Abstract
Plasma membrane (PM) H+-ATPases are the primary pumps responsible for the establishment of cellular membrane potential in plants. In addition to regulating basic aspects of plant cell function, these enzymes contribute to signaling events in response to diverse environmental stimuli. Here, we focus on the roles of the PM H+-ATPase during plant–pathogen interactions. PM H+-ATPases are dynamically regulated during plant immune responses and recent quantitative proteomics studies suggest complex spatial and temporal modulation of PM H+-ATPase activity during early pathogen recognition events. Additional data indicate that PM H+-ATPases cooperate with the plant immune signaling protein RIN4 to regulate stomatal apertures during bacterial invasion of leaf tissue. Furthermore, pathogens have evolved mechanisms to manipulate PM H+-ATPase activity during infection. Thus, these ubiquitous plant enzymes contribute to plant immune responses and are targeted by pathogens to increase plant susceptibility.
Keywords: Ion channels, ion transport, defense responses, disease responses, plant–microbe interactions
INTRODUCTION
Plants have evolved a multi-layered immune system that dynamically responds to pathogens. Most classes of plant pathogens remain outside the host cell membrane during their lifecycle. As a result, the plant plasma membrane (PM) is a key mediator of communication between plants and microbes. Initial pathogen recognition occurs at the PM and many of the earliest cellular responses to microbial invasion are controlled by PM-localized enzymes and ion channels (Boller and Felix, 2009). In addition, multiple downstream responses to pathogen stimuli are executed at the plasma membrane. Pathogens must manipulate host cells in order to suppress these defense responses and procure nutrients. It is therefore expected that membrane transport processes have numerous functions in compatible (susceptible) and incompatible (resistant) interactions (Hahn and Mendgen, 2001; Ward et al., 2009). Plant PM H+-ATPases use energy derived from ATP hydrolysis to pump protons from the cytosol to the extracellular space, thus creating and maintaining a negative membrane potential and a transmembrane pH gradient (acidic outside). This proton electrochemical gradient can control multiple aspects of transport across the PM. PM H+-ATPase activity is dynamically regulated during plant immune responses and multiple pathogens target this family of enzymes during infection. In this review, we focus on the roles of the PM H+-ATPase, a key regulator of membrane transport, in plant–microbe interactions.
The Plant Immune System
Pathogen threats are recognized by the plant as ‘non-self’ and/or ‘modified-self’ during microbial infection (Boller and Felix, 2009). There are two major branches of the plant innate immune system that have been described based on the types of pathogen molecules or proteins that are recognized (Chisholm et al., 2006). Pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) is thought to be one of the first lines of active plant defense. PAMPs are conserved microbial features, such as bacterial flagellin or fungal chitin, which plants have evolved to recognize as potential threats. The recognition of PAMPs is mediated by PM localized pattern recognition receptors, often belonging to the class of enzymes known as Receptor-Like protein Kinases (RLKs). The RLK FLS2 (FLAGELLIN SENSING 2) is the most well characterized pattern recognition receptor in Arabidopsis; it directly binds a conserved 22 amino acid epitope of flagellin termed flg22 (Boller and Felix, 2009). Upon recognition of flg22, FLS2 quickly heterodimerizes with its co-receptor BAK1 (BRI1-ASSOCIATED KINASE 1) to initiate intracellular signaling. This results in rapid changes in cellular homeostasis including alkalinization of the apoplast, increased intracellular calcium levels, other ion fluxes, generation of reactive oxygen species, and activation of mitogen-activated protein kinase cascades (Felix et al., 1999; Boller and Felix, 2009; Jeworutzki et al., 2010). PM H+-ATPases are dynamically regulated during early responses to flg22 treatment, suggesting a functional role for these enzymes during execution of PTI (Nuhse et al., 2007; Keinath et al., 2010). Ultimately, the plant cell mounts a defense response through production and secretion of antimicrobial compounds to the apoplast, reinforcement of the cell wall, hormone synthesis, and initiation of systemic resistance signaling to distant parts of the plant (Boller and Felix, 2009). It is hypothesized that PTI is critical in thwarting infection by many potentially pathogenic organisms (Chisholm et al., 2006).
Many host-adapted pathogens are capable of suppressing PTI signaling and downstream responses in large part through secretion of proteinaceous virulence factors, termed effectors, which commonly target host signaling components to evade or inactivate immune responses (Cui et al., 2009). To date, most characterized effectors from phytopathogens target intracellular host proteins and processes, but apoplastic effectors have been described from some fungal pathogens (Stergiopoulos and de Wit, 2009). The virulence contribution of the Type III Secretion System (T3SS) of Gram-negative bacteria has been well documented in mammalian and plant pathogens (Buttner and He, 2009). The bacterial T3SS functions as a conduit for effector delivery inside host cells (Buttner and He, 2009). Once inside the cell, characterized effectors generally operate by modifying key host proteins involved in immune responses or perception, thus enhancing pathogen virulence (Cui et al., 2009). In addition to effector proteins, many pathogens also secrete small molecule toxins that contribute to the pathogenic lifestyle of the microbe (Möbius and Hertweck, 2009). Modulation of plant PM H+-ATPase activity by pathogen effectors or toxins appears to be a relatively common event during infection.
Plants have also evolved a second branch of the immune system, termed Effector-triggered immunity (ETI) (Chisholm et al., 2006). This type of plant immunity enables plants to recognize particular pathogen-derived effector proteins in host genotypes expressing corresponding resistance (R) genes. In the majority of cases, ETI is initiated by intracellular R proteins with nucleotide-binding leucine-rich repeat domain architecture (Chisholm et al., 2006). Some apoplastic fungal effectors are recognized by PM-localized receptor-like proteins (Stergiopoulos and de Wit, 2009). Recognition of pathogen effectors can occur through direct binding. However, in the case of bacterial pathogens, effector recognition primarily occurs indirectly, where plant R proteins detect effector-induced modification of host targets as a form of modified self (Chisholm et al., 2006). ETI signaling limits pathogen establishment at infection sites and often culminates in a localized programmed cell death, termed the Hypersensitive Response (HR).
The situation described above (ETI leading to HR) is not necessarily effective against every pathogen a plant might encounter. There are two primary classes of plant pathogens: biotrophs (organisms that obtain nutrients from living host tissue) and necrotrophs (organisms that derive nutrients from dead tissue). Necrotrophs secrete toxins and lytic enzymes that induce cell death and cause tissue maceration; the pathogen then uses the cell degradation products as a nutrient source. In fact, many necrotrophic pathogens actively promote cell death processes and some toxins appear to target R-like proteins to induce cell death and plant susceptibility (Wolpert et al., 2002; Lorang et al., 2007). Conversely, obligate biotrophic pathogens absolutely require living host cells to procure nourishment and might be effectively contained during ETI. Some pathogens exhibit both biotrophic and necrotrophic phases of growth during infection and are termed hemibiotrophs. Therefore, cell death can have disparate roles in determining the outcome of plant–pathogen interactions, depending on both the pathogen and its stage of growth. Differential regulation of PM H+-ATPase activity during plant immune responses and direct manipulation by pathogens suggests that these enzymes perform multiple roles in susceptible and resistant interactions.
PM H+-ATPASES: CRITICAL ROLES IN PLANT PHYSIOLOGY
In plants, PM H+-ATPases are the primary pumps responsible for the establishment of cellular membrane potential. PM H+-ATPases use energy from ATP hydrolysis to pump protons from the cytosol to the extracellular space. This activity creates a proton electrochemical gradient across the plasma membrane that is utilized by channel and carrier proteins to facilitate ion and solute exchange across the membrane (Sondergaard et al., 2004). Furthermore, activation or inhibition of the PM H+-ATPase can modulate membrane potential, thereby changing the activities of voltage-gated channels and controlling ion flux at the PM (Ward et al., 2009). By regulating membrane transport processes that are necessary for life, PM H+-ATPases are absolutely essential for normal plant growth and development (Haruta et al., 2010).
PM H+-ATPases establish and maintain the major electrochemical gradient used for secondary transport at the plasma membrane and therefore control many basic processes in plant physiology (Sondergaard et al., 2004). Proton extrusion permits selective solute and ion uptake at the root periphery and at the endodermal cell layer within roots. Furthermore, PM H+-ATPase activity is essential for phloem loading and unloading of photosynthate and the maintenance of proper source/sink relationships within the plant (Zhao et al., 2000). Therefore, PM H+-ATPases power plant nutrient uptake and distribution throughout the plant (Sondergaard et al., 2004). The high levels of expression of PM H+-ATPases in transport cells supports the idea that these enzymes are critical regulators of membrane transport processes (Bouche-Pillon et al., 1994; DeWitt and Sussman, 1995). Cell turgor and stomatal apertures (see below) are similarly regulated by PM H+-ATPase via solute and ion accumulation within cells and subsequent changes in water potential (Sondergaard et al., 2004). Accumulating evidence suggests that PM H+-ATPases also function in long-distance signaling events via the modulation of membrane potential and cellular/apoplastic pH (Zimmermann et al., 2009). Not surprisingly, due to its importance in regulating basic aspects of plant cell function and nutrient transport, pathogens have evolved direct and indirect mechanisms to manipulate PM H+-ATPase activity during infection.
REGULATION OF PLASMA MEMBRANE H+-ATPASES
Due to its central role in regulating plant membrane transport processes, it is expected that (1) a subset of PM H+-ATPases is ubiquitously expressed and (2) PM H+-ATPase activity is tightly controlled (Duby and Boutry, 2009). PM H+-ATPases comprise a large gene family in plants, with 11 members in Arabidopsis thaliana (AHA1-11). A combination of gene expression and genetic analyses has revealed that multiple AHAs are expressed in many tissues and organs, but some differences exist in expression levels and tissue specificity of individual AHA isoforms (Gaxiola et al., 2007). Genetic analyses of AHA knockouts have revealed few phenotypes, suggesting that removal of a single isoform can be compensated for by other isoforms. PM H+-ATPases are members of a large gene family that exhibit expression overlap and functional redundancy that can mask key roles of individual isoforms, making straightforward genetic analyses difficult. AHA1 and AHA2 are highly expressed in almost all tissues and organs, indicating that they function as the key PM H+-ATPases in maintaining ion homeostasis (Haruta et al., 2010). Consistent with this hypothesis, single knockouts of AHA1 or AHA2 have no obvious phenotypes, but the double knockout is lethal (Liu et al., 2009; Haruta et al., 2010).
PM H+-ATPases do not undergo significant changes in transcript levels in response to most biotic or abiotic stresses, and changes in activity and solute fluxes are frequently uncoupled from transcriptional changes (Gaxiola et al., 2007). Therefore, the activity of PM H+-ATPases is likely under post-translational control. Phosphorylation is one well characterized mechanism that can act to regulate this pump's activity. Phosphorylation of the penultimate threonine residue within the auto-inhibitory C-terminal domain is critical for interaction with a 14-3-3 regulatory protein (Baunsgaard et al., 1998; Svennelid et al., 1999). 14-3-3 proteins recognize and bind to phosphorylated proteins with one of three canonical binding motifs. 14-3-3- proteins generally function as dimers and bind the C-terminus of PM H+-ATPases at a conserved mode III binding site (XYpTV-COOH) (Duby and Boutry, 2009). Binding of the 14-3-3 protein relieves C-terminal intramolecular inhibition and results in activation of the PM H+-ATPase. Furthermore, single particle analysis using electron cryomicroscopy indicates that the activated Nicotiana plumbaginifolia PM H+-ATPase PMA2 consists of six phosphorylated H+-ATPase molecules assembled in a hexameric structure along with six 14-3-3 molecules (Kanczewska et al., 2005; Ottmann et al., 2007). The penultimate threonine residue and accompanying mode III binding motif is widely conserved across H+-ATPases throughout the plant kingdom, suggesting that this mechanism of activation is highly conserved (Duby and Boutry, 2009). Additional phosphorylated residues within the C-terminal domain have been reported to affect enzyme activity incrementally, suggesting that enzyme activation can be gradual and tightly controlled (Speth et al., 2010). Accumulating experimental evidence also implies that PM H+-ATPases can be negatively regulated by phosphorylation in multiple plants, including tomato (Xing et al., 1996), beet (Lino et al., 1998), maize (De Nisi et al., 1999), tobacco (Desbrosses et al., 1998; Duby et al., 2009), and Arabidopsis (Fuglsang et al., 2007). Recently, the PKS5 kinase in Arabidopsis was shown to phosphorylate AHA2 at serine 931. Phosphorylation of serine 931 inhibits AHA2's interaction with 14-3-3 proteins even when the penultimate threonine is phosphorylated, indicating that multiple factors in addition to 14-3-3 binding coordinate activation of the pump (Fuglsang et al., 2007). Thus, PM H+-ATPase activity is positively and negatively influenced by phosphorylation at multiple sites and regulation is likely fine-tuned by distinct kinases and phosphatases.
In addition to regulation via phosphorylation, PM H+-ATPase protein abundance and/or localization within the plasma membrane may also play an important role in regulating enzyme activity. Auxin has been shown to induce exocytosis of a pool of pre-synthesized PM H+-ATPase, possibly providing a mechanism to regulate rapid focal accumulation of the enzyme (Hager et al., 1991). Multiple PM H+-ATPases in both Arabidopsis and tobacco have been detected in detergent resistant microdomains by mass spectrometry (Shahollari et al., 2004; Morel et al., 2006). Consistent with these findings, immunolocalization experiments have found PM H+-ATPases localize within defined patches on the plasma membrane (Bouche-Pillon et al., 1994; DeWitt and Sussman, 1995). These experiments suggest that PM H+-ATPase activity can be influenced by lateral compartmentalization within the plasma membrane and that localized activity of the pump could influence membrane transport processes.
PM H+-ATPASE REGULATION IN PLANT IMMUNE SIGNALING
PAMP-Triggered Immunity (PTI)
One of the earliest cellular responses during activation of plant immune responses is the modulation of extracellular pH. It has long been observed that various general pathogen elicitors/PAMPs induce ion fluxes and rapid alkalinization of the medium in cell culture suspensions of various plant species (Felix et al., 1993; Nurnberger et al., 1994; Kuchitsu et al., 1997; Felix et al., 1999; Boller and Felix, 2009). This extracellular alkalinization response could result from inhibition of PM H+-ATPase activity, opening of PM proton channels resulting in proton influx down the electrochemical gradient, and/or activation of proton-coupled secondary transporters. In Arabidopsis leaf mesophyll cells, perception of the PAMP flg22 by FLS2 results in rapid membrane depolarization and alkalinization of the apoplast (Jeworutzki et al., 2010). Although initially thought to be the consequence of inhibition of the PM H+-ATPase, data indicate that these responses are a result of high levels of proton influx and activation of anion channels that may not be triggered solely by PM H+-ATPase inhibition (Hagendoorn et al., 1991; Thain et al., 1995; Jeworutzki et al., 2010). Proton influx occurs in concert with an even larger anion (Cl– and NO3–) efflux and together these ion fluxes are the likely source of membrane depolarization during PTI (Jeworutzki et al., 2010). Exogenous application of fusicoccin (a strong PM H+-ATPase activator) hyperpolarized the membrane but did not prevent depolarization in response to subsequent flg22 treatment (Jeworutzki et al., 2010). However, under natural infection conditions, it is likely that the PM H+-ATPase contributes to PAMP-induced membrane depolarization and apoplast alkalization in concert with anion efflux and proton influx. Other experiments support a role for PM H+-ATPase inhibition during PTI (Nuhse et al., 2007; Amborabé et al., 2008) (see below). Consistent with the hypothesis that PM H+-ATPases function in early immune signaling events, Arabidopsis expressing constitutively active alleles of AHA1 exhibit a reduced burst of reactive oxygen species in response to flg22 (Keinath et al., 2010). These experiments suggest that down-regulation of PM H+-ATPase activity during PTI might be required for a subset of plant immune responses. Calcium and reactive oxygen species are known to influence PM H+-ATPase activity in guard cells (Kim et al., 2010). Calcium influx and production of reactive oxygen species at the plasma membrane occur very rapidly after PAMP treatment and may also contribute to PM H+-ATPase regulation in mesophyll cells during PTI (Boller and Felix, 2009).
Post-Translational Control of PM H+-ATPase during PAMP-Triggered Immunity
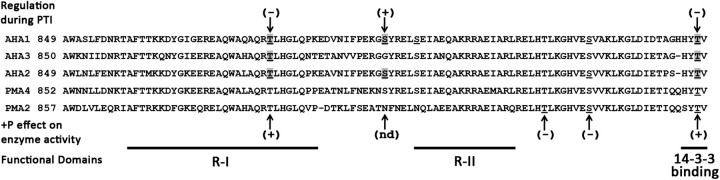
As described above, PM H+-ATPase activity can be positively and negatively influenced by phosphorylation and localization (Duby and Boutry, 2009). Recent proteomics experiments have contributed to a greater understanding of dynamic PM H+-ATPase phosphorylation during plant immune responses. The phosphorylation status of the Arabidopsis PM H+-ATPases (AHAs) changes dramatically during PTI signaling in response to flg22 (Benschop et al., 2007; Nuhse et al., 2007). Consistently, both groups observed a significant reduction in AHA1 and AHA2 peptides containing the phosphorylated penultimate threonine residue known to mediate 14-3-3 binding and enzyme activation (Olsson et al., 1998; Svennelid et al., 1999; Benschop et al., 2007; Nuhse et al., 2007) (Figure 1). These results indicate that dephosphorylation and inactivation of the enzyme occur during PTI, which is consistent with the observed alkalinization response of plant cell cultures to various PAMPs. In addition to the regulatory penultimate threonine residue, other sites in the C-terminal regulatory domain of AHA1-3 were identified whose phosphorylation status appears to change dynamically during PTI (Figure 1). Phosphorylation of a highly conserved threonine residue (Thr881 in AHA1/2) decreases nearly 10-fold in response to flg22 (Nuhse et al., 2007). Phosphomimetic mutations of this residue in AHA2 and the corresponding residue in PMA2 have been shown to activate PM H+-ATPase activity (Niittylä et al., 2007; Speth et al., 2010). Thus, phosphorylation of two residues known to promote AHA activity decreases during PTI. In contrast, phosphorylation of another residue, AHA1/2 Ser899, increases several-fold during PTI (Nuhse et al., 2007). It will be interesting to determine whether phosphorylation of this residue negatively impacts AHA activity as suggested by coordinate regulation of the other identified residues. Taken together, data suggest that PM H+-ATPases are under multiple layers of control during PTI, potentially through sequential regulation of phosphorylated residues.
Figure 1.
Regulation of PM H+-ATPases by Phosphorylation. ClustalW alignment of the C-terminal regulatory domain of Arabidopsis (AHA) and Nicotiana (PMA) PM H+-ATPases. Underlined residues have been identified in planta as phosphorylated. Phosphorylation status of shaded residues significantly changes during PTI responses (upper). Phosphorylation (+P) effect on enzyme activity is indicated below the alignment. Protein functional domains are indicated at the bottom. (+), increase in phosphorylation or activity. (−), decrease in phosphorylation or activity. (nd), not determined. R-I and R-II are inhibitory domains.
Additional post-translational control of PM H+-ATPase activity could result from dynamic changes in protein localization. Recent data indicate that Arabidopsis AHA1, AHA2, AHA3, and AHA4 localize to detergent resistant membrane (DRM) fractions within 5 min post flg22 application (Keinath et al., 2010). DRMs are characterized by insolubility in cold detergents, such as Triton X-100, after biochemical isolation. DRMs are enriched in sterols and saturated lipids (and associated proteins) and have been suggested to form functional membrane microdomains in vivo (Laloi et al., 2007). The rapid enrichment of AHAs in DRMs during PTI suggests that AHA activity might be modulated as a result of lateral compartmentalization within the plasma membrane. A number of regulatory proteins involved in plant innate immune signaling and perception, such as receptor-like protein kinases, are also enriched in DRMs (Shahollari et al., 2004; Morel et al., 2006). Association of the PM H+-ATPase within membrane microdomains indicates that these enzymes may be rapidly recruited to specific microsites within the PM in order to mediate responses immediately downstream of PAMP receptor activation. As the membrane sterol environment has been reported to affect PM H+-ATPase activity, demonstrating functional relevance between enzyme activity and DRM localization is an exciting area for research (Grandmougin-Ferjani et al., 1997). Taken together, these proteomics experiments suggest that the activity of Arabidopsis PM H+-ATPase isoforms is dynamically regulated via post-translational modification and membrane compartmentalization during plant immune responses.
Effector-Triggered Immunity (ETI)
Although alkalinization of the extracellular medium is common during PTI, acidification of the apoplast is associated with some cases of ETI. Cladosporium fulvum, the leaf mold fungus of tomato, secretes several effector proteins into the plant apoplast during infection (Stergiopoulos and de Wit, 2009). Recognition of the Avr5 effector from C. fulvum by the corresponding R protein Cf5 results in acidification of the extracellular medium in tomato cell lines (Vera-Estrella et al., 1994). This acidification response has been attributed to activation of the PM H+-ATPase (Vera-Estrella et al., 1994). Intriguingly, PM H+-ATPase activity in this system appears to be positively regulated by dephosphorylation, as phosphatase inhibitors blocked the increase in PM H+-ATPase activity. Furthermore, Avr5/Cf-5-dependent increases in PM H+-ATPase activity were correlated with enzyme dephosphorylation using γ-32P labeling of isolated PM fractions (Vera-Estrella et al., 1994; Xing et al., 1996). These observations are in agreement with recent data demonstrating negative regulation of PM H+-ATPase activity by phosphorylation (Fuglsang et al., 2007; Duby et al., 2009). Other studies in tomato report that stimulation of PM H+-ATPase activity causes the accumulation of the defense hormone salicylic acid and the transcription of pathogenesis-related (PR) genes (Schaller and Oecking, 1999). Taken together, these studies indicate that PM H+-ATPase activation is not only associated with, but can also trigger, immune responses in tomato.
Acidification of the apoplast was also observed in incompatible interactions of barley plants containing the Mla3 resistance gene with the powdery mildew fungus Blumeria graminis f.sp. hordei (Bgh) expressing the AvrMla3 effector (Zhou et al., 2000). This decrease in extracellular pH preceded HR development, and it was demonstrated that activation of the PM H+-ATPase increased the occurrence of cell death in compatible interactions (Zhou et al., 2000). However, additional studies indicate that other incompatible barley–Bgh interactions mediated by different R genes result in a long-term apoplastic alkalinization (Felle et al., 2004). Different experimental set-ups and timescales could have played a role in these disparate outcomes. It is possible that the PM H+-ATPase is differentially regulated, both spatially and temporally, within the plant during the manifestation of ETI. Furthermore, barley Mla resistance gene alleles activate resistance at different temporal stages of Bgh infection, which indicates some level of mechanistic divergence (Boyd et al., 1995). In any case, acidification of the apoplast via activation of the PM H+-ATPase appears to potentiate defense responses in some incompatible plant–microbe interactions mediated by R genes. As alkalinization of the apoplast precedes cell death in many elicitor- or pathogen-treated cell types, it remains to be determined just how the nature of the recognition event affects PM H+-ATPase activity and other cellular responses to pathogens (Blumwald et al., 1998).
PM H+ATPASE ROLE IN REGULATING STOMATAL APERTURES AND PATHOGEN ENTRY
All plant pathogens must invade host tissue during infection. Foliar pathogens frequently enter leaves through wounds or natural openings such as stomata. Stomata are microscopic pores formed by guard cells in the leaf epidermis that regulate gas exchange and transpiration during photosynthesis. However, stomata are not merely openings at the leaf surface, passively allowing pathogen access to the interior of leaves during infection. The guard cells that make up stomatal pores react dynamically to prevent pathogen entry (Melotto et al., 2006). Furthermore, recent data indicate a key role for the PM H+-ATPase working together with plant immune signaling components to regulate stomata movements during bacterial invasion (Liu et al., 2009).
The function of the PM H+-ATPase in stomatal opening is well established and has been reviewed in detail (Dietrich et al., 2001; Kim et al., 2010). During stomatal opening, PM H+-ATPases are activated, resulting in hyperpolarization of the guard cell membrane. This event activates inward rectifying voltage-gated K+ channels. The influx of K+ is accompanied by an influx of anions (Cl– and NO3–) and production of osmotically active solutes from starch. Ion and solute accumulation in the cell interior lowers water potential and drives the uptake of water into guard cells, increasing turgor and widening the stomatal aperture. Inhibition of the PM H+-ATPase is also important for ABA-mediated stomatal closure. Lines expressing constitutively active alleles of Arabidopsis AHA1 result in an open stomata phenotype and are completely insensitive to ABA-induced stomata closure but not other stimuli. This result suggests that down-regulation of PM H+-ATPase activity is an important component of the ABA-mediated stomatal closure pathway (Merlot et al., 2007).
Upon perception of pathogens, guard cells of diverse plant species rapidly lose turgor, resulting in stomatal closure that prevents pathogen invasion (Melotto et al., 2006). Importantly, plant mutants with stomata that do not close in response to bacterial inoculation or PAMP application are more susceptible than wild-type plants (Melotto et al., 2006; Liu et al., 2009). Inoculation of the Arabidopsis leaf surface with plant pathogenic Pseudomonas syringae pv. tomato DC3000 (Pto DC3000), human pathogenic E. coli, as well as purified fungal or bacterial PAMPs induces similar stomatal closure, indicating that this is a universal response during PTI. Data from Arabidopsis mutant lines indicate that PAMP-induced stomatal closure is dependent on both salicylic acid- and ABA-biosynthesis and associated signaling components (Melotto et al., 2006; Zhang et al., 2008; Zeng and He, 2010). Notably, epistasis analyses indicate that SA acts upstream of ABA during PAMP-triggered stomatal closure (Zeng and He, 2010). Taken together with data from constitutively active AHA1 lines, it appears that down-regulation of PM H+-ATPase activity via the phytohormone ABA contributes to PAMP-induced stomatal closure (Melotto et al., 2006; Merlot et al., 2007; Liu et al., 2009). Even though bacteria are expected to present multiple distinct PAMPs during infection, the PAMP receptor FLS2 has a critical role in stomatal closure during bacterial infection of Arabidopsis (Zeng and He, 2010). Bacterial motility is necessary for successful invasion into the leaf interior. Therefore, flagellin expression may be up-regulated on leaf surfaces, and correspond to a highly abundant bacterial PAMP. These experiments highlight the importance of pathogen surveillance and dynamic stomata-based immune responses that function during early stages of bacterial invasion.
Phytopathogenic bacteria have developed mechanisms to reverse PTI-induced stomata closure. Pto DC3000 inoculation induces stomata to close within 1 h, but stomata are re-opened 3 h post inoculation (Melotto et al., 2006). It was demonstrated that coronatine is the major virulence factor in Pto DC3000 that conditions stomata to re-open after PTI-induced closure (Melotto et al., 2006). Coronatine is a structural mimic of the bioactive jasmonic acid–isoleucine conjugate, and a potent inducer of COI1-dependent jasmonate responses in plants. Several strains of P. syringae produce coronatine, indicating that this might be a valuable mechanism to facilitate bacterial invasion. Coronatine has several functions during Pseudomonas pathogenesis (Underwood et al., 2007). During bacterial invasion, coronatine operates by reversing the flg22-induced inhibition of K+ inward-rectifying ion channels, and this contributes to stomatal re-opening (Zhang et al., 2008). In addition to coronatine, there are other examples of bacterial virulence factors that manipulate stomatal apertures. Xanthomonas campestris pv. campestris secretes a small molecule virulence factor that causes plants to re-open stomata (Gudesblat et al., 2009). Recently, another Pseudomonas syringae toxin, syringolin A, was shown to reverse PTI-associated stomata closure likely via its action as a proteasome inhibitor (Schellenberg et al., 2010). Thus, multiple bacterial pathogens have evolved distinct ways to counteract stomata closure during plant immune responses. Some fungal pathogens also secrete toxins that target PM H+-ATPase activity and affect stomatal apertures (see below). Therefore, active manipulation of stomata by phytopathogens appears to be a common way in which pathogens gain access to the interior of leaf tissue.
RIN4 Regulates PM H+-ATPase Activity during Pathogen Infection
RIN4 is an important plant protein that operates in both PTI and ETI. RIN4 is plasma membrane-localized and genetically acts as a negative regulator of disease resistance. RIN4 overexpression lines exhibit reduced PTI responses to PAMPs and nonpathogenic bacteria while rin4 null mutants exhibit enhanced disease resistance (Kim et al., 2005). Several pathogen effectors have been shown to target and modify RIN4 during bacterial pathogenesis (Mackey et al., 2002; Axtell and Staskawicz, 2003). Furthermore, RIN4 associates with and is ‘guarded’ by at least two R proteins—RPS2 and RPM1—which initiate rapid ETI responses upon sensing effector-mediated RIN4 modification (Mackey et al., 2002; Axtell and Staskawicz, 2003). In the absence of pathogen modification, RIN4 acts as a negative regulator of RPM1 and RPS2 proteins in order to prevent inappropriate defense activation (Belkhadir et al., 2004).
Recently, a new role for RIN4 during pathogen invasion was uncovered. RIN4 works in concert with Arabidopsis PM H+-ATPases to regulate stomatal opening during bacterial invasion of leaf tissue (Liu et al., 2009). The RIN4–AHA interaction was originally detected by co-immunoprecipitation of RIN4 and associated proteins in Arabidopsis. Both AHA1 and AHA2 peptides were identified by mass spectrometry. Furthermore, AHA1 and AHA2 C-terminal domains were shown to interact with RIN4 in vitro and in planta using yeast two-hybrid, protein pull-downs and bi-molecular fluorescence complementation assays (Liu et al., 2009). AHA activity was found to be positively regulated by RIN4 in planta using Arabidopsis RIN4 overexpression and knockout lines. Adding recombinant RIN4 protein to purified PM vesicles increased AHA activity, indicating that this effect is direct. Importantly, it was demonstrated through different bacterial inoculation techniques that RIN4 can operate at the level of pathogen invasion. Spray inoculation of bacteria onto the leaf surface of rin4/rpm1/rps2 plants resulted in significantly enhanced disease resistance when compared with rpm1/rps2 lines, while syringe infiltration of bacteria into the leaf interior had only a minor effect on bacterial growth, indicating that RIN4 acts as a negative regulator of defense during early stages of bacterial invasion. Furthermore, constitutively active AHA1 lines, whose stomata do not close in response to bacteria, were more susceptible to bacteria under spray inoculation conditions (Liu et al., 2009). Measurements of stomatal apertures revealed that RIN4 is required for stomata re-opening by virulent Pto DC3000 because the stomata of rin4/rpm1/rps2, but not rpm1/rps2, lines remain closed 3 h after inoculation with virulent bacteria. Taken together, these results demonstrate that RIN4 cooperates with AHA1/2 to influence stomatal behavior during pathogen attack. However, the molecular mechanism for how RIN4 acts to regulate AHA activity is currently unknown.
Interestingly, RIN4 contains a 14-3-3 mode II binding site (RADE[pS]P, G. Coaker, unpublished). This site is phosphorylated in planta based on phosphoproteomic studies, indicating that RIN4 might be regulated by 14-3-3 proteins (Benschop et al., 2007; Nuhse et al., 2007) (G. Coaker, unpublished). Therefore, it is tempting to speculate that RIN4 can modulate PM H+-ATPase activity through its interaction with AHA's C-terminal regulatory domain and 14-3-3 proteins. RIN4 is phosphorylated in response to the P. syringae effectors AvrB and AvrRpm1 in both resistant and susceptible plant genotypes, suggesting that post-translational modification of RIN4 by bacterial effectors could promote bacterial infection, possible at early stages of tissue invasion (Mackey et al., 2002). It is important to note that effector delivery into guard cells or epidermal cells has not yet been demonstrated. If effectors are indeed delivered into guard cells, their potential modification of PM H+-ATPase activity via RIN4 will be an important area of research. It will also be important to determine whether differential phosphorylation of RIN4 in response to bacterial effectors or plant immune signaling directly impacts AHA activity in guard cells as well as other cell types.
PM H+-ATPASE ACTIVITY IS TARGETED BY PATHOGENS
Given the numerous roles of PM H+-ATPase activity in plant cell physiology and induced responses to environmental stimuli, it is no wonder that pathogens have evolved mechanisms to target this enzyme (Table 1). Alterations in PM H+-ATPase activity, via stimulation or inhibition, can have drastic effects on plant cell function. PM H+-ATPases generate cellular membrane potential and therefore influence the transport of numerous molecules into and out of the plant cell (Sondergaard et al., 2004). Manipulation of membrane potential by pathogens could result in significant nutrient accumulation in host tissue (Zhao et al., 2000). Microbes may exploit host membrane energization to facilitate pathogen membrane transport processes in order to acquire nutrients from host tissue. Pathogen acidification of the apoplastic space could also promote plant cell wall loosening that may facilitate fungal movement in host tissue (Duby and Boutry, 2009). In addition, over-activation of PM H+-ATPases can lead to cell death (Möbius and Hertweck, 2009). Below, we highlight several instances in which pathogens directly or indirectly manipulate PM H+-ATPase activity during infection (Table 1).
Table 1.
Phytotoxins Known to Modulate PM H+-ATPase Activity.
| Toxin | Pathogen | PM H+-ATPase regulation | Mode | Toxin/ disease symptoms | References |
| Fusicoccin | Fusicoccum amygdale | Stimulation | Direct | Open stomata/ necrosis | Marre, 1979; Baunsgaard et al., 1998 |
| NIP1 | Rhynchosporium secalis | Stimulation | N/D | Open stomata/ necrosis | Wevelsiep et al., 1993; van't Slot et al., 2007 |
| Beticolin-1 | Cercospora beticola | Inhibition | Indirect | Necrosis | Simon-Plas et al., 1996 |
| Fumonisin B1 | Fusarium verticillioides | Inhibition | Indirect | Necrosis | Gutiérrez-Nájera et al., 2005 |
| Bacterial lipopeptides | Pseudomonas sp. | Inhibition | Indirect | Necrosis | Batoko et al., 1998 |
N/D, not determined.
Fusicoccin
Fusicoccin is the major phytotoxic metabolite produced by the fungal pathogen Fusicoccum (Phomopsis) amygdale, the causal agent of peach and almond canker (Ballio et al., 1976). This diterpene glucoside is perhaps the most well known modulator of PM H+-ATPase activity and has been used for decades as a tool in plant physiology research (Marre, 1979). Despite the limited host range of the pathogen from which it is derived, fusicoccin is bioactive in nearly all higher plant species (Marre, 1979). Fusicoccin treatment stimulates the virtually irreversible opening of stomata, increasing transpiration rates, causing uncontrolled water loss and eventual wilting of affected leaves and plants, which mimics disease symptoms caused by the fungus. The role of fusicoccin during fungal invasion of leaf tissue has not been established. Manipulation of stomata may allow the fungus to gain access to the interior of the leaf. In addition, fusicoccin treatment can lead to cell death and may contribute to canker formation, presumably through the over-stimulation of the PM H+-ATPase and subsequent pleiotropic effects (Möbius and Hertweck, 2009). Consistent with this hypothesis, transgenic lines transformed with a constitutively active PM H+-ATPase gene lacking the C-terminal auto-inhibitory region display necrotic lesions, severely altered plant architecture, and exhibit enhanced seedling mortality (Liu et al., 2009). Therefore, fusicoccin could be a virulence factor during the necrotrophic growth stage of the F. amygdale disease cycle.
Fusicoccin has been a critical tool for understanding the regulation of PM H+-ATPases (Jahn et al., 1997; Baunsgaard et al., 1998; Olsson et al., 1998). 14-3-3 protein binding to the PM H+-ATPase was originally uncovered in the presence of fusicoccin and was subsequently demonstrated to occur in its absence (Jahn et al., 1997). Fusicoccin functions to irreversibly stabilize the interaction of the C-terminal regulatory domain of PM H+-ATPases with 14-3-3 proteins, leading to relief of auto-inhibition and constitutive proton pumping (Jahn et al., 1997; Baunsgaard et al., 1998). With regard to stomatal opening, fusicoccin-induced hyperpolarization of the membrane of guard cells opens inwardly rectifying ion channels, lowering intracellular water potential, and driving uptake of water, increasing guard cell turgor and opening stomatal pores. The crystal structure of a ternary complex composed of a tobacco H+-ATPase PMA2 C-terminal phosphopeptide, fusicoccin, and 14-3-3 protein has been solved (Wurtele et al., 2003). Structural and thermodynamic analysis indicates that fusicoccin increases the binding affinity of the 14-3-3 protein to the PMA2 C-terminal phosphopeptide nearly 100-fold (Wurtele et al., 2003). Thus, fusicoccin functions as a molecular adhesive that promotes the interaction between PM H+-ATPases and 14-3-3 regulatory proteins, locking the PM H+-ATPase in an activated state.
NIP1 and NIP3
Infection of barley by the leaf scald fungus, Rhynchosporium secalis, a necrotroph, results in an increased incidence of open stomata during early stages of fungal growth (Ayres, 1972). Accordingly, the small, secreted, necrosis-inducing effectors NIP1 and NIP3 were shown to activate the PM H+-ATPase in isolated barley plasma membranes, probably through an indirect mechanism (Wevelsiep et al., 1993). Purified NIP1 and NIP3 induce necrosis on several cereal crops and bean, likely via sustained activation of the PM H+-ATPase (Wevelsiep et al., 1993).
NIP1 can act as a virulence factor on susceptible hosts, but is recognized in barley genotypes carrying the Rrs1 R gene (Rohe et al., 1995). Importantly, NIP1 stimulates PM H+-ATPase activity in both susceptible and resistant genotypes. NIP1 alleles from different R. secalis strains encode proteins with amino acid polymorphisms that confer quantitative differences in their ability to activate PM H+-ATPase activity. However, functional experiments were unable to separate H+-ATPase stimulating, necrosis-inducing, and avirulence activities, indicating that all function through the same mechanism (Fiegen and Knogge, 2002). Furthermore, NIP1 was shown to stimulate H+-ATPase activity and bind a single class of sites in isolated plasma membranes from isogenic resistant and susceptible barley lines (van't Slot et al., 2007). NIP1 binding sites were also found in other host and non-host cereal crops, but not Arabidopsis. As NIP1 induces necrosis in these cereal species, but not Arabidopsis, NIP1 binding was correlated with its necrosis-inducing activity in these species (van't Slot et al., 2007). This result suggests that Arabidopsis may lack an as yet undetermined factor present in cereals that modulates activation of the PM H+-ATPase. Strong, sustained stimulation of PM H+-ATPase activity is the likely cause of NIP1-mediated necrosis, enabling R. secalis to acquire nutrients from necrotic tissue. It is still unclear whether stimulation of PM H+-ATPase activity is the primary virulence function of this fungal effector or a consequence of a yet unknown mechanism.
Pathogen Toxins Disrupt Membrane Integrity and Impact PM H+-ATPase Activity
Cercospora beticola is a necrotrophic fungal pathogen that causes leaf spot of sugar beet. It produces beticolin toxins during infection and infiltration of leaf tissue with purified Beticolin toxins induces necrotic lesions that mimic disease symptoms (Ballio, 1991). CBT/beticolin-1 toxin can inhibit ATP hydrolysis and proton pumping activity of the PM H+-ATPase in vitro (Simon-Plas et al., 1996). However, other studies suggest that the primary function of Beticolin toxins is to disrupt membrane function by forming pores that could dissipate the proton electrochemical gradient across the membrane (Goudet et al., 2000). Thus, the observed inhibition of PM H+-ATPase activity is likely a secondary effect of these toxins.
Fumonisin B1 (FB1) is a phytotoxin produced by the hemibiotrophic fungus Fusarium verticillioides and other species of Fusarium. FB1 is a sphingoid toxin, structurally related to AAL-toxin produced by Alternaria alternata, and shares similar properties in its animal toxicity. FB1 has several reported toxic effects on cellular function, one of which is significant inhibition of the PM H+-ATPase and other P-type ATPases (Abbas et al., 1994; Gutiérrez-Nájera et al., 2005). Due to its amphiphilic chemical structure, FB1 can insert into lipid bilayers and enhance membrane fluidity (Ferrante et al., 2002; Gutiérrez-Nájera et al., 2005). Therefore, its inhibitory effect on the PM H+-ATPase is likely indirect and a consequence of membrane disruption that leads to aberrant enzyme function.
Bacterial pathogens in the genus Pseudomonas produce structurally related lipopeptide toxins (syringomycin, syringotoxin, syringopeptins, fuscopeptins). These amphiphilic molecules can form pores in the PM, disturbing membrane function and permitting ion flow (Serra et al., 1999). The toxins can inhibit PM H+-ATPase enzyme activity individually and in a synergistic manner, which is likely a consequence of membrane disruption (Batoko et al., 1998).
PERSPECTIVES AND CONCLUSIONS
In this review, we have summarized the roles of the PM H+-ATPase in plant–pathogen interactions. Recent proteomics studies have revealed that PM H+-ATPase enzymes are under elaborate regulation during plant immune responses. The phosphorylation status of Arabidopsis AHAs at multiple amino acid residues changes rapidly during PTI signaling, suggesting that plant cells exert multi-layered control over these important proteins during complex signaling events (Nuhse et al., 2007) (Figure 1). The identification of protein kinase and phosphatase enzymes that mediate the post-translational modifications of PM H+-ATPases during plant immune responses will enhance our understanding of PM H+-ATPase regulation in general and can be used as tools to investigate specific roles of the proton pumps during defense signaling. The spatial and temporal regulation of PM H+-ATPases in specific cell types and functional sites within the PM would permit the plant to fine-tune enzyme activity in order to achieve appropriate responses to pathogen attack. High-resolution in vivo imaging techniques to monitor protein localization and activity will likely be valuable tools for future investigations.
Due to its established importance in regulating stomatal apertures, it is not surprising that the PM H+-ATPase also functions in stomata-based immune responses. As in other cell types, PM H+-ATPase activity appears to be down-regulated during PTI responses in guard cells; this inhibition contributes to loss of turgor and causes stomata to close in order to prevent pathogen entry (Liu et al., 2009). Intriguingly, PM H+-ATPases appear to physically and spatially associate with plant immune signaling components and this may have direct consequences on how pathogens are able to re-open stomata during infection. Further study is needed to determine the molecular mechanisms of how virulent pathogens manipulate guard cell signaling, re-activate the PM H+-ATPase or downstream ion channels, and gain entry into the leaf interior. Manipulation of PM H+-ATPase activity in vivo by both plants and attacking pathogens likely occurs in a host cell type- and pathogen infection stage-specific manner. Understanding how these complex interactions lead to resistance or susceptibility will likely keep researchers engaged for years to come.
FUNDING
G.C. is supported by grants from the NIH (1R01GM092772-01) and the USDA (2010-65108-20527). J.M.E. is supported by an NSF IGERT graduate research training grant (DGE-0653984).
Acknowledgments
We apologize to our colleagues whose work we were unable to cite due to page limitations. We thank Jun Liu, Z. Daniel Lin, and Manjula Govindarajulu for their comments and suggestions on the manuscript. No conflict of interest declared.
References
- Abbas HK, Tanaka T, Duke SO, Porter JK, Wray EM, Hodges L, Sessions AE, Wang E, Merrill AH, Jr., Riley RT. Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 1994;106:1085–1093. doi: 10.1104/pp.106.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amborabé B-E, Bonmort J, Fleurat-Lessard P, Roblin G. Early events induced by chitosan on plant cells. J. Exp. Bot. 2008;59:2317–2324. doi: 10.1093/jxb/ern096. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Ayres PG. Abnormal behaviour of stomata in barley leaves infected with Rhynchosporium secalis (Oudem.) J.J.Davis. J Exp. Bot. 1972;23:683–691. [Google Scholar]
- Ballio A. Non-host-selective fungal phytotoxins: biochemical aspects of their mode of action. Experientia. 1991;47:783–790. [Google Scholar]
- Ballio A, D'Alessio V, Randazzo G, Bottalico A, Graniti A, Sparapano L, Bosnar B, Casinovi CG, Gribanovski-Sassu O. Occurrence of fusicoccin in plant tissues infected by Fusicoccum amygdalei Del. Physiol. Plant Pathol. 1976;8:163–169. [Google Scholar]
- Batoko H, de Kerchove d'Exaerde A, Kinet J-M, Bouharmont J, Gage RA, Maraite H, Boutry M. (1998) Modulation of plant plasma membrane H+-ATPase by phytotoxic lipodepsipeptides produced by the plant pathogen Pseudomonas fuscovaginae. Biochem. Biophys. Acta. 1372:216–226. doi: 10.1016/s0005-2736(98)00060-1. [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Fuglsang AT, Jahn T, Korthout HA, de Boer AH, Palmgren MG. The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 1998;13:661–671. doi: 10.1046/j.1365-313x.1998.00083.x. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O'Flaherty M, Heck AJ, Slijper M, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Lam BC-H. Early signal transduction pathways in plant–pathogen interactions. Trends Plant Sci. 1998;3:342–346. [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bouche-Pillon S, Fleurat-Lessard P, Fromont JC, Serrano R, Bonnemain JL. Immunolocalization of the plasma membrane H+-ATPase in minor veins of Vicia faba in relation to phloem loading. Plant Physiol. 1994;105:691–697. doi: 10.1104/pp.105.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd LA, Smith PH, Foster EM, Brown JK. The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J. 1995;7:959–968. [Google Scholar]
- Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–1664. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host–microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol. 2009;11:1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- De Nisi P, Dell'Orto M, Pirovano L, Zocchi G. Calcium-dependent phosphorylation regulates the plasma-membrane H+-ATPase activity of maize (Zea mays L.) roots. Planta. 1999;209:187–194. doi: 10.1007/s004250050621. [DOI] [PubMed] [Google Scholar]
- Desbrosses G, Stelling J, Renaudin JP. Dephosphorylation activates the purified plant plasma membrane H+-ATPase. Eur. J. Biochem. 1998;251:496–503. doi: 10.1046/j.1432-1327.1998.2510496.x. [DOI] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR. Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell. 1995;7:2053–2067. doi: 10.1105/tpc.7.12.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich P, Sanders D, Hedrich R. The role of ion channels in light-dependent stomatal opening. J. Exp. Bot. 2001;52:1959–1967. doi: 10.1093/jexbot/52.363.1959. [DOI] [PubMed] [Google Scholar]
- Duby G, Boutry M. The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Arch. 2009;457:645–655. doi: 10.1007/s00424-008-0457-x. [DOI] [PubMed] [Google Scholar]
- Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J. Biol. Chem. 2009;284:4213–4221. doi: 10.1074/jbc.M807311200. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 1993;4:307–316. [Google Scholar]
- Felle HH, Herrmann A, Hanstein S, Huckelhoven R, Kogel KH. Apoplastic pH signaling in barley leaves attacked by the powdery mildew fungus Blumeria graminis f. sp. hordei. Mol. Plant Microbe Interact. 2004;17:118–123. doi: 10.1094/MPMI.2004.17.1.118. [DOI] [PubMed] [Google Scholar]
- Ferrante MC, Meli R, Mattace Raso G, Esposito E, Severino L, Di Carlo G, Lucisano A. Effect of fumonisin B1 on structure and function of macrophage plasma membrane. Toxicol. Lett. 2002;129:181–187. doi: 10.1016/s0378-4274(01)00476-3. [DOI] [PubMed] [Google Scholar]
- Fiegen M, Knogge W. Amino acid alterations in isoforms of the effector protein NIP1 from Rhynchosporium secalis have similar effects on its avirulence- and virulence-associated activities on barley. Physiol. Mol. Plant Pathol. 2002;61:299–302. [Google Scholar]
- Fuglsang AT, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007;19:1617–1634. doi: 10.1105/tpc.105.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K. Plant proton pumps. FEBS Lett. 2007;581:2204–2214. doi: 10.1016/j.febslet.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Goudet C, Milat M-L, Sentenac H, Thibaud J-B. Beticolins, nonpeptidic, polycyclic molecules produced by the phytopathogenic fungus Cercospora beticola, as a new family of ion channel-forming toxins. Mol. Plant Microbe Interact. 2000;13:203–209. doi: 10.1094/MPMI.2000.13.2.203. [DOI] [PubMed] [Google Scholar]
- Grandmougin-Ferjani A, Schuler-Muller I, Hartmann MA. Sterol modulation of the plasma membrane H+-ATPase activity from corn roots reconstituted into soybean lipids. Plant Physiol. 1997;113:163–174. doi: 10.1104/pp.113.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Torres PS, Vojnov AA. Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 2009;149:1017–1027. doi: 10.1104/pp.108.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Nájera N, Muñoz-Clares RA, Palacios-Bahena S, Ramírez J, Sánchez-Nieto S, Plasencia J, Gavilanes-Ruíz M. Fumonisin B1, a sphingoid toxin, is a potent inhibitor of the plasma membrane H+-ATPase. Planta. 2005;221:589–596. doi: 10.1007/s00425-004-1469-1. [DOI] [PubMed] [Google Scholar]
- Hagendoorn MJM, Poortinga AM, Sang HWWF, van der Plas LHW, van Walraven HS. Effect of elicitors on the plasma membrane of Petunia hybrida cell suspensions: role of deltapH in signal transduction. Plant Physiol. 1991;96:1261–1267. doi: 10.1104/pp.96.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A, Debus G, Edel HG, Stransky H, Serrano R. Auxin induces exocytosis and the rapid synthesis of a high-turnover pool of plasma-membrane H+-ATPase. Planta. 1991;185:527–537. doi: 10.1007/BF00202963. [DOI] [PubMed] [Google Scholar]
- Hahn M, Mendgen K. Signal and nutrient exchange at biotrophic plant–fungus interfaces. Curr. Opin. Plant Biol. 2001;4:322–327. doi: 10.1016/s1369-5266(00)00180-1. [DOI] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010;285:17918–17929. doi: 10.1074/jbc.M110.101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Bruntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9:1805–1814. doi: 10.1105/tpc.9.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E, Roelfsema MR, Anschutz U, Krol E, Elzenga JT, Felix G, Boller T, Hedrich R, Becker D. Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca-associated opening of plasma membrane anion channels. Plant J. 2010;62:367–378. doi: 10.1111/j.1365-313X.2010.04155.x. [DOI] [PubMed] [Google Scholar]
- Kanczewska J, Marco S, Vandermeeren C, Maudoux O, Rigaud J-L, Boutry M. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl Acad. Sci. U S A. 2005;102:11675–11680. doi: 10.1073/pnas.0504498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Asano H, Grossniklaus U, Schulze W, Robatzek S, Panstruga R. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 2010;285:39140–39149. doi: 10.1074/jbc.M110.160531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall A.J., Belkhadir Y., DebRoy S., Dangl J.L., Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchitsu K, Yazaki Y, Sakano K, Shibuya N. Transient cytoplasmic pH change and ion fluxes through the plasma membrane in suspension-cultured rice cells triggered by N-acetylchitooligosaccharide elicitor. Plant Cell Physiol. 1997;38:1012–1018. [Google Scholar]
- Laloi M, et al. Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 2007;143:461–472. doi: 10.1104/pp.106.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino B, Baizabal-Aguirre VM, González de la Vara LE. The plasma-membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta. 1998;204:352–359. doi: 10.1007/s004250050266. [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren M, Staskawicz B, Coaker G. RIN4 functions with the plasma membrane H+-ATPase to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang JM, Sweat TA, Wolpert TJ. Plant disease susceptibility conferred by a ‘resistance’ gene. Proc. Natl Acad. Sci. U S A. 2007;104:14861–14866. doi: 10.1073/pnas.0702572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Marre E. Fusicoccin: a tool in plant physiology. Annu. Rev. Plant Physiol. 1979;30:273–288. [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Merlot S, et al. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007;26:3216–3226. doi: 10.1038/sj.emboj.7601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius N, Hertweck C. Fungal phytotoxins as mediators of virulence. Curr. Opin. Plant Biol. 2009;12:390–398. doi: 10.1016/j.pbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Morel J, Claverol S, Mongrand S, Furt F, Fromentin J, Bessoule J-J, Blein J-P, Simon-Plas F. Proteomics of plant detergent-resistant membranes. Mol. Cell Proteomics. 2006;5:1396–1411. doi: 10.1074/mcp.M600044-MCP200. [DOI] [PubMed] [Google Scholar]
- Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell Proteomics. 2007;6:1711–1726. doi: 10.1074/mcp.M700164-MCP200. [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–940. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger T, Nennstiel D, Jabs T, Sacks WR, Hahlbrock K, Scheel D. High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell. 1994;78:449–460. doi: 10.1016/0092-8674(94)90423-5. [DOI] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 1998;118:551–555. doi: 10.1104/pp.118.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell. 2007;25:427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Rohe M, Gierlich A, Hermann H, Hahn M, Schmidt B, Rosahl S, Knogge W. The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 1995;14:4168–4177. doi: 10.1002/j.1460-2075.1995.tb00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell. 1999;11:263–272. doi: 10.1105/tpc.11.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg B, Ramel C, Dudler R. Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol. Plant Microbe Interact. 2010;23:1287–1293. doi: 10.1094/MPMI-04-10-0094. [DOI] [PubMed] [Google Scholar]
- Serra MD, Fagiuoli G, Nordera P, Bernhart I, Volpe CD, Di Giorgio D, Ballio A, Menestrina G. The interaction of lipodepsipeptide toxins from Pseudomonas syringae pv. syringae with biological and model membranes: comparison of syringotoxin, syringomycin, and two syringopeptins. Mol. Plant Microbe Interact. 1999;12:391–400. doi: 10.1094/MPMI.1999.12.5.391. [DOI] [PubMed] [Google Scholar]
- Shahollari B, Peskan-Berghöfer T, Oelmüller R. Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiol. Plant. 2004;122:397–403. [Google Scholar]
- Simon-Plas F, Gomes E, Milat ML, Pugin A, Blein JP. Cercospora beticola toxins (X. inhibition of plasma membrane H+-ATPase by beticolin-1) Plant Physiol. 1996;111:773–779. doi: 10.1104/pp.111.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard TE, Schulz A, Palmgren MG. Energization of transport processes in plants. roles of the plasma membrane H+-ATPase. Plant Physiol. 2004;136:2475–2482. doi: 10.1104/pp.104.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C, Jaspert N, Marcon C, Oecking C. Regulation of the plant plasma membrane H+-ATPase by its C-terminal domain: what do we know for sure? Eur. J. Cell Biol. 2010;89:145–151. doi: 10.1016/j.ejcb.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I, de Wit PJGM. Fungal effector proteins. Annu. Rev. Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell. 1999;11:2379–2391. doi: 10.1105/tpc.11.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain J, Gubb I, Wildon DC. Depolarization of tomato leaf cells by oligogalacturonide elicitors. Plant Cell Environ. 1995;18:211–214. [Google Scholar]
- Underwood W, Melotto M, He SY. Role of plant stomata in bacterial invasion. Cell Microbiol. 2007;9:1621–1629. doi: 10.1111/j.1462-5822.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- van't Slot KAE, Gierlich A, Knogge W. A single binding site mediates resistance- and disease-associated activities of the effector protein NIP1 from the barley pathogen Rhynchosporium secalis. Plant Physiol. 2007;144:1654–1666. doi: 10.1104/pp.106.094912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Higgins VJ, Blumwald E. Plant defense response to fungal pathogens (activation of host-plasma membrane H+-ATPase by elicitor-induced enzyme dephosphorylation) Plant Physiol. 1994;104:209–215. doi: 10.1104/pp.104.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Maser P, Schroeder JI. Plant ion channels: gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 2009;71:59–82. doi: 10.1146/annurev.physiol.010908.163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevelsiep L, Rupping E, Knogge W. Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol. 1993;101:297–301. doi: 10.1104/pp.101.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert TJ, Dunkle LD, Ciuffetti LM. Host-selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 2002;40:251–285. doi: 10.1146/annurev.phyto.40.011402.114210. [DOI] [PubMed] [Google Scholar]
- Wurtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003;22:987–994. doi: 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing T, Higgins VJ, Blumwald E. Regulation of plant defense response to fungal pathogens: two types of protein kinases in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell. 1996;8:555–564. doi: 10.1105/tpc.8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153:1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 2008;56:984–996. doi: 10.1111/j.1365-313X.2008.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Dielen V, Kinet J-M, Boutry M. Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell. 2000;12:535–546. doi: 10.1105/tpc.12.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Andersen CH, Burhenne K, Fischer PH, Collinge DB, Thordal-Christensen H. Proton extrusion is an essential signalling component in the HR of epidermal single cells in the barley–powdery mildew interaction. Plant J. 2000;23:245–254. doi: 10.1046/j.1365-313x.2000.00777.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann MR, Maischak H, Mithofer A, Boland W, Felle HH. System potentials, a novel electrical long-distance apoplastic signal in plants, induced by wounding. Plant Physiol. 2009;149:1593–1600. doi: 10.1104/pp.108.133884. [DOI] [PMC free article] [PubMed] [Google Scholar]