Abstract
Understanding the structure and function of neural circuits is central is neuroscience research. To address the associated questions, new genetically encoded tools have been developed for mapping, monitoring, and manipulating neurons. Essential to implementation of these tools is their selective delivery to defined neuronal populations in the brain. This has been facilitated by recent improvements in cell type–specific transgene expression using recombinant adeno-associated viral vectors. Here, we highlight these developments and discuss areas for improvement that could further expand capabilities for neural circuit analysis.
Recombinant adeno-associated virus (rAAV) is widely used for gene delivery to the brain, because it permits nontoxic transduction of postmitotic cells and long-term gene expression in neurons. These basic properties, which make rAAV promising for human gene therapy, also facilitate research into the structure and function of neural circuits. Here, Drs. Betley and Sternson review recent developments in the application of rAAV vectors to (1) achieve cell type—selective transgene expression and (2) deliver molecular tools for mapping, monitoring, and manipulating neural circuits.
Introduction
The central nervous system orchestrates exceptionally diverse and complex biological processes, including learning, emotion, and decision-making. These and other brain functions are mediated by collections of neurons that are intricately wired into circuits through synaptic connections (Shepherd, 2004). Understanding the structure and function of these neural circuits is a key goal of neuroscience research. The combination of recently developed genetically encoded molecular tools for visualizing and perturbing circuits (Luo et al., 2008) and precisely targeted cell type–selective gene delivery approaches is dramatically expanding capabilities for neural circuit analysis.
Recombinant adeno-associated virus (rAAV) (Samulski et al., 1989) is widely used for gene delivery to the brain because it permits nontoxic transduction of postmitotic cells and long-term gene expression in neurons (Klein et al., 1998; McCown et al., 1996). These basic properties, which make rAAV promising for human gene therapy, also facilitate research into the structure and function of neural circuits (McCown 2005; Buning et al., 2008). Key improvements in rAAV vector design have advanced selective gene delivery to molecularly-defined neuron populations. Here, we review these recent developments with a focus on the application of rAAV vectors to (1) achieve cell type–specific transgene expression and (2) to deliver molecular tools for mapping, monitoring, and manipulating neural circuits.
Cell-Type Specificity and Neural Transduction
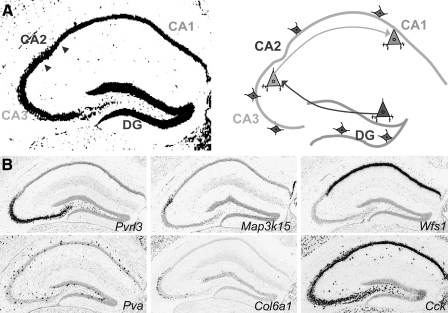
Within neural circuits, neurons with similar connectivity and functional roles are treated as circuit nodes (Fig. 1A). A major challenge has been identifying and manipulating these nodes. Neuronal subtypes are categorized based on morphological, physiological, and molecular characteristics (Gupta et al., 2000). However, investigation of individual cell types within circuits is often complicated by their intermingled distribution within the same anatomical domain (Klausberger and Somogyi 2008), making them neither readily distinguishable nor accessible for selective manipulation.
FIG. 1.
Marker genes specify circuit nodes in hippocampal circuitry. (A) Connections between anatomical regions of the hippocampus. In hippocampal circuits, granule cells located in the dentate gyrus (DG) send axonal projections to the CA3 region. The pyramidal cells in CA3 send axonal projections to CA1 neurons. Interspersed throughout the hippocampus are multiple classes of local inhibitory interneurons (diamond shaped in scheme). (B) Gene expression patterns label neurons selectively in the three main anatomical domains of the hippocampus. Pvrl3 marks CA3; Col6a1 marks a subset of CA3 neurons; Map3k15 marks CA2; Wfs1 marks CA1. Two inhibitory interneuron populations interspersed throughout the hippocampus can be distinguished by the expression of Pva or Cck. Within the hippocampal region, these genes mark neurons in distinct anatomical domains, however the expressed genes are not exclusive to these hippocampal regions, as they are also expressed in other anatomical domains of the brain. Images taken from: Allen Mouse Brain Atlas. Seattle (WA): Allen Institute for Brain Science. Copyright, 2009. (Lein et al., 2007) Available from: http://mouse.brain-map.org.
The divergent molecular characteristics of neurons provide a solution to this problem in which unique gene expression patterns define distinct neuronal cell types (Fig. 1B). These expressed marker genes can serve as entry points for delivery of genetically encoded molecular tools by using the corresponding gene promoter region to drive transgene expression (Tsien et al., 1996). Because of the diversity of possible genetic modifications, including expression of transgenes that permit neuron visualization and perturbation, gene expression patterns are being increasingly used to specify neuronal populations for neural circuit analysis (Arenkiel and Ehlers, 2009; Zagoraiou et al., 2009).
One limitation of most molecular marker genes is that often they are not restricted in expression to a unique neuronal population throughout the brain. Nevertheless, many markers are specific for neuronal subtypes within circumscribed brain regions (Lein et al., 2007) (Fig. 1B). Therefore, one challenge in applying this genetic approach is to restrict transgene expression regionally. rAAV vectors enable regional gene expression by local injection of the virus. Despite this, the application of rAAV for cell type–specific gene expression has been limited by technical challenges associated with insufficient transgene expression or lack of specificity. Recent advances employing cell type–specific promoters, selective viral infectivity, and intersections of viral techniques with genetically modified organisms have largely overcome these limitations, opening new possibilities for widespread use of rAAV vectors in neural circuit research.
Cell type specificity for AAV vector expression: promoters
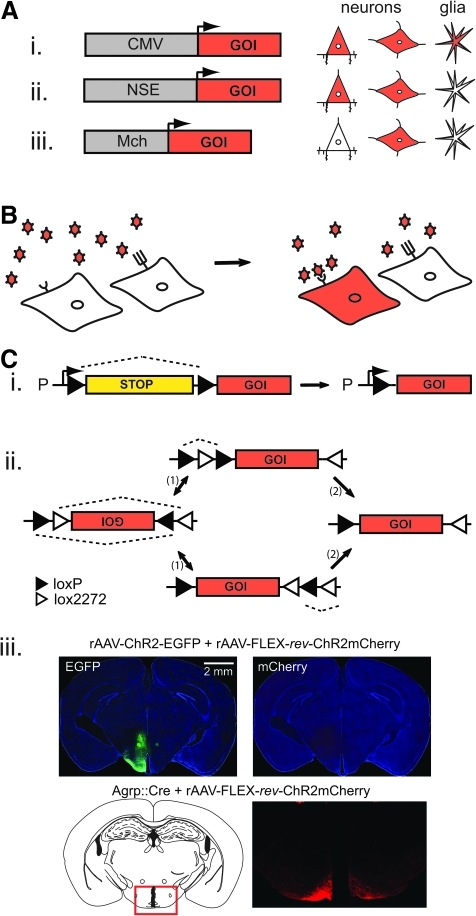
Promoters are gene regulatory elements that determine the strength and the cell-type specificity of expression. Initial studies using rAAV vectors in neurons focused on stable expression of a transgene. While expression driven by the cytomegalovirus (CMV) promoter decreased with time and led to transgene expression in neurons as well as some astrocytes, rAAV vectors with the neuron-specific enolase (NSE) promoter were shown to be suitable for long-term, neuron-selective expression of brain-derived neurotrophic factor (Fig. 2A) (Klein et al., 1998).
FIG. 2.
Strategies for cell type–selective transgene expression with recombinant adeno-associated virus (rAAV). (A) Promoter specificity. Schematic showing cell-type specificity of short promoter fragments in rAAV. The cytomegalovirus (CMV) promoter drives expression nonselectively in neurons and glia, while the neuron-specific enolase (NSE) promoter is selective for neurons, and the melanin-concentrating hormone (MCH) promoter targets expression to a subpopulation of neurons in the lateral hypothalamus. (B) Tropism specificity. AAV capsid proteins lead to differential infectivity between cell populations based on recognition between capsids and cell surface receptors. (C) Cre-dependent intersectional genetic switches. (i) Insertion of a 1–2 kb stop cassette after the promoter (P) and upstream of a gene of interest (GOI). The stop cassette is flanked by two loxP sites which are the recognition sequences that recombine in the presence of Cre. After recombination, the stop cassette is excised (one loxP site remains) and transcription of the GOI proceeds. (ii) Use of a flip-excision (FLEX) switch to activate gene expression specifically in the presence of Cre-recombinase. The GOI is inverted between two pairs of heterotypic, antiparallel lox-type recombination sites, which first undergo an inversion of the coding sequence, followed by excision of two sites, leading to one of each orthogonal recombination site oppositely oriented and incapable of further recombination. (iii) Demonstration of Cre-dependent transgene expression using a FLEX switch. The top panels show tissue from a wild-type mouse brain co-injected with both a rAAV2/1-ChR2-EGFP virus and a rAAV2/1-FLEX-rev-ChR2-mCherry virus. Expression of EGFP is broadly observed while mCherry is not expressed in the absence of Cre. Injection of rAAV2/1-FLEX-rev-ChR2-mCherry into an Agrp::Cre mouse (bottom panel) shows expression of the mCherry signal only in the arcuate nucleus, which is the sole location of AGRP neurons in the brain. Adapted from Journal of Neuroscience (Atasoy et al., 2008), copyright 2008.
Because promoter regions in mammals are often large (tens to hundreds of kilobases), promoter size is a key challenge for using rAAV due to its small packaging capacity (up to 4.7–5.0 kb) (Dong et al., 1996). A truncated human synapsin-1 promoter has proven effective for neuron-selective gene expression (Glover et al., 2002; Kugler et al., 2003) and only occupies 10% of the rAAV genome. Development of rAAV-compatible promoters that are selective for well-defined neuronal subsets was first demonstrated in neurons expressing the gene for melanin-concentrating hormone (MCH) (van den Pol et al., 2004). rAAV containing a short MCH promoter fragment selectively drove expression of green fluorescent protein (GFP) in this subpopulation of lateral hypothalamic neurons and allowed investigation of their physiological properties (Fig. 2A).
This approach sparked efforts to identify other promoter fragments that drive expression in defined neural subtypes. However, determination of short cell type–specific promoters has proven challenging. A systematic attempt to identify such regulatory elements for mouse cortical inhibitory interneurons highlights the difficulties (Nathanson et al., 2009a, 2009b). In an elegant study, conserved promoter elements from inhibitory neuron marker genes were derived from the pufferfish genome, which has a compact gene regulatory structure that is compatible with size limitations of rAAV. Unfortunately, these promoters were not sufficient to control transgene expression in previously defined subtypes of mammalian cortical inhibitory neurons (Nathanson et al., 2009a). This is likely due to the importance of multiple regulatory elements for recapitulating gene expression patterns in the mammalian brain (Gong et al., 2003), which often are not known and, further, would not be compatible with the limited packaging size of rAAV. More research into identification of the DNA regulatory sequences that underlie cell type–specific gene expression is clearly required to facilitate design of small synthetic promoter fragments. The primary limitation to the faithful reproduction of an endogenous gene expression pattern by a rAAV-expressed transgene continues to be viral genome packaging constraints.
Cell-type specificity for AAV vector expression: tropism
The tropism (cell type–selective infectivity) of AAV serotypes has also been investigated for targeting transgenes to specific cell populations. AAV serotypes differ in capsid protein sequence (Rutledge et al., 1998). These sequence differences affect receptor-mediated endocytosis of AAV particles, which are based on interactions of capsid proteins with receptors on the neuronal surface (Fig. 2B). Multiple recombinant serotypes have been identified that have improved the potency and broadened the tropism for rAAVs (Cearley and Wolfe 2006; Gao et al., 2005; Taymans et al., 2007). Some of these molecular interactions have been characterized for AAV serotypes (Buning et al., 2008), and a comparative analysis of neuronal infectivity for different serotypes showed different patterns of transduction that varied by brain region (Taymans et al., 2007); however, cell-type selectivity was not investigated.
Recently, promising strategies have been developed to generate and select for novel capsid sequences using ligand-mediated receptor targeting, mutagenesis, and DNA shuffling coupled with selection techniques (for a review see [Michelfelder and Trepel, 2009]). Ultimately, such techniques may be used to generate cell type–specific rAAV vectors for use in neural circuits. In one case, the ultimate goal of generating cell type–specific rAAV tropism was achieved by screening a series of mutant AAV6 capsids for astrocyte selectivity. One of these mutant capsids efficiently transduced Muller glial cells in the retina with high selectivity over other cell types (Klimczak et al., 2009). Additional work will show if this can be generalized to engineer capsids with selectivity for specific neuronal cell types.
Cell-type specificity for AAV vector expression: intersectional genetic switches
Currently, the most effective approaches for cell type–specific transgene expression are based on intersectional genetic manipulations (Tsien et al., 1996; Dymecki et al., 2010). These approaches have been adapted recently for use with rAAV vectors containing a strong, ubiquitously expressed promoter that is only active in the presence of a second, selectively expressed genetic component such as Cre-recombinase (Cre) (Atasoy et al., 2008; Kuhlman and Huang, 2008). Selective expression of Cre is achieved by genetic modification of the mouse genome using large upstream promoter regulatory regions that cannot be accommodated in the AAV genome (Gong et al., 2007). To render rAAV expression vectors Cre-dependent requires a compact genetic element for achieving conditional expression. One method uses a Cre-dependent transcriptional stop cassette placed between the promoter and the transgene (Fig. 2Ci). However, stop cassettes are large and typically occupy a significant portion of the AAV genome, which restricts the size of the promoter and the transgene (Kuhlman and Huang, 2008). An alternative strategy is a flip-excision (FLEX) switch, which involves stable Cre-dependent transgene inversion (Fig. 2Cii) (Atasoy et al., 2008). This approach is especially well-suited for use in rAAV due to the high efficiency and compactness (only ∼5% of AAV packaging size) of the FLEX switch. Furthermore, in the absence of Cre, the transgene is inverted with respect to the promoter, and thus expression is very tightly regulated (Fig. 2Ciii), unlike with the transcriptional stop cassette, which experiences noticeable levels of “read-through” of the transgene (Kuhlman and Huang, 2008). Most importantly, intersectional approaches transfer the challenging problem of using cell type–specific promoters to the realm of mouse genetics, where manipulation of large DNA fragments is not a limitation. With the appropriate Cre-expressing mouse line, Cre-dependent rAAV allows facile, cell type–specific targeting of any transgene (within the size constraints of the rAAV genome). This capability has expanded the application of rAAV vectors for neural circuit analysis.
Use of rAAV With Genetically Encoded Tools for Mapping, Monitoring, and Manipulating Neural Circuits
rAAV for circuit mapping
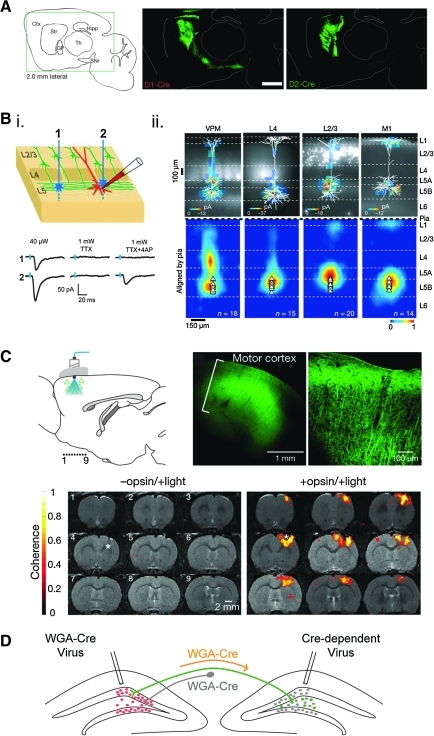
Circuit mapping aims to identify and characterize synaptic connections between neuronal cell types. Anatomical and functional circuit mapping strategies are being used in conjunction with cell type–specific rAAV vectors to dissect neural circuits with unprecedented precision. Targeted injection of rAAV vectors expressing fluorescent proteins drives high expression levels in localized brain regions, which allows visualization of long-range axon projections (Chamberlin et al., 1998). Until recently, limited progress was made by using rAAV for cell type–specific circuit mapping, because specific promoters typically did not deliver the high levels of fluorescent protein expression necessary to label axons. However, the development of Cre-dependent viral vectors with strong and ubiquitous promoters has greatly enhanced the ability to label axons that project throughout the brain. In one example, this approach was used to distinguish the projection patterns of two intermingled striatal projection neuron populations defined by expression of either dopamine receptor type 1 or type 2 (D1R or D2R) (Kravitz et al., 2010). Using a Cre-dependent viral vector with a strong promoter (EF-1α) driving a GFP transgene in combination with genetically modified mice expressing Cre under the D1R and D2R promoter revealed the strikingly distinct striato-nigral and striato-pallidal projection patterns for the intermingled D1R and D2R cell populations, respectively (Fig. 3A). While these cell type–specific anatomical techniques show the regional targets of neuron projections, they often do not give definitive confirmation of synaptic connectivity to a particular cell type. Because anatomical techniques also provide limited insight into the properties of a synaptic connection, functional circuit mapping techniques in combination with rAAV transgene targeting are proving especially useful for establishing connections and simultaneously characterizing their strength, sign, and other properties.
FIG. 3.
Applications of rAAV for neural circuit analysis. (A) Selective, Cre-dependent viral-mediated expression of YFP in two distinct populations of striatal neurons. D1R-expressing neurons project to the substantia nigra pars reticulata (SNr), D2R-expressing neurons project to the globus pallidus (GP). These experiments highlight discrete projection patterns for molecularly defined neurons occupying the same anatomical, indicating distinct neuronal functions. Reprinted by permission from Macmillan Publishers Ltd: Nature (Kravitz et al., 2010), copyright 2010. www.nature.com. (B) Mapping the subcellular distribution of inputs onto pyramidal neurons in the cortex using sCRACM. (i) The top panel shows two photostimulation configurations, where the postsynaptic neuron (red) is being recorded from and the blue stars indicate spots where the ChR2 expressing axons (green) are stimulated. The bottom panel provides representative traces of excitatory postsynaptic currents evoked by photostimulation corresponding to the locations indicated in the sketch. In the absence of the voltage-gated sodium channel blocker tetrodotoxin (TTX), both stimulation sites evoke synaptic responses, but these are both blocked with TTX, which prevents action potentials. Addition of the potassium channel blocker 4-aminopyridine (4-AP) renders ChR2 activation at a synaptic connection sufficient to drive release and evoke a synaptic response. Blue tickmarks indicate the laser pulse. Laser power is also shown for these conditions. (ii) Shown in the top panel are examples of sCRACM maps overlaid on somato-dendritic reconstructions. The fluorescent neurons are ChR2-positive neurons that are being mapped onto a pyramidal neuron. The bottom panel shows group averages of the inputs onto the post-synaptic neuron. Reprinted by permission from Macmillan Publishers Ltd: Nature (Petreanu et al., 2009), copyright 2009. www.nature.com. (C) Functional magnetic resonance imaging (fMRI) with blood oxygenation level detection (BOLD) signal detects activation of neurons resulting from cortical neuron stimulation. Diagram shows cortical neurons transduced by a virus expressing the ChR2-EYFP transgene as triangles and the path of the photostimulation in blue. Adjacent images are of ChR2-EYFP expressing neurons in the motor cortex. In brain slices that lack the ChR2 opsin, negligible levels of BOLD activation are observed; however, when AAV5-ChR2 is expressed in the cortex (+opsin), BOLD activation is observed. Reprinted by permission from Macmillan Publishers Ltd: Nature (Lee et al., 2010), copyright 2010. www.nature.com (D) Target region specificity. Injection of a Cre-dependent virus to infect neurons in one region of the central nervous system can be combined with an injection of a virus expressing wheat germ agglutinin (WGA)-Cre in a known anatomical target region of a subset of neurons in the original infection region. The fusion of WGA to the Cre recombinase promotes retrograde transport from the axon terminals of all neurons contributing to that projection field back to the cell body. Once at the nucleus, the Cre-recombinase unmasks the expression of the gene of interest (green fluorescent protein [GFP] in this case) of only the neurons in the original infection field that project to the appropriate target region. Reprinted by permission from Elsevier: Cell (Gradinaru et al., 2010), copyright 2010. www.sciencedirect.com/science/journal/00928674
A powerful new functional circuit mapping technique combines electrophysiological recordings of synaptic responses with neuronal expression of the algal light-activated ion channel, channelrhodopsin-2 (ChR2). ChR2 depolarizes neurons after a light flash and can selectively induce action potential firing in a molecularly defined cell type with millisecond precision (Boyden et al., 2005; Li et al., 2005). This genetically encoded tool is the basis for ChR2-assisted circuit mapping (CRACM) (Petreanu et al., 2007; Wang et al., 2007). ChR2 distributes throughout a neuron, including the axonal arbor, and allows selective activation of axons from a defined neuron population, in a background of axons originating from other cell types. Electrophysiology in brain slices containing these ChR2-expressing axons permits synaptic connections to be identified by light activation of the presynaptic axon while recording the evoked synaptic current in the postsynaptic neuron. rAAV-mediated expression of ChR2 has been critical for generalizing the use of this tool because ChR2 has a very low ionic conductance and, thus, large quantities of the channel must be expressed to render neurons and their axons reliably photoexcitable. In combination with Cre-dependent rAAV vectors, CRACM has been used to dissect intermingled circuits in the hypothalamus that are involved in regulating feeding behavior (Atasoy et al., 2008). The simplicity of this approach, requiring standard electrophysiological equipment and a relatively inexpensive fiber-coupled laser, makes it a rapid technique for definitively establishing and characterizing synaptic connections with cell type–specificity.
Cell type–specific neuron activation has enabled two further refinements, opposite in scale, for neural circuit mapping: (1) the subcellular distribution of synaptic inputs and (2) the influence of a localized neuron population on activity throughout the whole brain. Common to both approaches is the use of rAAV vectors to achieve high levels of ChR2 expression.
CRACM has been extended to identify the subcellular domains of synaptic contacts (sCRACM). In CRACM, an action potential propagates to all of the synapses made by a given axon, which makes it difficult to identify the origin of a synaptic input (Fig. 3Bi). However, under conditions in which action potentials are blocked by the voltage-gated sodium channel blocker tetrodotoxin and potassium channels are inhibited by 4-aminopyridine, ChR2 activation with a focused beam of laser light directly drives synaptic vesicle release but only in the vicinity of the laser spot (Fig. 3Bi). Thus, the position at which photostimulation evokes a synaptic response correlates with the region of synaptic contact. The location of the photostimulus and amplitude of the response are used to generate a map that reflects the subcellular distribution of synaptic connection strengths from inputs that originate from a defined ChR2-expressing presynaptic cell population, which can be overlaid with the morphological reconstruction of the postsynaptic neuron. This technique has been applied to map the spatial segregation of intracortical and cortico-thalamic inputs to cortical neuron dendritic arbors (Fig. 3Bii) (Petreanu et al., 2009). This information contributes to understanding neural computations in the postsynaptic neuron.
Functional connectivity has also been examined on a whole-brain scale by activating a subset of neurons while monitoring the resultant activity throughout the brain. In this approach, ChR2 is used to selectively drive action potentials in a cell population while using functional magnetic resonance imaging (fMRI) with blood oxygenation level detection (BOLD), which is thought to correlate with electrical activity. Using a CaMKIIa promoter in AAV2/5, ChR2 was expressed selectively in excitatory neurons in rat motor cortex. Optical activation of these neurons in anesthetized animals induced a BOLD signal in discrete regions throughout the brain (Lee et al., 2010) (Fig. 3C) (for a related approach in awake animals, see Desai et al., 2010). Specific ChR2 expression in motor cortex coupled with optical stimulation allowed unambiguous determination of the activated neuron population that served as the origin of the stimulus, in contrast to traditional electrical stimulation techniques, in which neurons as well as axon fibers of passage are both excited. Nevertheless, this technique involves exogenously stimulating populations of neurons with trains of action potentials and aims to measure the consequences of activating a neuronal network instead of identifying monosynaptic connections.
AAV for monitoring neuron activity
The relationship of neuron electrical activity to sensory inputs or ongoing behavior is widely investigated for exploring the function of neurons, and more recently, neuronal networks. Monitoring neuron activity in a network is a challenge that is increasingly being approached by large-scale, multi-neuronal imaging of calcium levels, which has been shown to reflect electrical activity (Tian et al., 2009). rAAV vectors can deliver genetically encoded calcium indicators for imaging calcium dynamics in vivo with sufficient expression levels for efficient detection. rAAV expressing the camgaroo-2 indicator under the control of a tetracycline transactivator (tTA) responsive promoter element was injected into the olfactory bulb of CaMKIIa-tTA transgenic mice (Hasan et al., 2004). This permitted expression in olfactory sensory axons and granule cells, and calcium dynamics were monitored in response to different odorants. More recently, another calcium indicator, GCaMP3, was targeted to the mouse motor cortex using rAAV2/5, which allowed dynamic activity in multiple neurons to be simultaneously monitored in the running mouse (Tian et al., 2009). These tools could be further extended to molecularly defined subpopulations with the cell type–specific expression strategies described above.
Manipulation of neural circuitry
Another recent advance has been the development of genetically encoded tools for reversibly manipulating neuron activity, which permits the causal relationship between neural activity and behavior to be explored. These tools are rapidly (millisecond to minute timescales) switchable in response to ligand binding or a light flash. Thus, they can be expressed for long periods without perturbing cellular function until they are engaged under the conditions chosen for a particular experiment. Because rAAV provides strong, cell type–specific transgene expression, it is increasingly being used for studies that involve activating or silencing molecularly defined neural populations.
Genetically encoded neuronal silencers are used to test the necessity of a neuron population for a behavioral function. For example, somatostatin (SST)-expressing cells in the pre-Botzinger complex, a hindbrain structure that regulates breathing, were selectively silenced to test their necessity in respiration. This study used the G-protein–coupled allatostatin receptor (AlstR) from Drosophila that can silence mammalian neurons when complexed to its ligand, allatostatin, by activating a potassium conductance (Tan et al., 2006). Separately, the ligand and its receptor are thought not to perturb mammalian neurons. Using rAAV2/2, AlstR was specifically expressed in the subpopulation of SST cells in the medulla under the control of a somatostatin promoter fragment (Tan et al., 2008). SST cells could be reversibly silenced by application of allatostatin, which eliminated the respiratory rhythm until allatostatin washout. These experiments demonstrated the necessity of the SST neuron subpopulation in the pre-Botzinger complex for respiration. Recently, other reversible neuronal silencers using either pharmacological (Armbruster et al., 2007; Karpova et al., 2005; Lerchner et al., 2007) or optical (Zhang et al., 2007) triggers have been developed and could be employed in a similar manner.
For selective neuron activation, ChR2 has emerged as an effective tool for investigating the sufficiency of specific neuronal cell types to contribute to behavior or physiology. Stereotaxic injection of a Cre-dependent rAAV2/1 vector using the EFα-1 promoter was used to target ChR2 to a midbrain-localized subset of dopamine-expressing neurons in transgenic mice expressing Cre under the control of the tyrosine hydroxylase promoter (Tsai et al., 2009). Stimulation of midbrain dopamine neurons induced a preference for the environment in which stimulation was delivered. Cell type–specific photostimulation of these neurons was used to probe preferred firing patterns for forming a place preference. These experiments showed that a burst of dopamine neuron firing was more effective for inducing a preference than a tonic stimulation pattern (Tsai et al., 2009). Similar cell type–specific viral tools have also been used for investigation of the causal relationship between activity patterns and behaviors such as sleep (Adamantidis et al., 2007), locomotion (Kravitz et al., 2010), addiction (Lobo et al., 2010), and feeding (Aponte et al., 2011).
Future directions for rAAV in neural circuit research
As reviewed here, improvements in cell type–specific gene expression strategies have increased the use of rAAV for neural circuit research. However, these techniques are best developed for use in genetically modified organisms. Future advances for rAAV vectors in neural circuit research are likely to come from new approaches to access functionally useful neuron populations in genetically unmodified organisms.
One attractive approach would be to further develop methods to efficiently engineer viral tropism for selective infection of a desired cell population. For this, a promising research direction is the development of pseudotyped rAAV vectors. Lentiviral vectors have been pseudotyped with the envelope glycoproteins of other viruses to alter or enhance the tropism (Stitz et al., 2000). The envelope glycoproteins, which bind cell surface receptors, are necessary for entry into the target cell, and changing the recognition properties of these ligands may enhance cell type selectivity (reviewed in Bischof and Cornetta, 2010). Pseudotyped AAV capsids with single chain antibodies (Yang et al., 1998) or ligands for cell surface receptors (Wu et al., 2000) have been investigated; however, the endogenous tropism of the virus can be a complication to engineering high specificity.
Another consideration for neural circuit dissection is to distinguish cell types, in part, by their projection patterns. Neurons in the same brain region show divergent axon projections, indicating distinct functions. Thus, an additional area for further development is the capacity for rAAV to selectively infect axon terminals. Several viral vectors have this property including pseudorabies virus (Braz et al., 2009), rabies virus (Wickersham et al., 2007), herpes virus (Lima et al., 2009), and canine adenovirus (Darvas and Palmiter 2009); however, the efficiency of axon transduction is variable and some of these vectors can be toxic to neurons. Recent reports suggested that self-complementary AAV vectors, specifically AAV1, exhibit efficient retrograde transduction properties from the neuromuscular junction back to motor neurons (Hollis et al., 2008). Also, other pseudotypes of AAV, including AAV6, have been shown to have retrograde transduction properties (Towne et al., 2010); however, the efficiency of these vectors supplied at central targets remains to be determined. Improvements may come from the design and/or guided evolution of novel rAAV serotypes with efficient retrograde transduction properties.
A complementary strategy that may prove useful for targeting neurons based on axonal projections involves the intersection of two viral vectors. rAAV expressing Cre fused to the plant lectin wheat germ agglutinin (WGA-Cre) (Gradinaru et al., 2010) is targeted to a brain region receiving axon projections from a neuron population of interest. Expression of WGA-Cre at the injection site leads to its being taken up by axon terminals in this region and transported back to cell bodies, for example on the contralateral side of the brain. If a second rAAV with a Cre-dependent transgene is injected into this contralateral side, then only the subset of neurons with those projections will have the two components required for selective activation of the Cre-dependent transgene (Fig. 3D). Such an approach also has the advantage of not requiring a genetically modified organism, potentially increasing general applicability.
With advances in new molecular and genetic properties, rAAV vectors will continue to be a prominent tool for illuminating the circuit-based underpinnings of brain function. Additional progress could be maximized by close interaction of virologists that are discovering and designing new rAAV properties and neuroscience researchers with specific neural circuit applications. In light of the critical role of viral tools for dissecting neural circuits and the challenges for testing and optimizing new tools, such cross-disciplinary approaches will likely be important for advances in both fields.
Author Disclosure Statement
No competing financial interests exist.
References
- Adamantidis A.R. Zhang F. Aravanis A.M., et al. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte Y. Atasoy D. Sternson S.M. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat. Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel B.R. Ehlers M.D. Molecular genetics and imaging technologies for circuit-based neuroanatomy. Nature. 2009;461:900–907. doi: 10.1038/nature08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster B.N. Li X. Pausch M.H., et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D. Aponte Y. Su H.H. Sternson S.M. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof D. Cornetta K. Flexibility in cell targeting by pseudotyping lentiviral vectors. Methods Mol. Biol. 2010;614:53–68. doi: 10.1007/978-1-60761-533-0_3. [DOI] [PubMed] [Google Scholar]
- Boyden E.S. Zhang F. Bamberg E., et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Braz J.M. Enquist L.W. Basbaum A.I. Inputs to serotonergic neurons revealed by conditional viral transneuronal tracing. J. Comp. Neurol. 2009;514:145–160. doi: 10.1002/cne.22003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buning H. Perabo L. Coutelle O., et al. Recent developments in adeno-associated virus vector technology. J. Gene Med. 2008;10:717–733. doi: 10.1002/jgm.1205. [DOI] [PubMed] [Google Scholar]
- Cearley C.N. Wolfe J.H. Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol. Ther. 2006;13:528–537. doi: 10.1016/j.ymthe.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Chamberlin N.L. Du B. De Lacalle S. Saper C.B. Recombinant adeno-associated virus vector: use for transgene expression and anterograde tract tracing in the CNS. Brain Res. 1998;793:169–175. doi: 10.1016/s0006-8993(98)00169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvas M. Palmiter R.D. Restriction of dopamine signaling to the dorsolateral striatum is sufficient for many cognitive behaviors. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14664–14669. doi: 10.1073/pnas.0907299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. Kahn I. Knoblich U., et al. Mapping brain networks in awake mice using combined optical neural control and fMRI. J. Neurophysiol. 2010. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Dong J.Y. Fan P.D. Frizzell R.A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- Dymecki S.M. Ray R.S. Kim J.C. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477C:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- Gao G. Vandenberghe L.H. Wilson J.M. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- Glover C.P. Bienemann A.S. Heywood D.J., et al. Adenoviral-mediated, high-level, cell-specific transgene expression: a SYN1-WPRE cassette mediates increased transgene expression with no loss of neuron specificity. Mol. Ther. 2002;5:509–516. doi: 10.1006/mthe.2002.0588. [DOI] [PubMed] [Google Scholar]
- Gong S. Doughty M. Harbaugh C.R., et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S. Zheng C. Doughty M.L., et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gradinaru V. Zhang F. Ramakrishnan C., et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Wang Y. Markram H. Organizing principles for a diversity of GABAergic interneurons and synapses in the neocortex. Science. 2000;287:273–278. doi: 10.1126/science.287.5451.273. [DOI] [PubMed] [Google Scholar]
- Hasan M.T. Friedrich R.W. Euler T., et al. Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS Biol. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis E.R., 2nd Kadoya K. Hirsch M., et al. Efficient retrograde neuronal transduction utilizing self-complementary AAV1. Mol. Ther. 2008;16:296–301. doi: 10.1038/sj.mt.6300367. [DOI] [PubMed] [Google Scholar]
- Karpova A.Y. Tervo D.G. Gray N.W. Svoboda K. Rapid and reversible chemical inactivation of synaptic transmission in genetically targeted neurons. Neuron. 2005;48:727–735. doi: 10.1016/j.neuron.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Klausberger T. Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.L. Meyer E.M. Peel A.L., et al. Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp. Neurol. 1998;150:183–194. doi: 10.1006/exnr.1997.6736. [DOI] [PubMed] [Google Scholar]
- Klimczak R.R. Koerber J.T. Dalkara D., et al. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Muller cells. PLoS One. 2009;4:e7467. doi: 10.1371/journal.pone.0007467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz A.V. Freeze B.S. Parker P.R., et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler S. Kilic E. Bahr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- Kuhlman S.J. Huang Z.J. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS ONE. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H. Durand R. Gradinaru V., et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S. Hawrylycz M.J. Ao N., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lerchner W. Xiao C. Nashmi R., et al. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron. 2007;54:35–49. doi: 10.1016/j.neuron.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Li X. Gutierrez D.V. Hanson M.G., et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima S.Q. Hromadka T. Znamenskiy P. Zador A.M. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo M.K. Covington H.E., 3rd Chaudhury D., et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Callaway E.M. Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown T.J. Adeno-associated virus (AAV) vectors in the CNS. Curr. Gene Ther. 2005;5:333–338. doi: 10.2174/1566523054064995. [DOI] [PubMed] [Google Scholar]
- McCown T.J. Xiao X. Li J., et al. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 1996;713:99–107. doi: 10.1016/0006-8993(95)01488-8. [DOI] [PubMed] [Google Scholar]
- Michelfelder S. Trepel M. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv. Genet. 2009;67:29–60. doi: 10.1016/S0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- Nathanson J.L. Jappelli R. Scheeff E.D., et al. Short promoters in viral vectors drive selective expression in mammalian inhibitory neurons, but do not restrict activity to specific inhibitory cell-types. Front. Neural Circuits. 2009a;3:19. doi: 10.3389/neuro.04.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson J.L. Yanagawa Y. Obata K. Callaway E.M. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009b;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L. Huber D. Sobczyk A. Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Petreanu L. Mao T. Sternson S.M. Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge E.A. Halbert C.L. Russell D.W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R.J. Chang L.S. Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd G.M. The synaptic organization of the brain. Oxford University Press; New York: 2004. p. 719. [Google Scholar]
- Stitz J. Buchholz C.J. Engelstadter M., et al. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology. 2000;273:16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
- Tan E.M. Yamaguchi Y. Horwitz G.D., et al. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51:157–170. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Tan W. Janczewski W.A. Yang P., et al. Silencing preBotzinger complex somatostatin-expressing neurons induces persistent apnea in awake rat. Nat. Neurosci. 2008;11:538–540. doi: 10.1038/nn.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans J.M. Vandenberghe L.H. Haute C.V., et al. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum. Gene Ther. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- Tian L. Hires S.A. Mao T., et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towne C. Schneider B.L. Kieran D., et al. Efficient transduction of non-human primate motor neurons after intramuscular delivery of recombinant AAV serotype 6. Gene Ther. 2010;17:141–146. doi: 10.1038/gt.2009.119. [DOI] [PubMed] [Google Scholar]
- Tsai H.C. Zhang F. Adamantidis A., et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien J.Z. Chen D.F. Gerber D., et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- van den Pol A.N. Acuna-Goycolea C. Clark K.R. Ghosh P.K. Physiological properties of hypothalamic MCH neurons identified with selective expression of reporter gene after recombinant virus infection. Neuron. 2004;42:635–652. doi: 10.1016/s0896-6273(04)00251-x. [DOI] [PubMed] [Google Scholar]
- Wang H. Peca J. Matsuzaki M., et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham I.R. Lyon D.C. Barnard R.J., et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. Xiao W. Conlon T., et al. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 2000;74:8635–8647. doi: 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. Mamounas M. Yu G., et al. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Hum. Gene Ther. 1998;9:1929–1937. doi: 10.1089/hum.1998.9.13-1929. [DOI] [PubMed] [Google Scholar]
- Zagoraiou L. Akay T. Martin J.F., et al. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64:645–662. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Wang L.P. Brauner M., et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]