxygenase activating protein |
Alox5ap | −2.11 | 0.0001 |
| Gene model 1960, (NCBI) | Gm1960 | 3.75 | 0.0004 | Phosphorylase kinase beta | Phkb | −2.04 | 0.0001 |
| C-type lectin domain family 4, member e |
Clec4e | 2.69 | 0.0017 | Phospholipase C-like 1 | Plcl1 | −2.01 | 0.0432 |
| Toll-like receptor 1 | Tlr1 | 2.65 | 0.0001 | Development | |||
| CD40 antigen | Cd40 | 2.62 | 0.0176 | Exostoses (multiple) 1 | Ext1 | −2.90 | 0.0072 |
| 2′–5′ oligoadenylate synthetase-like 1 |
Oasl1 | 2.61 | 0.0046 | Sloan–Kettering viral oncogene homolog |
Ski | −2.65 | 0.0016 |
| Histocompatibility 2, Q region locus 8 |
H2-Q8 | 2.49 | 0.0004 | Angiogenin, ribonuclease A family, member 1 |
Ang1 | −2.22 | 0.0120 |
| Colony-stimulating factor 2 (granulocyte–macrophage) |
Csf2 | 2.42 | 0.0333 | Motile sperm domain containing 3 |
Mospd3 | −2.13 | 0.0485 |
| Immunoresponsive gene 1 | Irg1 | 2.34 | 0.0014 | Ribonuclease, RNase A family 4 |
Rnase4 | −2.04 | 0.0004 |
| T-cell specific GTPase | Tgtp | 2.28 | 0.0008 | Protein biosystesis and modification | |||
| Repeats 1 | LOC667373 | 2.28 | 0.0002 | Transforming growth factor, beta receptor I |
Tgfbr1 | −2.95 | 0.0236 |
| Histocompatibility 2, K1, K region |
H2-K1 | 2.03 | 0.0014 | FMS-like tyrosine kinase 1 | Flt1 | −2.70 | 0.0128 |
| Macrophage activation 2 like | Mpa2l | 2.03 | 0.0009 | Zinc and ring finger 4 | Znrf4 | −2.58 | 0.0113 |
| Apoptosis | Ribosomal protein S13 | Rps13 | −2.48 | 0.0147 | |||
| Receptor (TNFRSF)- interacting serine–threonine kinase 2 |
Ripk2 | 3.27 | 0.0012 | E3 ubiquitin protein ligase, HECT domain containing, 1 |
Edd1 | −2.46 | 0.0115 |
| Stam-binding protein | Stambp | 2.15 | 0.0090 | Chymotrypsin like | Ctrl | −2.17 | 0.0108 |
| Caspase recruitment domain 4 | Card4 | 2.03 | 0.0009 | SUMO/sentrin specific peptidase 6 |
Senp6 | −2.10 | 0.0080 |
| Tumor necrosis factor, alpha-induced protein 3 |
Tnfaip3 | 2.01 | 0.0017 | Protein kinase C, alpha | Prkca | −2.06 | 0.0362 |
| Ion transport | Immune response | ||||||
| Cytochrome P450, family 7, subfamily b, polypeptide 1 |
Cyp7b1 | 2.42 | 0.00003 | Complement component factor h |
Cfh | −2.89 | 0.0332 |
| Potassium voltage-gated channel |
Kcnf1 | 2.29 | 0.0013 | Histocompatibility 2, class II, locus Mb1 |
H2-DMb1 | −2.26 | 0.0113 |
| Thioredoxin-like 1 | Txnl1 | 2.11 | 0.0114 | Defensin beta 1 | Defb1 | −2.03 | 0.0091 |
| Polycystic kidney disease 1 like 3 |
Pkd1l3 | 2.04 | 0.0346 | Ion transport | |||
| Signal transduction | Solute carrier anion transporter family, member 2b1 |
Slco2b1 | −2.67 | 0.0105 | |||
| Phosphodiesterase 2A, cGMP-stimulated |
Pde2a | 3.36 | 0.0030 | Potassium channel, subfamily J, member 16 |
Kcnj16 | −2.17 | 0.0082 |
| Thrombospondin type 1 motif, 4 |
Adamts4 | 3.33 | 0.0005 | Solute carrier family 4 (anion exchanger), member 4 |
Slc4a4 | −2.12 | 0.001 |
| Membrane-spanning 4 domains | Ms4a4d | 2.87 | 0.0006 | Myelin basic protein expression factor 2, repressor |
Myef2 | −2.02 | 0.0249 |
| Regulator of G-protein signaling 4 |
Rgs4 | 2.60 | 0.0053 | Transcription | |||
| Like sequence 4 | Emr4 | 2.52 | 0.0003 | Myeloid ecotropic viral integration site-related gene 1 |
Mrg1 | −2.36 | 0.0204 |
| Prostaglandin I receptor (IP) | Ptgir | 2.34 | 0.0055 | Cyclin L2 | Ccnl2 | −2.02 | 0.0294 |
| Spi-B transcription factor (Spi-1/PU.1 related) |
Spib | 2.27 | 0.0322 | Cell cycle | |||
| BMP-binding endothelial regulator |
Bmper | 2.19 | 0.0015 | SMC4 structural maintenance of chromosomes 4-like 1 |
Smc4l1 | −2.15 | 0.0003 |
| Adenosine A2b receptor | Adora2b | 2.15 | 0.0324 | Establishment of cohesion 1 homolog 2 (S. cerevisiae) |
Esco2 | −2.06 | 0.0301 |
| Rho family GTPase 1 | Rnd1 | 2.09 | 0.0000001 | ||||
| Kinase suppressor of ras 1 | Ksr1 | 2.08 | 0.0105 | ||||
| Somatostatin receptor 3 | Sstr3 | 2.07 | 0.0291 | ||||
| Transglutaminase 2, C polypeptide |
Tgm2 | 2.02 | 0.0083 | ||||
| Formyl peptide receptor 1 | Fpr1 | 2.01 | 0.0002 | ||||
| Regulation of transcription | Other functions | ||||||
| NK2 transcription factor related, locus 3 (Drosophila) |
Nkx2-3 | 2.39 | 0.0074 | Insulin-like growth factor binding protein 5 |
Igfbp5 | −3.11 | 0.0070 |
| Forkhead-like 18 (Drosophila) | Fkhl18 | 2.32 | 0.0100 | Cortactin-binding protein 2 | Cttnbp2 | −2.36 | 0.00002 |
| Doublesex and mab-3-related transcription factor |
Dmrta1 | 2.10 | 0.0103 | RAD51-associated protein 1 | Rad51ap1 | −2.24 | 0.0022 |
| Cells 2, p49/p100 | Nfkb2 | 2.05 | 0.0020 | HIV-1 Rev-binding protein like |
Hrbl | −2.10 | 0.0017 |
| Nuclear receptor interacting protein1 |
Nrip1 | 2.03 | 0.0057 | RNA-binding region (RNP1, RRM) containing 3 |
Rnpc3 | −2.07 | 0.0123 |
| Cells inhibitor, epsilon | Nfkbie | 2.01 | 0.0019 | Heat shock protein 90 kDa alpha, class A member 1 |
Hsp90aa1 | −2.06 | 0.0024 |
| Metabolism | Mannose receptor, C type 1 | Mrc1 | −2.02 | 0.0028 | |||
| Vanin 3 | Vnn3 | 3.59 | 0.0001 | WD repeat domain 1 | Wdr1 | −2.01 | 0.0100 |
| Chitinase 3-like 1 | Chi3l1 | 3.20 | 0.00003 | Not classified function | |||
| Prostaglandin E synthase | Ptges | 2.89 | 0.0016 | Tripartite motif protein 34 | Trim34 | −2.81 | 0.0195 |
| Carbonic anhydrase 13 | Car13 | 2.66 | 0.0038 | CTD small phosphatase like 2 | Ctdspl2 | −2.59 | 0.0031 |
| Prostaglandin-endoperoxide synthase 2 |
Ptgs2 | 2.47 | 0.0065 | Membrane protein, palmitoylated 6 |
Mpp6 | −2.52 | 0.0149 |
| Urocanase domain containing 1 |
Uroc1 | 2.29 | 0.0050 | DNA2 DNA replication helicase 2-like (yeast) |
Dna2l | −2.52 | 0.0257 |
| AMP deaminase 3 | Ampd3 | 2.11 | 0.0012 | Meningioma-expressed antigen 5 (hyaluronidase) |
Mgea5 | −2.46 | 0.0326 |
| Matrix metallopeptidase 13 | Mmp13 | 2.03 | 0.0214 | Cytotoxic T lymphocyte- associated protein 2 beta |
Ctla2b | −2.37 | 0.0011 |
| Development | Leukocyte-associated Ig-like receptor 1 |
Lair1 | −2.36 | 0.0025 | |||
| Tumor necrosis factor, alpha-induced protein 2 |
Tnfaip2 | 3.10 | 0.0001 | Glucosaminyl (N-acetyl) transferase 1, core 2 |
Gcnt1 | −2.35 | 0.0004 |
| (Semaphorin) 4A | Sema4a | 2.88 | 0.0002 | Cytoplasmic polyadenylation element binding protein 2 |
Cpeb2 | −2.21 | 0.0161 |
| VGF nerve growth factor inducible |
Vgf | 2.43 | 0.0006 | SERTA domain containing 4 | Sertad4 | −2.20 | 0.0011 |
| Adrenomedullin | Adm | 2.00 | 0.0381 | RAB3A interacting protein (rabin3)-like 1 |
Rab3il1 | −2.18 | 0.0026 |
| Cell adhesion | Navigator 3 | LOC676640 | −2.16 | 0.0092 | |||
| Thrombospondin 2 | Thbs2 | 2.27 | 0.0189 | Radial spokehead-like 3 | Rshl3 | −2.13 | 0.0030 |
| CD44 antigen | Cd44 | 2.22 | 0.0004 | Leucine zipper protein 2 | Luzp2 | −2.13 | 0.0262 |
| Transport | Retinitis pigmentosa 9 homolog (human) |
Rp9h | −2.10 | 0.0006 | |||
| Solute carrier family 7, member 11 |
Slc7a11 | 4.25 | 0.0003 | Nudix-type motif 10 | Nudt10 | −2.10 | 0.0053 |
| Solute carrier family 1, member 2 |
Slc1a2 | 2.15 | 0.0053 | Tetratricopeptide repeat domain 14 |
Ttc14 | −2.10 | 0.0360 |
| Syntaxin 11 | Stx11 | 2.05 | 0.0071 | Tudor domain containing 3 | Tdrd3 | −2.09 | 0.0232 |
| Other functions | Coiled-coil domain containing 39 |
Ccdc39 | −2.03 | 0.0090 | |||
| Pentraxin-related gene | Ptx3 | 3.12 | 0.00002 | DNA segment, Chr 3, ERATO Doi 300, expressed |
D3Ertd300e | −2.02 | 0.0261 |
| Tissue inhibitor of metalloproteinase 1 |
Timp1 | 3.10 | 0.00001 | Cytotoxic T lymphocyte- associated protein 2 alpha |
Ctla2a | −2.01 | 0.0033 |
| Coagulation factor C homolog (Limulus polyphemus) |
Coch | 2.71 | 0.0221 | Glypican 6 | Gpc6 | −2.00 | 0.0035 |
| Superoxide dismutase 2, mitochondrial |
Sod2 | 2.61 | 0.0002 | ||||
| Actin-binding LIM protein 1 | Ablim1 | 2.22 | 0.0405 | ||||
| Protein C receptor, endothelial | Procr | 2.09 | 0.00003 | ||||
| Schlafen 1 | Slfn1 | 2.04 | 0.0039 | ||||
| Histone deacetylase 2 | Hdac2 | 2.01 | 0.0060 | ||||
| Not classified function | |||||||
| leucine-rich repeat LGI family, member 2 |
Lgi2 | 5.31 | 0.0011 | ||||
| Nuclear protein 1 | Nupr1 | 3.40 | 0.0080 | ||||
| Ladinin | Lad1 | 3.30 | 0.0012 | ||||
| Z-DNA binding protein 1 | Zbp1 | 2.95 | 0.0064 | ||||
| Cysteine-rich secretory protein LCCL |
Crispld2 | 2.78 | 0.0073 | ||||
| Serine (or cysteine) peptidase inhibitor |
Serpinb5 | 2.78 | 0.0143 | ||||
| Bassoon | Bsn | 2.44 | 0.0140 | ||||
| Additional sex combs like 2 (Drosophila) |
Asxl2 | 2.42 | 0.0305 | ||||
| Ventricular zone expressed PH domain homolog 1 |
Veph1 | 2.38 | 0.0048 | ||||
| Purkinje cell protein 4 | Pcp4 | 2.37 | 0.0068 | ||||
| SLAM family member 8 | Slamf8 | 2.27 | 0.0018 | ||||
| Transmembrane protein 25 | Tmem25 | 2.22 | 0.0148 | ||||
| DENN/MADD domain containing 3 |
Dennd3 | 2.20 | 0.0293 | ||||
| Zinc finger CCCH type containing 12A |
Zc3h12a | 2.13 | 0.0017 | ||||
| Schlafen 4 | Slfn4 | 2.13 | 0.0013 | ||||
| Transmembrane protein 74 | Tmem74 | 2.13 | 0.0282 | ||||
| TNFAIP3 interacting protein 1 | Tnip1 | 2.09 | 0.0011 | ||||
| Ataxin 2 | Atxn2 | 2.06 | 0.0304 | ||||
| Glycophorin C | Gypc | 2.04 | 0.0305 | ||||
| ELOVL family member 7 | Elovl7 | 2.03 | 0.00005 | ||||
Up- and down-regulated transcripts in primary mouse astrocytes exposed to HIV-1. Results are selected by the criteria: FC>2, t test p value <0.05. Genes are classified by biological pathways using NetAffx Analysis Center (Affymetrix)
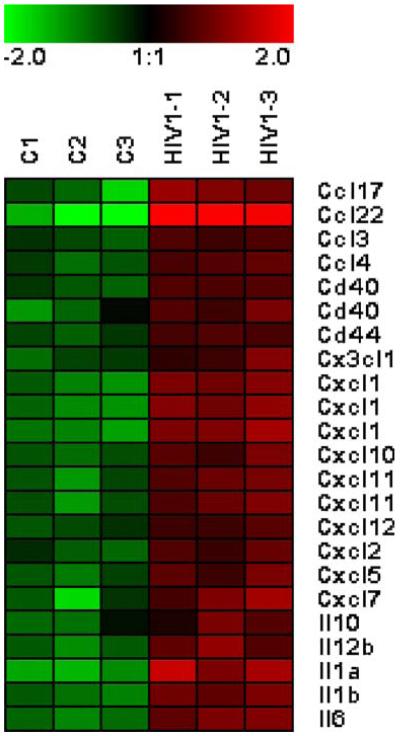
Fig. 1.
Heatmap representing the signal values of chemokines and cytokines up-regulated in mouse astrocytes exposed to HIV-1. Red color represents up-regulation, and green color represents down-regulation. Uninfected controls are represented by C1, C2, and C3 and HIV-1-exposed mouse astrocytes by HIV1-1, HIV1-2, and HIV1-3. The selected chemokines and cytokines in the figure were significant with a t test p value of <0.05 and a fold change >2. The figure was generated using the values obtained from the ArrayAssist 5.5.1. software (Stratagene). Briefly, the RMA data values were calculated and transformed by variance stabilization, logarithmic scale, and baseline transformation. The final values were represented in a heatmap using Genesis software (http://genome.tugraz.at/)
All the studies we discuss (Table 1) compare cellular RNA expression in a ground state to an infected or treated state. As discussed in previous sections, the methods chosen to obtain and interpret the findings are diverse. Several different microarray platforms, from spotted microarrays using two-channel detection (i.e., Cy3–Cy5 microarrays) to oligonucleotide microarray using one-channel detection (i.e., Affymetrix GeneChip), were employed. Data analysis and statistical methods also differed among analysis platforms and among biological materials, and in some cases, statistical method was not listed. Assignment of biological significance to the changes in gene expression observed also varied from study to study. To some extent, the acceptable cut-off of gene fold changes (FCs) and significance analysis depend on the uniformity of the material used and robustness of results. For example, there is less variability in gene expression patterns among replicate samples of cultured cells, especially cell clones, than among brain tissue samples from different individuals, even if they present with similar pathology. The number of replicas and statistical analysis approaches need to adjust to the nature of the biological source of data. Finally, most of the studies listed in Table 1 provided some functional classification of significantly regulated transcripts. Since there are a large number of gene ontology analysis programs and one gene can participate in several different pathways (Pavlidis et al. 2004; Werner 2008), the functional classification of the genes listed can differ from one study to another. Because not all the studies listed provided raw data in print or online, our comparison was limited to the final sets of significantly changed genes presented in each publication. Some studies could not be included in Table 1 because of insufficient information about the dataset discussed. Overall, with the caveats listed in the previous section, many commonalities regarding HIV-1 associated pathogenic changes in gene expression in astrocytes can be discerned.
The most general observation from the three studies with human astrocytes listed in Table 1 is that the cells respond profoundly to HIV-1 or HIV-1 proteins with broad and significant changes in cellular gene expression. The extent of the changes detected depended on the gene chip platform used and viral stimulus employed. Galey et al. analyzed gene expression pattern of human fetal astrocytes exposed to HIV-1 or to gp120 using lineage-specific immune- or neuro-microarrays containing 1,153 transcripts each (Galey et al. 2003; Table 1). These analytical platforms query a limited number of transcripts but address the expression of genes that are either functionally coordinated or act within a broad biological category. Of the target 2,306 genes in both arrays, 295 transcripts in total (12%) were found modulated by HIV-1 and/or gp120. Most of the transcripts were altered in expression either by HIV-1 or by gp120, with only 17 genes regulated similarly by both agents. Expression of 10 genes was altered by both stimuli, but in opposite directions. Chemokine and cytokine induction occurred predominantly in HIV-1-infected cells and not gp120 exposed cells (Galey et al. 2003). This report shows that HIV-1 induces a number of well-defined immune response genes in astrocytes as well as alters expression of a broader category of genes involved in nervous system functions. The differential signaling by HIV-1 and gp120 in astrocytes in this work indicates that gp120 is not the only component of intact virus that signals to astrocytes in long-term exposure. Indeed, since astrocytes in this study were tested 3 days after HIV-1 infection or gp120 exposure (Galey et al. 2003), the gene changes in HIV-1-infected cells likely reflected the activity of intracellular Tat or Nef proteins.
A more comprehensive gene expression profiling study in human fetal astrocytes challenged with HIV-1 employed the Affymetrix human GeneChip U133 A/B, which detected 44,000 transcripts (Kim et al. 2004). Distinct from a natural exposure of astrocytes to HIV-1, when infection occurs in a minority of cells (Li et al. 2007), this study investigated the pattern of cellular gene expression under the condition of widespread expression of HIV-1 throughout the astrocyte population (Canki et al. 2001). Human fetal astrocytes were infected with HIV-1 pseudotyped with the envelope glycoprotein G of vesicular stomatitis virus (VSV-G) permitting virus expression in 60–80% of human fetal astrocytes and subjected to microarray analysis at the peak of infection (Kim et al. 2004). Expression of over 730 genes was significantly altered. The most significantly up-regulated genes in these analysis included interferon-related genes as STAT1, IFNα21, IRF7, OAS1, IFIT1, MX1, and other genes involved in intercellular contacts, cell adhesion, intracellular signaling, transcriptional regulation, physiological processes, and others. Also, the complement C3 component was significantly up-regulated. Down-regulated genes cluster the most significant in cell cycle, DNA replication, and cell proliferation pathways (Kim et al. 2004).
In the third astrocyte study listed in Table 1, Kramer-Hämmerle analyzed the transcriptome of human astrocytes stably expressing HIV-1 Nef. Nef is an early viral protein that increases the pathogenicity of the virus (Greenway et al. 2003). Nef protein expression was also found in astrocytes in brains of HIV-1-infected children with encephalitis (Saito et al. 1994). The most represented up-regulated pathways by native Nef protein in contrast to myristoylation-defective Nef-variant were implicated in small GTPase signaling, apoptosis, and lipid metabolism. Many of the genes identified were connected to the JAK/STAT and the MAPK signaling pathways.
Gene expression profile in murine astrocytes exposed to HIV-1
Although rodent cells lack HIV-1 receptors and are therefore not susceptible to native HIV-1 infection, cultured rodent astrocytes, neurons, and animals themselves have proven to be useful as model systems for studying neuropathogenic effects of HIV-1 virions or individual viral proteins (Benos et al. 1994; Toggas et al. 1994; Sui et al. 2006; Alirezaei et al. 2007; El-Hage et al. 2008; Wang et al. 2008). Here, we present for the first time global gene expression profile of primary mouse astrocytes exposed to intact HIV-1 in culture. Day2 neonatal mouse astrocytes in a third passage in culture were exposed in triplicates to cell-free HIV-1/NL4-3 at m.o.i. of 1 and cultured for 24 h. As control, cells were treated in triplicates with a mock virus concentrate at the same dilution (vol/vol) as the wild-type virus. As an example of large-scale gene expression analysis, we shall provide some details of the method; full description of the procedure is in Kim et al. (2004). Total RNA was prepared from cell cultures using the RNAeasy total RNA extraction kit (Qiagen, CA, USA). RNA quality was assessed by electrophoresis and spectrophotometric analysis; 2μg of total RNA was used to generate a cDNA, and then 1μg of cDNA product was used in an in vitro transcription reaction that contained biotinylated UTP and CTP. Twenty micrograms of full-length cRNA was fragmented and was subjected to gene expression analysis on the Affymetrix U74 mouse array chip. Scanner results were analyzed using ArrayAssist software (Stratagene; http://www.stratagene.com). Probe level analysis was performed using RMA (Robust Micro Array) algorithm. After verification of data quality by Affymetrix internal controls and signal distribution analysis as described in Stratagene Array Assist Protocol, data were transformed using variance stabilization and logarithm transformation with a base of 2. Values were normalized by mean intensities of all chip samples. Means of normalized expression values were calculated for triplicates of individual systems, and these values were used to calculate FC in the transcript. Genes showing FC values above 2 or below −2 and unpaired t test p values of <0.05 were defined as significantly changed. By these cut-off criteria, we identified 124 up-regulated and 87 down-regulated transcripts. Table 2 lists the majority of the significantly changed genes in this experiment grouped into functional categories using gene ontology tools available at the NetAffx™ Analysis Center (http://www.affymetrix.com/analysis/index.affx). The up-regulated pathways included immune and inflammatory responses and signal transduction whereas cell adhesion, signal transduction and protein modification were down-regulated (Table 2). A heatmap representing expression of up-regulated inflammatory cytokines in this system is showed in Fig. 1.
These results indicate that exposure of mouse astrocytes to HIV-1 in vitro causes broad and significant changes in the gene expression profile of these cells and that many of the changes resemble those observed by microarray analyses in human astrocytes exposed to HIV-1 or gp120 (Galey et al. 2003; Kim et al. 2004). It should be noted that such transcriptional profiles, although comprehensive in terms of the number of cellular transcripts analyzed, do not provide full picture of the biological changes that may occur in stimulus-responding cells. Many cellular functions are subject to post-transcriptional regulation, for example by alternative splicing of mRNA (Maragakis et al. 2004), translational regulation of protein expression by untranslated regions of mRNA (Kim et al. 2003; Irier et al. 2009), changes in protein stability (Arancibio-Cárcamo and Kittler 2009; Park et al. 2009), or post-translational protein modifications through acetylation or phosphorylation (Maestre et al. 2008; MacDonald and Roskams 2009), and such responses would not be detected by microarray analysis. Furthermore, the large amount of gene expression data generated in these experiments necessitates an arbitrary definition, based on a statistical paradigm chosen, of what constitutes a significant change in gene expression. Customarily, gene changes are considered significant if fold changes are above 2.0 and unpaired t test p values are <0.05 (Cui and Churchill 2003; Jafari and Azuaje 2006; De Muth 2009). This may result in missing biologically meaningful processes if the genes involved changed less than 2-fold (or more than −2.0 fold). Conversely, with a p value of 0.05, 5% of the observed changes may be due to chance. The p values are a statistical measure of reproducibility of replicate systems in an experiment or reproducibility between replicate experiments (De Muth 2009), and thus, lower p values instill better confidence in an array experiment. In the mouse astrocyte microarray study shown in this work, the majority of test results were lower than p=0.01 with some reaching p=10−5 to 10−7 (Table 2), indicating that few gene changes observed here were due to chance. However, regardless of the statistical treatment and the stringency of microarray results, the lists of significantly altered genes placed within functional pathways such as shown in Table 2 and Fig. 1 are most valuable as a molecular map for posing hypotheses that integrate knowledge from many diverse experiments with a given system.
Comparison of gene profiling studies in astrocytes exposed to HIV-1 or HIV-1 proteins
Despite the different microarrays approaches and systems used, many specific changes in gene expression were found in two or more studies (Table 3). The highest correlation was observed between the Galey and Kim studies. Both used primary human fetal astrocytes. Galey et al. exposed astrocytes to either gp120 or HIV-1, whereas Kim et al. uses HIV-1 pseudotyped with VSV-protein G to increase the frequency of infection. Four gene changes in Kim’s work correlated with Galey’s HIV system and eight with Galey’s gp120 (Table 3). The fact that there is no greater correlation in gene expression patterns in both models can be explained in part by different microarray approaches used or by differences in virus strains. Two different ways to alter astrocytic function by HIV-1 were previously described (Wang et al. 2004): Although only a small fraction of astrocytes are infected in vivo (Nuovo et al. 1994; Takahashi et al. 1996; Trillo-Pazos et al. 2003) and in vitro (Tornatore et al. 1991; Nath et al. 1995), the infection is not cytolytic, allowing cells to survive during perturbation by virus. Independent of productive infection, astrocytes respond to binding HIV-1 or envelope protein gp120 with functional changes consistent with changes in gene expression (Wang et al. 2004). Those two mechanisms to alter astrocyte function by HIV-1 may explain in part the differences observed in the two studies of HIV-1 exposure to astrocytes in culture that were compared.
Table 3.
Comparison of significantly regulated genes in the different studies listed in Table 1
| Astrocytes |
Brain tissue |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Galey HIV |
Galey gp120 | Kim | Kramer- Hämmerle |
Borjabad and Volsky |
Sui and Buch |
Stephen | Roberts | Masliah | |
| Astrocytes | Galey HIV |
STAT1 ITGAM GFAP AKAP12 SLC9A3R1 PPP1CA BTF3 COL3A1 ALCAM MME PIP5KIC EFNB2 GRB2 GGCX RPS6 FPR1 CYC1 |
CUL4A STAT1 TRIP13 FPR1 |
EPAC RRAD IGFBP3 ITGB1 PSMD2 |
RGS4 TIMP1 PTPRO |
STAT1 SPARC |
0 | BAX VIM TIMP1 STAT1 |
TGFBR3 |
| Galey gp120 |
CHL1 ICAM1 STAT1 ERCC2 IGFBP3 UGP2 FPR1 APEH |
CTNNA1 ERCC2 SREBF1 |
IL10 FLT1 | STAT1 IGFBP3 |
0 | STAT1 HLADRB1 CX3CR1 IFIT4 |
IFIT4 STAT1 GPR37 | ||
| Kim | IFITM1 SLC2A3 MGST1 SEMA4D ERCC2 |
SLC7A11 CD44 CHI3L1 CXCL1 CXCL2 ESCO2 IGFBP5 NCAM1 RAD51AP1 |
IRF1 ZIC1 CD44 STAT1 ERBB3 IGFBP3 |
IFI6 | MX1 IFIT1 IFITM1 CHI3L1 ISG15 STAT1 BST2 LLGL1 DTNA |
MX1 IFIT1 IFITM1 ISG15 BST2 STAT1 DTNA FXYD2 |
|||
| Kramer- Hämmerle |
ABLIM1 C1R LAD1 TNFAIP2 |
0 | PTN | LGALS3BP GIP3 IFITM1 STAT3 MDK ITGB4 RAF1 IGFBP2 |
IFITM1 GFAP LGALS3BP |
||||
| Borjabad and Volsky |
IL1A CXCL10 CD44 SOD2 |
IL6 | CHI3L1 NUPR1 TGM2 TIMP1 |
0 | |||||
| Brain tissue |
Sui and Buch |
0 | ISGF3G SPP1 FN1 GIP2 FCER1G STAT1 |
STAT1 EPHB1 | |||||
| Sthepens | 0 | 0 | |||||||
| Roberts | ISG15 IFITM1 IFIT1 IFIT4 IFI44 MX1 IGL@ BST2 HLA-A HLA-C HLA-F B2M STAT1 DTNA LGALS3BP |
||||||||
| Masliah | |||||||||
Bold font indicates down-regulated genes
The approach of Kramer-Hämmerle et al. using an astrocyma cell line expressing Nef protein shows many genes in common with Galey and Kim approaches. Common genes between HIV-1 and Nef expressing cells likely represent the expression changes induced by Nef protein after astrocyte infection by HIV-1. In addition, CTNNA1, ERCC2, and SREBF1 were found altered in astrocytes exposed to gp120 alone or expressing Nef (Table 3).
In general, genes identified as up-regulated by more than one analytical approach discussed here include IFN-related genes (STAT1 and IFITM1), cell adhesion molecules (CHL1, ICAM1, ITGAM, ITGB1, and CTNNA1), and transcription factors (STAT1 and SREBF1; Table 3). The DNA repair gene ERCC2 that can play a role in degradation of retroviral cDNA (Yoder et al. 2006) and CUL4A gene were also induced. Expression of this gene is necessary for HIV-1 Vpr-induced cell cycle arrest (Tan et al. 2007), and its induction is consistent with the down-regulation of cell-cycle-related genes observed in other studies (Kim et al. 2004; Masliah et al. 2004). Down-regulated genes identified by Galey and Kim included the transcription factor TRIP13, the gene FPR1 that is implicated in some inflammatory processes and chemotaxis, IGFBP3 involved in cell proliferation and apoptotic processes, and the metabolic genes UGP2 and APEH.
We also compared cellular transcripts altered in HIV-1-exposed mouse astrocytes to those obtained in gene profiling studies in HIV-1-infected human astrocytes (Table 3). Surprisingly, transcriptomes from HIV-1-treated mouse and human astrocytes showed many commonalities despite the different species and microarray technologies used (Table 3). We found correlation for several genes in mouse astrocytes and Galey and Kim studies. Many of these genes are implicated in immune responses (IL-10, CD44, CXCL1, and CXCL2). Some of the significantly changed genes common to mouse and human astrocytes are of interest in the context of the neuropathology observed in HIV-associated dementia. For instance, the gene regulator of G-protein signaling 4 (RGS4) modulates signal transduction through several neurotransmitter receptor systems and has been reported as being implicated in many neurological disorders like schizophrenia, bipolar disorder, Huntington disease, or behavioral abnormalities (Ding and Hegde 2008; McOmish et al. 2008; Runne et al. 2008). Down-regulated genes FLT1 and PTPRO may be implicated in neurogenesis and axon guidance respectively and may participate in memory/learning processes (Cao et al. 2004; Chen and Bixby 2005; Zhang et al. 2005; Krum et al. 2008). This analysis confirms at a functional genomics level that mouse astrocytes, similar to mouse neurons and the animals themselves, are suitable models for investigation of many aspects of HIV-1 pathogenesis.
Comparison with infected brain tissues
One of the models used to study HIV-associated dementia has been the infection with SIV in rhesus macaques. This model reproduces many aspects of HIV brain disease in people, including CNS infection and dysfunction (Roberts et al. 2003) with cognitive and motor problems and the development of SIV encephalitis. This experimental infection has also been analyzed for changes in cellular gene expression associated with immunodeficiency virus infection in the brain. Sui et al. (2003) and Buch et al. (2004) used SHIV (virus bearing the envelope of HIV-1 on the background of SIV) as a model of neurological disease. Using microarrays, they analyzed the gene changes induced in the brains of SHIV-infected macaques with encephalitis when compared to those without encephalitis. Another comparison was made by Roberts et al. (Roberts et al. 2003, 2004) using SIV-infected macaque brains to study SIVE and acute SIV infection. They performed microarray analysis in the frontal lobe of the infected macaques with acute SIV infection, different time points after infection, and SIVE and compared their expression levels with the uninfected control cases. Many of the genes show an increased expression during the acute phase of infection, long-term infection, and SIVE (IFN- and IL-6-related genes). Both approaches show an up-regulation of genes implicated in the inflammatory response, including promoting monocyte infiltration, differentiation, and activation in the brain. Focusing upon cytokine expression, Stephens et al. used cytokine microarrays and identified eight genes induced in brains of macaques infected with SHIV compared to uninfected controls among them IFI6, PTN, and IL-6 that were also identified in other studies (Table 3).
A study comparing brains from HIV-1-infected patients with and without HIVE conducted by Masliah et al. 2004 show that interferon-related genes and neuro-inflammatory responses were up-regulated in patients with HIVE. These data are consistent with the results observed in SIVE and in cultured astrocytes. Down-regulated pathways included synaptic plasticity and transmission, cell cycle, and signaling molecules. Gelman et al. (2004) focused upon specific gene families and identified many dysregulated ionic conductance carriers in HAD or mild cognitive motor disorder compared with brains from uninfected people. Such neuronal channelopathies can alter membrane excitation and are consistent with some of the features of HAD. A study focusing on the expression of toll-like receptors (TLR)-related genes conducted by Salaria et al. (2007) found a correlation between expression levels of TLR-related genes and neuropathological parameters indicators of neurodegeneration.
Looking across the studies cited and comparing the individual genes dysregulated in human astrocytes and in macaque and human brain, one finds a significant number of genes that are similarly regulated (Table 3). Those genes are mostly implicated in immune response, including interferon-related genes (STAT1, IFIT1, IFIT4, IFITM1, IFI6, IRF1, MX1, and ISG15), other immune-related genes (HLADRB1, LGALS3BP, and BST2), and other genes implicated in brain function (DTNA, GFAP, MDK, and ZIC1). Two genes were down-regulated in macaque brain and human astrocytes (IGFBP3 and ERBB3), and only one gene was found down-regulated in human brain and the astrocyte model, FXYD2, a gene probably implicated in glutamate uptake by EAAT1 in astrocytes (Gegelashvili et al. 2007). Many other genes up-regulated in mouse astrocytes were also up-regulated in brain tissue of macaque SIV or SHIV model of encephalitis. They include immune response genes (IL1A, CXCL10, CD44, and IL-6), oxidative stress genes (SOD2), and others as CHI3L1. Alteration in CHI3L1 expression was associated with schizophrenia (Chung et al. 2003; Table 3).
Together, these results suggest that HIV-1-exposed and/or infected astrocytes may participate in the inflammatory responses observed in patients with HIVE. Because many of the transcriptional changes observed in HIV-1-exposed astrocytes in vitro can also be found in other cells involved in HIV-1 brain disease, particularly microglial cells and macrophages (Lipton and Gendelman 1995; Kolson and Pomerantz 1996), it is important to determine whether any of these molecular markers are also (or exclusively) altered in astrocytes in vivo. Thus far, besides routine detection of GFAP as a marker of gliosis (Budka 2005), there are only a few reports on expression of other cellular markers by astrocytes in brains from HIV-1-infected patients. Using in situ hybridization, Nuovo and Alfieri (1996) showed elevated chemokine MIP-1α and MIP-1β RNA in astrocytes in brain tissues of an AIDS patient (Nuovo and Alfieri 1996). More recently, Torres-Muñoz and colleagues identified elevated glial clusterin in astrocytes in tissue sections from brains of patients with HIV-1 dementia and suggested that clusterin may serve as a marker of neuroprotective function of astrocytes independent of GFAP expression (Torres-Muñoz et al. 2001b). Interestingly, MIP-1α RNA was significantly up-regulated (FC of 2.22; p=6×10−6) in HIV-1-exposed astrocytes in vitro (Table 2). Clusterin gene was also up-regulated in our astrocyte study (FC of 1.30 and p=0.027) but was not included in Table 2 because of our cut-off criteria. Other factors in the brain besides HIV-1, such as cytokines or viral proteins secreted by infected/activated microglia, could trigger induction of MIP-1α and clusterin in astrocytes in the cited studies. Nevertheless, analysis of glial transcriptome profiles in vitro together with rigorous correlative studies in patients’ tissues or animal models can be useful for elucidating the role of astrocytes in HIV-associated dementia.
Other studies in astrocytes using functional genomics with potential implications for HAD
Astrocytes actively respond to a wide array of environmental stimuli and can participate in local innate immune responses in the brain. It has been demonstrated that astrocytes express receptors of the TLR family and that following activation, astrocytes secrete chemokines and cytokines that can not only mediate immune responses but also contribute to neuroinflammation (for review, see Farina et al. 2007). Rivieccio et al. (2005, 2006) employed microarray analysis to determine global gene expression profiles of human fetal astrocytes stimulated with IL-1β or with polyriboinosinic polyribocytidylic acid (pIC), a TLR3 ligand analog. IL-1β and pIC induced extensive gene transcription changes in astrocytes, including up-regulation of IFN-stimulated genes, chemokines, and other genes participating in inflammation. pIC was more effective than IL-1β in activating antiviral response genes. Many of the genes induced in astrocytes by IL-1β and/or pIC, including the antiviral response genes MX1, IFIT1, OAS1, IRF7, or the chemokine CXCL1, were also induced in HIV-1-infected astrocytes (Kim et al. 2004; Table 1). It is noteworthy that the chemokines CXCL1 and CXCL10 and the cytokine IL-6 were also up-regulated in mouse astrocytes exposed to HIV-1 (Table 2). These results indicate that HIV-1 can be considered a mediator of inflammatory responses in astrocytes on par with natural inflammatory stimuli such as IL-1β. In another example, Meeuwsen et al. (2003) compared gene expression profiles of astrocytes exposed to proinflammatory cytokines TNF-α and IL-1β. Up-regulated genes included chemokines, growth factors, and receptors. Some of the genes induced in astrocytes by TNF-α or IL-1β described in this work were up-regulated by HIV-1 or SIV in vivo (Table 1). Other microarray analyses have shown that exposure of astrocytes in culture to different viruses (Cai et al. 2003; Radhakrishnan et al. 2003; Aravalli et al. 2006; Rubio and Sanz-Rodriguez 2007), conditions of hypoxia (Mense et al. 2006), and environmental toxins (Everall et al. 2005; Mense et al. 2006; Sengupta et al. 2007) can all significantly alter gene expression profiles of these cells. While the results of such studies cannot be directly extrapolated to pathogenic situations in HIV-1-infected brains, they clearly indicate that astrocytes are highly reactive cells and that their physiological functions can be profoundly altered by contact with a wide variety of natural and pathogenic stimuli, including HIV-1.
Conclusions
We have attempted to integrate findings from several experimental platforms using microarray analysis that reveal HIV-1 effects on gene expression in astrocytes. HIV-1 or HIV-1 proteins gp120, Nef, or Tat can induce innate immune effectors, interferon-stimulated genes, and other gene families in human astrocytes. Many of the individual genes with altered expression were found in common in more than one analysis, pointing to their potential significance in the HIV-1 pathogenic process. Similar gene expression changes were found in HIV-1-exposed mouse astrocytes in vitro, underscoring the usefulness of the mouse for study of HIV-1 pathogenesis. At another level of comparison, we also found commonalities in the changes induced by HIV-1 in culture with those observed in the brain of SIV-infected macaques or HIV-1-infected patients. Many up-regulated genes were found both in effectively pure astrocyte cultures and in mixed-cell tissue extracts from infected brains. This strongly suggests that changes in gene expression of astrocytes are a major component of the overall molecular profile of disease in the brains of HIV-1-infected people. It further suggests that exposure of astrocytes to HIV-1 in culture can be a useful tool for investigating the molecular and functional changes, leading to the development of HIV-associated dementia.
Acknowledgments
The authors would like to thank Ms. Wannasiri Lapcharoensap for her help in experiments with mouse astrocytes, Dr. Mary Jane Potash for reviewing the manuscript, and Ms. Ilene M. Totillo for manuscript preparation. This work was supported by grants NS31492, DA017618, MH083627, and NS061646 from the National Institutes of Health, Public Health Service. The mouse astrocyte gene expression data described in this work are available in the NCBI Gene Expression Omnibus (GEO) database using accession number GSE17383.
Contributor Information
Alejandra Borjabad, Molecular Virology Division, St. Luke’s-Roosevelt Hospital Center, 432 West 58th Street, Antenucci Building, Room 709, New York, NY 10019, USA; Department of Pathology & Cell Biology, Columbia University, New York, NY 10019, USA.
Andrew I. Brooks, Environmental and Occupational Health Science Institute, Department of Genetics, Rutgers University, Piscataway, NJ 08854, USA
David J. Volsky, Molecular Virology Division, St. Luke’s-Roosevelt Hospital Center, 432 West 58th Street, Antenucci Building, Room 709, New York, NY 10019, USA; Department of Pathology & Cell Biology, Columbia University, New York, NY 10019, USA
References
- Alirezaei M, Watry DD, Flynn CF, Kiosses WB, Masliah E, Williams BR, Kaul M, Lipton SA, Fox HS. Human immunodeficiency virus-1/surface glycoprotein 120 induces apoptosis through RNA-activated protein kinase signaling in neurons. J Neurosci. 2007;27:11047–11055. doi: 10.1523/JNEUROSCI.2733-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SF, Groves M, Giometto B, Beckett AAJ, Scaravilli F. Detection and localisation of HIV-1 DNA and RNA in fixed adult AIDS brain by polymerase chain reaction/in situ hybridisation technique. Acta Neuropathol. 1999;98:481–487. doi: 10.1007/s004010051113. [DOI] [PubMed] [Google Scholar]
- Anthony IC, Ramage SN, Carnie FW, Simmonds P, Bell JE. Influence of HAART on HIV-related CNS disease and neuroinflammation. J Neuropathol Exp Neurol. 2005;64:529–536. doi: 10.1093/jnen/64.6.529. [DOI] [PubMed] [Google Scholar]
- Antinori A, et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arancibio-Cárcamo IL, Kittler JT. Regulation of GABA(A) receptor membrane trafficking and synaptic localization. Pharmacol Ther. 2009;123:17–31. doi: 10.1016/j.pharmthera.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Aravalli RN, Hu S, Rowen TN, Gekker G, Lokensgard JR. Differential apoptotic signaling in primary glial cells infected with herpes simplex virus 1. J Neurovirology. 2006;12:501–510. doi: 10.1080/13550280601064921. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, DePinho RA. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci USA. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos DJ, Hahn BH, Bubien JK, Ghosh SK, Mashburn NA, Chaikin MA, Shaw GM, Benveniste EN. Envelope glycoprotein gp120 of human immunodeficiency virus type 1 alters ion transport in astrocytes: implications for AIDS dementia complex. Proc Natl Acad Sci USA. 1994;91:494–498. doi: 10.1073/pnas.91.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste EN, Shrikant P, Patton HK, Benos DJ. Neuroimmunologic mechanisms for disease in AIDS: The role of the astrocyte. In: Gendelman HE, Lipton AS, Epstein L, Swindells S, editors. The neurology of AIDS. Chapman & Hall; New York: 1998. pp. 130–146. [Google Scholar]
- Bezzi P, Volterra A. A neuron-glia signalling network in the active brain. Curr Opin Neurobiol. 2001;11:387–394. doi: 10.1016/s0959-4388(00)00223-3. [DOI] [PubMed] [Google Scholar]
- Bezzi P, Domercq M, Brambilla L, Galli R, Schols D, De Clercq E, Vescovi A, Bagetta G, Kollias G, Meldolesi J, Volterra A. CXCR4-activated astrocyte glutamate release via TNFa: amplification by microglia triggers neurotoxicity. Nat Neurosci. 2001;4:702–710. doi: 10.1038/89490. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukerche H, Su ZZ, Kang DC, Fisher PB. Cloning differentially expressed genes using rapid subtraction hybridization (RaSH) Methods Mol Biol. 2007;383:15–29. doi: 10.1007/978-1-59745-335-6_2. [DOI] [PubMed] [Google Scholar]
- Brack-Werner R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS. 1999;13:1–22. doi: 10.1097/00002030-199901140-00003. [DOI] [PubMed] [Google Scholar]
- Brazma A, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Buch S, Sui Y, Dhillon N, Potula R, Zien C, Pinson D, Li S, Dhillon S, Nicolay B, Sidelnik A, Li C, Villinger T, Bisarriya K, Narayan O. Investigations on four host response factors whose expression is enhanced in X4 SHIV encephalitis. J Neuroimmunol. 2004;157:71–80. doi: 10.1016/j.jneuroim.2004.08.034. [DOI] [PubMed] [Google Scholar]
- Budka H. Neuropathology of human immunodeficiency virus infection. Brain Pathol. 1991;1:163–175. doi: 10.1111/j.1750-3639.1991.tb00656.x. [DOI] [PubMed] [Google Scholar]
- Budka H. The neuropathology of HIV-associated brain disease. In: Gendelman HE, Grant I, Everall IP, Lipton SA, Swindells S, editors. The Neurology of AIDS. 2nd edn Oxford University Press; New York: 2005. pp. 375–391. [Google Scholar]
- Cai Y, Liu Y, Yu D, Zhang X. Down-regulation of transcription of the proapoptotic gene BNip3 in cultured astrocytes by murine coronavirus infection. Virology. 2003;316:104–115. doi: 10.1016/j.virol.2003.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canki M, Thai JNF, Chao W, Ghorpade A, Potash MJ, Volsky DJ. Highly productive infection with pseudotyped human immunodeficiency virus type 1 (HIV-1) indicates no intracellular restrictions to HIV-1 Replication in primary human astrocytes. J Virol. 2001;75:7925–7933. doi: 10.1128/JVI.75.17.7925-7933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Jiao X, Zuzga DS, Liu Y, Fong DM, Young D, During MJ. VEGF links hippocampal activity with neurogenesis, learning and memory. Nat Genet. 2004;36:827–835. doi: 10.1038/ng1395. [DOI] [PubMed] [Google Scholar]
- Chen B, Bixby JL. A novel substrate of receptor tyrosine phosphatase PTPRO is required for nerve growth factor-induced process outgrowth. J Neurosci. 2005;25:880–888. doi: 10.1523/JNEUROSCI.4365-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Hsueh HM, Delongchamp RR, Lin CJ, Tsai CA. Reproducibility of microarray data: a further analysis of microarray quality control (MAQC) data. BMC Bioinformatics. 2007;8:412. doi: 10.1186/1471-2105-8-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C, Tallerico T, Seeman P. Schizophrenia hippocampus has elevated expression of chondrex glycoprotein gene. Synapse. 2003;50:29–34. doi: 10.1002/syn.10228. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Cobb JP, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci USA. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant K, Major EO. Astrocytes as mediators of CNS injury in AIDS. In: Gendelman HE, Lipton AS, Epstein L, Swindells S, editors. The neurology of AIDS. New York; Chapman & Hall: 1998. pp. 147–155. [Google Scholar]
- Conant K, Garzino-Demo A, Nath A, McArthur JC, Halliday W, Power C, Gallo RC, Major EO. Induction of monocyte chemoattractant protein-1 in HIV-1 Tat-stimulated astrocytes and elevation in AIDS. Proc Natl Acad Sci USA. 1998;95:3117–3121. doi: 10.1073/pnas.95.6.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosenza-Nashat MA, Si Q, Zhao ML, Lee SC. Modulation of astrocyte proliferation by HIV-1: differential effects in productively infected, uninfected, and Nef-expressing cells. J Neuroimmunol. 2006;178:87–99. doi: 10.1016/j.jneuroim.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Cota M, Kleinschmidt A, Ceccherini-Silberstein F, Aloisi F, Mengozzi M, Mantovani A, Brack-Werner R, Poli G. Upregulated expression of interleukin-8, RANTES, and chemokine receptors in human astrocytic cells infected with HIV-1. J Neurovirology. 2000;6:75–83. doi: 10.3109/13550280009006384. [DOI] [PubMed] [Google Scholar]
- Cui X, Churchill GA. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agata V, Cavallaro S. Genomic portraits of the nervous system in health and disease. Neurochem Res. 2004;29:1201–1212. doi: 10.1023/b:nere.0000023607.84480.34. [DOI] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Davies DL, Niesman IR, Boop FA, Phelan KD. Heterogeneity of astroglia cultured from adult human temporal lobe. Int J Dev Neurosci. 2000;18:151–160. doi: 10.1016/s0736-5748(99)00083-0. [DOI] [PubMed] [Google Scholar]
- De Muth JE. Overview of biostatistics used in clinical research. Am J Health Syst Pharm. 2009;66:70–81. doi: 10.2146/ajhp070006. [DOI] [PubMed] [Google Scholar]
- Ding L, Hegde AN. Expression of RGS4 splice variants in dorsolateral prefrontal cortex of schizophrenic and bipolar disorder patients. Biol Psychiatry. 2008;65:541–545. doi: 10.1016/j.biopsych.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using gene ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Morehead J, Bradley J, Gorantla S, Ellison B, Kingsley J, Smith LM, Chao W, Bentsman G, Volsky DJ, Gendelman HE. Neuropathologic and neuroinflammatory activities of HIV-1-infected human astrocytes in murine brain. Glia. 2006;54:81–93. doi: 10.1002/glia.20358. [DOI] [PubMed] [Google Scholar]
- Dufva M, Flodin J, Nerstedt A, Ruetschi U, Rymo L. Epstein–Barr virus nuclear antigen 5 inhibits pre-mRNA cleavage and polyadenylation. Nucleic Acids Res. 2002;30:2131–2143. doi: 10.1093/nar/30.10.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Gurwell JA, Singh IN, Knapp PE, Nath A, Hauser KF. Synergistic increases in intracellular Ca2+, and the release of MCP-1, RANTES, and IL-6 by astrocytes treated with opiates and HIV-1 Tat. Glia. 2005;50:91–106. doi: 10.1002/glia.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Bruce-Keller AJ, Yakovleva T, Bazov I, Bakalkin G, Knapp PE, Hauser KF. Morphine exacerbates HIV-1 Tat-induced cytokine production in astrocytes through convergent effects on [Ca2+]i, NF-kB trafficking and transcription. PLoS ONE. 2008;3:e4093. doi: 10.1371/journal.pone.0004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R, Langford D, Masliah E. HIV and antiretroviral therapy in the brain: neuronal injury and repair. Nat Rev Neurosci. 2007;8:33–44. doi: 10.1038/nrn2040. [DOI] [PubMed] [Google Scholar]
- Everall I, Luthert P, Lantos P. A review of neuronal damage in human immunodeficiency virus infection: its assessment, possible mechanism and relationship to dementia. J Neuropathol Exp Neurol. 1993;52:561–566. doi: 10.1097/00005072-199311000-00002. [DOI] [PubMed] [Google Scholar]
- Everall I, Salaria S, Roberts E, Corbeil J, Sasik R, Fox H, Grant I, Masliah E. Methamphetamine stimulates interferon inducible genes in HIV infected brain. J Neuroimmunol. 2005;170:158–171. doi: 10.1016/j.jneuroim.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Everall IP, Salaria S, Atkinson JH, Young C, Corbeil J, Grant I, Masliah E. Diminished somatostatin gene expression in individuals with HIV and major depressive disorder. Neurology. 2006;67:1867–1869. doi: 10.1212/01.wnl.0000244436.04036.a2. [DOI] [PubMed] [Google Scholar]
- Farina C, Aloisi F, Meinl E. Astrocytes are active players in cerebral innate immunity. Trends Immunol. 2007;28:138–145. doi: 10.1016/j.it.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Fine SM, Angel RA, Perry SW, Epstein LG, Rothstein JD, Dewhurst S, Gelbard HA. Tumor necrosis factor a inhibits glutamate uptake by primary human astrocytes. J Biol Chem. 1996;271:15303–15306. doi: 10.1074/jbc.271.26.15303. [DOI] [PubMed] [Google Scholar]
- Galey D, Becker K, Haughey N, Kalehua A, Taub D, Woodward J, Mattson M, Nath A. Differential transcriptional regulation by human immunodeficiency virus type 1 and gp120 in human astrocytes. J Neurovirology. 2003;9:358–371. doi: 10.1080/13550280390201119. [DOI] [PubMed] [Google Scholar]
- Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- Gebicke-Haerter PJ. Microarrays and expression profiling in microglia research and in inflammatory brain disorders. J Neurosci Res. 2005;81:327–341. doi: 10.1002/jnr.20479. [DOI] [PubMed] [Google Scholar]
- Gegelashvili M, Rodriguez-Kern A, Sung L, Shimamoto K, Gegelashvili G. Glutamate transporter GLAST/EAAT1 directs cell surface expression of FXYD2/gamma subunit of Na, K-ATPase in human fetal astrocytes. Neurochem Int. 2007;50:916–920. doi: 10.1016/j.neuint.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Gelman BB, Soukup VM, Schuenke KW, Keherly MJ, Holzer C, 3rd, Richey FJ, Lahart CJ. Acquired neuronal channelopathies in HIV-associated dementia. J Neuroimmunol. 2004;157:111–119. doi: 10.1016/j.jneuroim.2004.08.044. [DOI] [PubMed] [Google Scholar]
- Genis P, Jett M, Bernton EW, Boyle T, Gelbard HA, Dzenko K, Keane RW, Resnick L, Mizrachi Y, Volsky DJ, Epstein LG, Gendelman HE. Cytokines and arachidonic metabolites produced during human immunodeficiency virus (HIV)-infected macrophage–astroglia interactions: implications for the neuropathogenesis of HIV disease. J Exp Med. 1992;176:1703–1718. doi: 10.1084/jem.176.6.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glabinski AR, Ransohoff RM. Chemokines and chemokine receptors in CNS pathology. J Neurovirology. 1999;5:3–12. doi: 10.3109/13550289909029740. [DOI] [PubMed] [Google Scholar]
- Glanzer JG, Haydon PG, Eberwine JH. Expression profile analysis of neurodegenerative disease: advances in specificity and resolution. Neurochem Res. 2004;29:1161–1168. doi: 10.1023/b:nere.0000023603.17615.8c. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- González-Scarano F, Martín-García J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Greenway AL, Holloway G, McPhee DA, Ellis P, Cornall A, Lidman M. HIV-1 Nef control of cell signalling molecules: multiple strategies to promote virus replication. J Biosci. 2003;28:323–335. doi: 10.1007/BF02970151. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman G. Microarray platforms—comparisons and contrasts. Pharmacogenomics. 2004;5:487–502. doi: 10.1517/14622416.5.5.487. [DOI] [PubMed] [Google Scholar]
- Hesselgesser J, Horuk R. Chemokine and chemokine receptor expression in the central nervous system. J Neurovirology. 1999;5:13–26. doi: 10.3109/13550289909029741. [DOI] [PubMed] [Google Scholar]
- Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- Holden CP, Haughey NJ, Nath A, Geiger JD. Role of Na+/H+ exchangers, excitatory amino acid receptors and voltage-operated Ca2+ channels in human immunodeficiency virus type 1 gp120-mediated increases in intracellular Ca2+ in human neurons and astrocytes. Neuroscience. 1999;91:1369–1378. doi: 10.1016/s0306-4522(98)00714-3. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irier HA, Shaw R, Lau A, Feng Y, Dingledine R. Translational regulation of GluR2 mRNAs in rat hippocampus by alternative 3′ untranslated regions. J Neurochem. 2009;109:584–594. doi: 10.1111/j.1471-4159.2009.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari P, Azuaje F. An assessment of recently published gene expression data analyses: reporting experimental design and statistical factors. BMC Med Inform Decis Mak. 2006;6:27. doi: 10.1186/1472-6947-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen AK, Hautaniemi S, Edgren H, Auvinen P, Saarela J, Kallioniemi OP, Monni O. Are data from different gene expression microarray platforms comparable? Genomics. 2004;83:1164–1168. doi: 10.1016/j.ygeno.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky DJ, Fisher PB. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene. 2005;353:8–15. doi: 10.1016/j.gene.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Kapetanovic IM, Rosenfeld S, Izmirlian G. Overview of commonly used bioinformatics methods and their applications. Ann N Y Acad Sci. 2004;1020:10–21. doi: 10.1196/annals.1310.003. [DOI] [PubMed] [Google Scholar]
- Kerr G, Ruskin HJ, Crane M, Doolan P. Techniques for clustering gene expression data. Comput Biol Med. 2008;38:283–293. doi: 10.1016/j.compbiomed.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Chao W, Choi SY, Volsky DJ. Cloning and characterization of the 3′-untranslated region of the human excitatory amino acid transporter 2 transcript. J Neurochem. 2003;86:1458–1467. doi: 10.1046/j.1471-4159.2003.01958.x. [DOI] [PubMed] [Google Scholar]
- Kim S-Y, Li J, Bentsman G, Brooks AI, Volsky DJ. Microarray analysis of changes in cellular gene expression induced by productive infection of primary human astrocytes: implications for HAD. J Neuroimmunol. 2004;157:17–26. doi: 10.1016/j.jneuroim.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Kolson DL, Pomerantz RJ. AIDS dementia and HIV-1-induced neurotoxicity: possible pathogenic associations and mechanisms. J Biomed Sci. 1996;3:389–414. doi: 10.1007/BF02258044. [DOI] [PubMed] [Google Scholar]
- Korn EL, Habermann JK, Upender MB, Ried T, McShane LM. Objective method of comparing DNA microarray image analysis systems. Biotechniques. 2004;36:960–967. doi: 10.2144/04366BI01. [DOI] [PubMed] [Google Scholar]
- Kramer-Hämmerle S, Hahn A, Brack-Werner R, Werner T. Elucidating effects of long-term expression of HIV-1 Nef on astrocytes by microarray, promoter, and literature analyses. Gene. 2005;358:31–38. doi: 10.1016/j.gene.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Krum JM, Mani N, Rosenstein JM. Roles of the endogenous VEGF receptors flt-1 and flk-1 in astroglial and vascular remodeling after brain injury. Exp Neurol. 2008;212:108–117. doi: 10.1016/j.expneurol.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch O, Oh J-W, Nath A, Benveniste EN. Induction of the chemokines interleukin-8 and IP-10 by human immunodeficiency virus type 1 Tat in astrocytes. J Virology. 2000;74:9214–9221. doi: 10.1128/jvi.74.19.9214-9221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi E, Kolson LD, Ulrich MA, Fu L, González-Scarano F. Chemokine receptors in the human brain and their relationship to HIV infection. J Neurovirology. 1998;4:301–311. doi: 10.3109/13550289809114531. [DOI] [PubMed] [Google Scholar]
- Li J, Bentsman G, Potash MJ, Volsky DJ. Human immunodeficiency virus type 1 efficiently binds to human fetal astrocytes and induces neuroinflammatory responses independent of infection. BMC Neurosci. 2007;8:31. doi: 10.1186/1471-2202-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton S, Gendelman HE. Dementia associated with the acquired immunodeficiency syndrome. New Engl J Med. 1995;233:934–940. doi: 10.1056/NEJM199504063321407. [DOI] [PubMed] [Google Scholar]
- Liu X, Jana M, Dasgupta S, Koka S, He J, Wood C, Pahan K. Human immunodeficiency virus type 1 (HIV-1) Tat induces nitric-oxide synthase in human astroglia. J Biol Chem. 2002;277:39312–39319. doi: 10.1074/jbc.M205107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, Klonowski P, Austin A, Lad N, Kaminski N, Galli SJ, Oksenberg JR, Raine CS, Heller R, Steinman L. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Gene expression profiling in fetal, aged, and Alzheimer hippocampus: a continuum of stress-related signaling. Neurochem Res. 2004;29:1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88:170–183. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Maestre C, Delgado-Esteban M, Gomez-Sanchez JC, Bolaños JP, Almeida A. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J. 2008;27:2736–2745. doi: 10.1038/emboj.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragakis NJ, Dykes-Hoberg M, Rothstein JD. Altered expression of the glutamate transporter EAAT2b in neurological disease. Ann Neurol. 2004;55:469–477. doi: 10.1002/ana.20003. [DOI] [PubMed] [Google Scholar]
- Martin KJ, Graner E, Li Y, Price LM, Kritzman BM, Fournier MV, Rhei E, Pardee AB. High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proc Natl Acad Sci USA. 2001;98:2646–2651. doi: 10.1073/pnas.041622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Ge N, Achim CL, Hansen LA, Wiley CA. Selective neuronal vulnerability in HIV encephalitis. J Neuropathol Exp Neurol. 1992;51:585–593. doi: 10.1097/00005072-199211000-00003. [DOI] [PubMed] [Google Scholar]
- Masliah E, Roberts ES, Langford D, Everall I, Crews L, Adame A, Rockenstein E, Fox HS. Patterns of gene dysregulation in the frontal cortex of patients with HIV encephalitis. J Neuroimmunol. 2004;157:163–175. doi: 10.1016/j.jneuroim.2004.08.026. [DOI] [PubMed] [Google Scholar]
- McArthur JC. HIV dementia: an evolving disease. J Neuroimmunol. 2004;157:3–10. doi: 10.1016/j.jneuroim.2004.08.042. [DOI] [PubMed] [Google Scholar]
- McManus CM, Weidenheim K, Woodman SE, Nunez J, Hesselgesser J, Nath A, Berman JW. Chemokine and chemokine-receptor expression in human glial elements: induction by the HIV protein, Tat, and chemokine autoregulation. Am J Pathol. 2000;156:1441–1453. doi: 10.1016/S0002-9440(10)65013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McOmish CE, Burrows EL, Howard M, Hannan AJ. PLC-beta1 knockout mice as a model of disrupted cortical development and plasticity: behavioral endophenotypes and dysregulation of RGS4 gene expression. Hippocampus. 2008;18:824–834. doi: 10.1002/hipo.20443. [DOI] [PubMed] [Google Scholar]
- Meeuwsen S, Persoon-Deen C, Bsibsi M, Ravid R, van Noort JM. Cytokine, chemokine and growth factor gene profiling of cultured human astrocytes after exposure to proinflammatory stimuli. Glia. 2003;43(3):243–253. doi: 10.1002/glia.10259. [DOI] [PubMed] [Google Scholar]
- Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ, Zhang L. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics. 2006;25:435–449. doi: 10.1152/physiolgenomics.00315.2005. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Schwartz JP. Gene expression patterns in in vivo normal adult astrocytes compared with cultured neonatal and normal adult astrocytes. Neurochem Int. 2004;45:203–242. doi: 10.1016/j.neuint.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- Nath A, Hartloper V, Furer M, Fowke KR. Infection of human fetal astrocytes with HIV-1: viral tropism and the role of cell to cell contact in viral transmission. J Neuropathol Exp Neurol. 1995;54:320–330. doi: 10.1097/00005072-199505000-00005. [DOI] [PubMed] [Google Scholar]
- Navia BA, Price RW. Clinical and biologic features of the AIDS dementia complex. In: Gendelman HE, Lipton SA, Epstein L, Swindells S, editors. The neurology of AIDS. Chapman & Hall; New York: 1998. pp. 229–240. [Google Scholar]
- Nuovo GJ, Alfieri ML. AIDS dementia is associated with massive, activated HIV-1 infection and concomitant expression of several cytokines. Mol Med. 1996;2:358–366. [PMC free article] [PubMed] [Google Scholar]
- Nuovo GJ, Gallery F, MacConnell P, Braun A. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acid and tumor necrosis-alpha RNA in the central nervous system. Am J Pathol. 1994;144:659–666. [PMC free article] [PubMed] [Google Scholar]
- Park JS, Seo J, Kim YO, Lee HS, Jo I. Coordinated regulation of angiopoietin-1 and vascular endothelial growth factor by arsenite in human brain microvascular pericytes: implications of arsenite-induced vascular dysfunction. Toxicol. 2009 doi: 10.1016/j.tox.2009.07.008. (in press) [DOI] [PubMed] [Google Scholar]
- Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol. 2003;77:10638–10644. doi: 10.1128/JVI.77.19.10638-10644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Ranki A, Nyberg M, Ovod V, Haltia M, Elovaara I, Raininko R, Haapasolo H, Krohn K. Abundant expression of HIV Nef and Rev proteins in brain astrocytes in vivo is associated with dementia. AIDS. 1995;9:1001–1008. doi: 10.1097/00002030-199509000-00004. [DOI] [PubMed] [Google Scholar]
- Rivieccio MA, John GR, Song X, Suh HS, Zhao Y, Lee SC, Brosnan CF. The cytokine IL-1beta activates IFN response factor 3 in human fetal astrocytes in culture. J Immunol. 2005;174:3719–3726. doi: 10.4049/jimmunol.174.6.3719. [DOI] [PubMed] [Google Scholar]
- Rivieccio MA, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee SC, Brosnan CF. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. 2006;177:4735–4741. doi: 10.4049/jimmunol.177.7.4735. [DOI] [PubMed] [Google Scholar]
- Roberts ES, Zandonatti MA, Watry DD, Madden LJ, Henriksen SJ, Taffe MA, Fox HS. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003;162:2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ES, Burudi EME, Flynn C, Madden LJ, Roinick KL, Watry DD, Zandonatti MA, Taffe MA, Fox HS. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: expression patterns throughout disease progression and impact on neuroAIDS. J Neuroimmunol. 2004;157:81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Ronaldson PT, Bendayan R. HIV-1 viral envelope glycoprotein gp120 triggers an inflammatory response in cultured rat astrocytes and regulates the functional expression of P-glycoprotein. Mol Pharmacol. 2006;70:1087–1098. doi: 10.1124/mol.106.025973. [DOI] [PubMed] [Google Scholar]
- Rubio N, Sanz-Rodriguez F. Induction of the CXCL1 (KC) chemokine in mouse astrocytes by infection with the murine encephalomyelitis virus of Theiler. Virology. 2007;358:98–108. doi: 10.1016/j.virol.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Runne H, Régulier E, Kuhn A, Zala D, Gokce O, Perrin V, Sick B, Aebischer P, Déglon N, Luthi-Carter R. Dysregulation of gene expression in primary neuron models of Huntington’s disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci. 2008;28:9723–9731. doi: 10.1523/JNEUROSCI.3044-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Sharer LR, Epstein LG, Michaels J, Mintz M, Louder M, Golding K, Cvetkovich TA, Blumberg BM. Overexpression of nef as a marker for restricted HIV-1 infection of astrocytes in postmortem pediatric central nervous tissues. Neurology. 1994;44:474–481. doi: 10.1212/wnl.44.3_part_1.474. [DOI] [PubMed] [Google Scholar]
- Salaria S, Badkoobehi H, Rockenstein E, Crews L, Chana G, Masliah E, Everall IP. Toll-like receptor pathway gene expression is associated with human immunodeficiency virus-associated neurodegeneration. J Neurovirol. 2007;13:496–503. doi: 10.1080/13550280701558616. [DOI] [PubMed] [Google Scholar]
- Scharpf R, Garrett ES, Hu J, Parmigiani G. Statistical modeling and visualization of molecular profiles in cancer. Biotechniques. 2003;(Suppl.):22–29. [PubMed] [Google Scholar]
- Schubert D. Developmental biology of cultured nerve, muscle, and glia. Wiley; New York: 1984. [Google Scholar]
- Sengupta A, Mense SM, Lan C, Zhou M, Mauro RE, Kellerman L, Bentsman G, Volsky DJ, Louis ED, Graziano JH, Zhang L. Gene expression profiling of human primary astrocytes exposed to manganese chloride indicates selective effects on several functions of the cells. Neurotoxicol. 2007;28:478–489. doi: 10.1016/j.neuro.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P, Koul N. Astrocyte, the star avatar: redefined. J Biosci. 2008;33:405–421. doi: 10.1007/s12038-008-0060-5. [DOI] [PubMed] [Google Scholar]
- Shapshak P, Duncan R, Torres-Muñoz JE, Duran EM, Minagar A, Petito CK. Analytic approaches to differential gene expression in AIDS versus control brains. Front Biosci. 2004;9:2935–2946. doi: 10.2741/1449. [DOI] [PubMed] [Google Scholar]
- Sharer LR, Epstein LG, Cho ES, Joshi VV, Meyenhofer MF, Rankin LF, Petito CK. Pathologic features of AIDS encephalopathy in children: evidence for LAV/HTLV-III infection of brain. Hum Pathol. 1986;17:271–284. doi: 10.1016/s0046-8177(83)80220-2. [DOI] [PubMed] [Google Scholar]
- Silver DJ, Steindler DA. Common astrocytic programs during brain development, injury and cancer. Trends Neurosci. 2009;32:303–311. doi: 10.1016/j.tins.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoff RP, Knapp PE. The origins and lineages of macroglial cells. In: Kettenmann H, Ransom BR, editors. Neuroglia. Oxford University Press; New York: 1995. pp. 135–148. [Google Scholar]
- Stephens EB, Jackson M, Cui L, Pacyniak E, Choudhuri R, Liverman CS, Salomon DS, Berman NE. Early dysregulation of cripto-1 and immunomodulatory genes in the cerebral cortex in a macaque model of neuroAIDS. Neurosci Lett. 2006;410(2):94–99. doi: 10.1016/j.neulet.2006.07.066. [DOI] [PubMed] [Google Scholar]
- Streit WJ. Microglial cells. In: Kettenmann H, Ransom BR, editors. Neuroglia. Oxford University Press; New York: 1995. pp. 85–96. [Google Scholar]
- Su Z-z, K D-c, Chen Y, Pekarskaya O, Chao W, Volsky DJ, Fisher PB. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- Su Z-Z, K D-c, Chen Y, Pekarskaya O, Chao W, Volsky DJ, Fisher PB. Identification of gene products suppressed by human immunodeficiency virus type 1 infection or gp120 exposure of primary human astrocytes by rapid subtraction hybridization. J Neurovirology. 2003;9:372–389. doi: 10.1080/13550280390201263. [DOI] [PubMed] [Google Scholar]
- Sui Y, Potula R, Pinson D, Adany I, Li Z, Day J, Buch E, Segebrecht J, Villinger F, Liu Z, Huang M, Narayan O, Buch S. Microarray analysis of cytokine and chemokine genes in the brains of macaques with SHIV-encephalitis. J Med Primatol. 2003;32:229–239. doi: 10.1034/j.1600-0684.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Sui Z, Fan S, Sniderhan L, Reisinger E, Litzburg A, Schifitto G, Gelbard HA, Dewhurst S, Maggirwar SB. Inhibition of mixed lineage kinase 3 prevents HIV-1 Tat-mediated neurotoxicity and monocyte activation. J Immunol. 2006;177:702–711. doi: 10.4049/jimmunol.177.1.702. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Wesselingh SL, Griffin DE, McArthur JC, Johnson RT, Glass JD. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39:705–711. doi: 10.1002/ana.410390606. [DOI] [PubMed] [Google Scholar]
- Tan PK, Downey TJ, Spitznagel EL, Jr, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Ehrlich E, Yu XF. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J Virol. 2007;81:10822–10830. doi: 10.1128/JVI.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Eisen M. Technical report. Stanford University; Stanford, CA: 1999. Clustering methods for the analysis of DNA microarray data. al. e. [Google Scholar]
- Toggas SM, Masliah E, Rockenstein EM, Rall GF, Abraham CR, Mucke L. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature. 1994;367:188–193. doi: 10.1038/367188a0. [DOI] [PubMed] [Google Scholar]
- Tornatore C, Nath A, Amemiya K, Major EO. Persistent human immunodeficiency virus type 1 infection in human fetal glial cells reactivated by T-cell factor(s) or by the cytokines tumor necrosis factor alpha and interleukin-1 beta. J Virol. 1991;65:6094–6100. doi: 10.1128/jvi.65.11.6094-6100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornatore C, Chandra R, Berger JR, Major EO. HIV-1 infection of subcortical astrocytes in the pediatric central nervous system. Neurology. 1994;44:481–487. doi: 10.1212/wnl.44.3_part_1.481. [DOI] [PubMed] [Google Scholar]
- Torres-Muñoz J, Stockton P, Tacoronte N, Roberts B, Maronpot RR, Petito CK. Detection of HIV-1 gene sequences in hippocampal neurons isolated from postmortem AIDS brains by laser capture microdissection. J Neuropathol Exp Neur. 2001a;60:885–892. doi: 10.1093/jnen/60.9.885. [DOI] [PubMed] [Google Scholar]
- Torres-Muñoz JE, Redondo M, Czeisler C, Roberts B, Tacoronte N, Petito CK. Upregulation of glial clusterin in brains of patients with AIDs. Brain Res. 2001b;888:297–301. doi: 10.1016/s0006-8993(00)03052-3. [DOI] [PubMed] [Google Scholar]
- Trillo-Pazos G, Diamanturos A, Rislove L, Menza T, Chao W, Belem P, Sadiq S, Morgello S, Sharer L, Volsky DJ. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 2003;13:144–154. doi: 10.1111/j.1750-3639.2003.tb00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pekarskaya O, Bencheikh M, Chao W, Gelbard H, Ghorpade A, Rothstein JD, Volsky DJ. Reduced expression of glutamate transporter EAAT2 and impaired glutamate transport in human primary astrocytes exposed to HIV-1 or gp120. Virology. 2003;312:60–73. doi: 10.1016/s0042-6822(03)00181-8. [DOI] [PubMed] [Google Scholar]
- Wang Z, Trillo-Pazos G, Kim S-Y, Canki M, Morgello S, Sharer LR, Gelbard HA, Su Z-Z, Kang D-C, Brooks AI, Fisher PB, Volsky DJ. Effects of human immunodeficiency virus type 1 on astrocyte gene expression and function: potential role in neuropathogenesis. J Neurovirology. 2004;10:25–32. doi: 10.1080/753312749. [DOI] [PubMed] [Google Scholar]
- Wang T, Gong N, Liu J, Kadiu I, Kraft-Terry SD, Mosley RL, Volsky DJ, Ciborowski P, Gendelman HE. Proteomic modeling for HIV-1 infected microglia-astrocyte crosstalk. PLoS ONE. 2008;3:e2507. doi: 10.1371/journal.pone.0002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T. Bioinformatics applications for pathway analysis of microarray data. Curr Opin Biotechnol. 2008;19:50–54. doi: 10.1016/j.copbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Wiley CA, Schrier RD, Nelson JA, Lampert PW, Oldstone MBA. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung MC, Pulliam L, Lau AS. The HIVenvelope protein gp120 is toxic to human brain-cell cultures through the induction of interleukin-6 and tumor necrosis factor-alpha. AIDS. 1995;9:137–143. [PubMed] [Google Scholar]
- Yoder K, Sarasin A, Kraemer K, McIlhatton M, Bushman F, Fishel R. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc Natl Acad Sci USA. 2006;103:4622–4627. doi: 10.1073/pnas.0509828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Amstein T, Shen J, Brush FR, Gershenfeld HK. Molecular correlates of emotional learning using genetically selected rat lines. Genes Brain Behav. 2005;4:99–109. doi: 10.1111/j.1601-183X.2004.00099.x. [DOI] [PubMed] [Google Scholar]
- Zhou BY, Liu Y, Kim B, Xiao Y, He JJ. Astrocyte activation and dysfunction and neuron death by HIV-1 Tat expression in astrocytes. Mol Cell Neurosci. 2004;27:296–305. doi: 10.1016/j.mcn.2004.07.003. [DOI] [PubMed] [Google Scholar]