Abstract
The extracellular matrix (ECM) is a complex, multiprotein network that has essential roles in tissue integrity and intercellular signaling in the metazoa. Thrombospondins (TSPs) are extracellular, calcium-binding glycoproteins that have biologically important roles in mammals in angiogenesis, vascular biology, connective tissues, immune response, and synaptogenesis. The evolution of these complex functional properties is poorly understood. We report here on the evolution of TSPs and their ligand-binding capacities, from comparative genomics of species representing the major phyla of metazoa and experimental analyses of the oligomerization properties of noncanonical TSPs of basal deuterostomes. Monomeric, dimeric, trimeric, and pentameric TSPs have arisen through separate evolutionary events involving gain, loss, or modification of a coiled-coil domain or distinct domains at the amino-terminus. The relative transience of monomeric forms under evolution implicates a biological importance for multivalency of the C-terminal region of TSPs. Most protostomes have a single TSP gene encoding a pentameric TSP. The pentameric form is also present in deuterostomes, and gene duplications at the origin of deuterostomes and gene loss and further gene duplication events in the vertebrate lineage gave rise to distinct forms and novel domain architectures. Parallel analysis of the major ligands of mammalian TSPs revealed that many binding activities are neofunctions representing either coevolutionary innovations in the deuterostome lineage or neofunctions of ancient molecules such as CD36. Contrasting widely conserved capacities include binding to heparan glycosaminoglycans, fibrillar collagen, or RGD-dependent integrins. These findings identify TSPs as fundamental components of the extracellular interaction systems of metazoa and thus impact understanding of the evolution of ECM networks. The widely conserved activities of TSPs in binding to ECM components or PS2 clade integrins will be relevant to use of TSPs in synthetic extracellular matrices or tissue engineering. In contrast, the neofunctions of vertebrate TSPs likely include interactions suitable for therapeutic targeting without general disruption of ECM.
Keywords: thrombospondins, integrins, extracellular matrix, coiled coil, collagen
Introduction
Mechanisms for stable interactions between cells have been of central importance in the evolution of multicellular organisms (Rokas 2008). In metazoa, these mechanisms include the secretion of a specialized extracellular matrix (ECM) containing collagens, proteoglycans, and multidomain glycoproteins that assemble into multimolecular networks and fibrils (Vakonakis and Campbell 2007). The ECM of vertebrates has increased structural and molecular complexity relative to the ECM of basal metazoa or protostomes due to expansion of gene families, increased numbers of alternatively spliced variants, and the evolution of glycoprotein innovations such as fibrinogen, fibronectin, and tenascin within the chordate lineage (Schéele et al. 2007; Doolittle 2009; Tucker and Chiquet Ehrismann 2009). Thus, understanding the evolution of ECM components that are conserved between invertebrates and vertebrates offers great scope for distinguishing core and ancillary mechanisms by which ECM is built as a multimolecular system. This knowledge is important for understanding the fundamental roles of ECM in tissue development and homeostasis, stem cell niches, wound repair, and regeneration and has potential applications in the design of synthetic ECM environments for tissue engineering.
Among the many ECM components of mammals, thrombospondins (TSPs), a five-gene family of calcium-binding glycoproteins, are of considerable interest because of their roles in the regulation of angiogenesis, vascular biology, connective tissue organization, synaptogenesis, and immune response (reviewed by Adams 2001; Bornstein et al. 2004; Zhang and Lawler 2007). Mammalian TSPs, designated TSP-1 through TSP-5 (TSP-5 is also known as cartilage oliogomeric matrix protein (COMP), for cartilage oligomeric matrix protein), are considered to act as accessories to the structural ECM because of their nonfibrillar nature, restricted and/or tissue-specific expression patterns, and because mouse gene knockouts have demonstrated that family members are not essential for viability (Lawler et al. 1998, Kyriakides et al. 1998, Adams 2001; Bornstein et al. 2004; Posey et al. 2008). Many activities of the trimeric TSP-1 and TSP-2 (subgroup A) are mediated by their thrombospondin type 1 repeat domains (TSRs) (Zhang and Lawler 2007). These domains are not present in the pentameric TSP-3, TSP-4, and TSP-5/COMP (subgroup B) and do not mediate incorporation of TSP-1 into ECM (Adams et al. 2008). However, disorganization of collagen fibrils has been reported in both Thbs2 and Thbs5/COMP knockout mice, implicating a broad role of TSPs in ECM assembly processes (Kyriakides et al. 1998, Posey et al. 2008). This is likely mediated by the highly conserved C-terminal region that is common to both trimeric and pentameric TSPs and that comprises tandem epidermal growth factor (EGF) domains, a series of calcium-binding TSP type 3 repeats and a C-terminal L-type lectin-like domain (Adams 2004). These domains undergo multiple intramolecular interactions to form a single structural unit with a calcium-dependent tertiary structure (Kvansakul et al. 2004; Carlson et al. 2005; Tan et al. 2009). Engineered C-terminal trimers of TSP-1 were identified to have ECM incorporation activity in cell culture (Adams et al. 2008). The characterization of a single TSP in Drosophila melanogaster, D-TSP, provided the first evidence for an extended evolutionary history of TSPs (Adams et al. 2003). D-TSP is pentameric and is essential for embryo viability and for maintenance of muscle–tendon attachments throughout embryogenesis, where it contributes to local ECM integrity as a PS2 integrin ligand (Adams et al. 2003; Chanana et al. 2007; Subramanian et al. 2007). These data further implicate TSPs in ECM assembly processes.
To better understand the complex activities of TSPs, we surveyed their evolution by analysis of sequenced genomes and expressed sequence tag (EST) databases of organisms representative of the major metazoan phyla. In conjunction with a complementary analysis of the evolution of the most well-characterized ligands of mammalian TSPs, we identify neofunctions resulting from gene family expansion and adaptive radiation of TSPs in deuterostomes, put forward a model for the major transitions in the evolution of the TSP family, and propose likely ancestral roles.
Materials and Methods
Phylogenetic Analysis
TSP-coding sequences were identified by BlastP and TBlastN searches of GenBank, dbest, and genome-specific databases. The following genomes were analyzed at DOE JGI: Branchiostoma floridae v1.0 (Putnam et al. 2008), Capitella sp. I v1, Ciona intestinalis v2 (Dehal et al. 2002), Daphnia pulex v1 (Bauer 2007), Dictyostelium purpureum QSDP1; Helobdella robusta v1, Lottia gigantea v1, Monosiga brevicollis v1 (King et al. 2008), Nematostella vectensis v1 (Putnam et al. 2007), and Trichoplax adhaerens v1 (Srivastava et al. 2008). Other genomes analyzed included Schmidtea mediterranea (Robb et al. 2008) and Ciona savignyi assembly release 1 (Small et al. 2007) at the Broad Institute, Callorhinchus milii (elephant shark) v1 genome assembly at http://esharkgenome.imcb.a-star.edu.sg/ (Venkatesh et al. 2007), and D. discoidium (Eichinger et al. 2005) and Petromyzon marinus preliminary assembly at ENSembl/Sanger Centre; at National Center for Biotechnology Information, the database of ESTs (dbest) and the genomes of Apis mellifera (Honeybee Genome Sequencing Consortium 2006), Caenorhabditis elegans (C. elegans Sequencing Consortium 1998), C. briggsae (Stein et al. 2003), D. melanogaster release 5.2 (Adams et al. 2000), Hydra magnipapillata, Strongylocentrotus purpuratus build 2.1 (Sea Urchin Genome Sequencing Consortium 2006), and Tribolium castena (Tribolium Genome Sequencing Consortium 2008). Preliminary sequence data from Saccoglossus kowalevskii draft assembly (Skow_1.0) were obtained from Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) Web site (http://www.hgsc.bcm.tmc.edu/project-species-o-Acorn%20worm.hgsc?pageLocation=Acorn%20worm). Crustacean TSP sequences from cDNA included Fenneropenaeus chinesensis (Sun et al. 2006), Marsupenaeous japonica (Yamano et al. 2004), and Penaeus monodon (Adams 2004). Vertebrate TSP sequences were as in McKenzie et al. (2006).
BlastP and TblastN searches for the following ligands of TSP-1, TSP-4, or TSP-5 were conducted against GenBank and the genomes listed above using the following protein sequences from Homo sapiens as queries: calreticulin, CD36, CD47, collagens I and IX, fibrinogen, integrin subunits α4, αv, β1, and β3, matrilin-2 and matrilin-3, and transforming growth factor β1 (TGFβ1).
Multiple sequence alignments were prepared in TCOFFEE (Notredame et al. 2000). For the phylogenetic analysis, alignments were prepared in MUSCLE (Edgar, 2004) using 450 residues including the type 3 repeats and L-lectin–like domain of 33 TSP sequences, representing TSPs from basal metazoa, protostomes, and the deuterostome gene family. Alignments were curated in Gblocks, with allowance of gaps in the final blocks. Phylogenetic trees were constructed by maximum likelihood method, PhyML (Guindon and Gascuel 2003), with 100 cycles of bootstrapping. The phylogenetic diagram is presented in TreeDyn using resources of Phylogeny.fr (http://phylogeny.lirmm.fr) (Dereeper et al. 2008).
Analysis of TSP Oligomerization Capacity
Ciona intestinalis cDNA clones ciad010b04, citb019a10, citb55k19, and cibd032g21 were obtained from Dr Noriyuki Satoh, Kyoto University, and the Academica DNA Sequencing Center, Center for Genetic Resource Information, Japan (Satou et al. 2002), and were sequenced in their entirety on both strands by automated DNA sequencing, carried out by CCF Molecular Biotechnology Core, using vector and cDNA-specific primers. Nucleotide sequences were deposited in GenBank (accession numbers: TSP-A, AY465896; TSP-Bi, DQ369966; TSP-Bii, DQ369968; TSP-DD, GU230168, an explanation for these names is given in the Results). The epitope-tagged construct for expression of C. intestinalis TSP-A was as described (Adams et al. 2003). Epitope-tagged constructs of cDNAs for expression of TSP-A or TSP-DD domains were prepared by polymerase chain reaction (PCR) amplification with the primer pairs listed in supplementary table S1 (Supplementary Material online) and ligation of PCR products into pcDNA3.1V5His expression plasmid by TOPO cloning (Invitrogen) or conventional ligation into pCEP expression plasmid (Kvansakul et al. 2004). Clones with cDNAs in the sense orientation were identified by restriction enzyme digestion and confirmed by DNA sequencing.
Transient expression in COS-7 cells was carried out by plasmid transfection as described (Adams et al. 2003). Conditioned media were collected 30–48 h later, clarified by centrifugation, and protease inhibitor cocktail (Roche) was added; 1.5 ml aliquots were mixed by rotation at 4 °C with 30 μl of 50:50 (v/v) NiNTA-agarose beads (Qiagen) or Talon metal affinity resin (Clontech), with identical results. Beads were washed four times in 50 mM Tris-HCl, pH 7.8, containing 300 mM NaCl and resuspended in SDS-PAGE sample buffer without or with 100 mM Dithiothreitol (DTT). Samples were resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels, transferred to Polyvinylidene fluoride membrane, and probed with mouse monoclonal antibody to V5 epitope-tag (Invitrogen) or polyhistidine (GenScript Corp.). Enhanced chemiluminescence detection (Applied Biosystems) was as described (Adams et al. 2003).
For bacterial expression of single domains of C. intestinalis TSP-A or TSP-DD, PCR products were amplified from cDNAs encoding the relevant domains with the indicated primers (supplementary table S1, Supplementary Material online) and ligated into pET19b plasmid (Novagen) in frame with a 10-histidine tag. Proteins were expressed in Escherichia coli BL21 Codon Plus (Stratagene) after induction with 0.1 mM Isopropyl β-D-1-thiogalactopyranoside for 45 min at 28 °C. Bacterial lysates were prepared for protein purification with Talon metal affinity resin as per manufacturer's instructions for native proteins, with Triton X-100 added to a final concentration of 1% after sonication. Lysates were rotated with resin for 1 h at 4 °C, washed extensively in Tris-buffered saline, and bound proteins released by boiling in SDS-PAGE sample buffer without or with 100 mM DTT, to test for disulfide bond formation. Proteins were resolved on 15% polyacrylamide gels and immunoblots were probed with antibody to polyhistidine as described above.
Results
Evolutionary Origin of TSPs in Basal Metazoa
Sequence regions including the type 3 repeats and L-lectin–like domain of human TSP-1, TSP-5/COMP, and Drosophila TSP were selected as divergent representatives of trimeric and pentameric TSPs (having approximately 54% sequence identity) for TblastN queries of genome and EST databases of species from the major phyla of metazoa. Initial hits were extended by reference to the genome sequence, ESTs, and additional searches with domains from the N-terminal regions of mammalian or Drosophila TSPs.
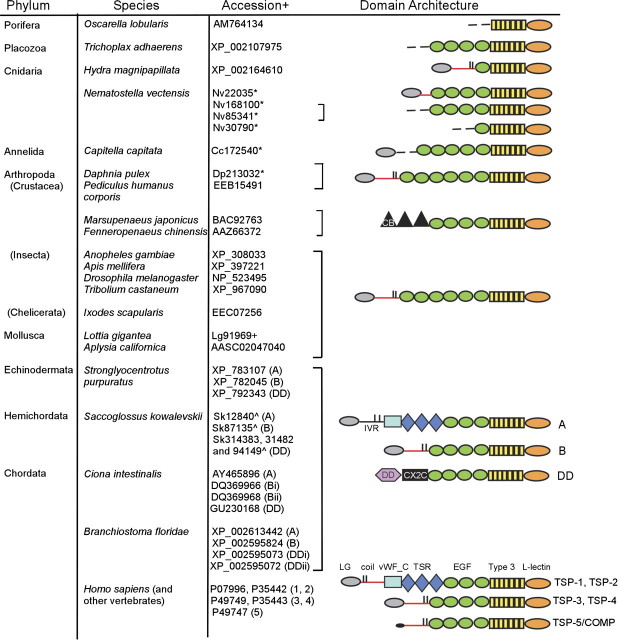
Protein-coding sequences with the domain architecture of the TSP C-terminal sequence region were identified in almost all metazoan species examined (fig. 1). No equivalent sequences were identified in the genome sequences or ESTs of the choanoflagellate M. brevicollis, the amoebozoa D. discoidium or D. purpureum, or several Apicomplexa genomes, all examined as outgroups to the metazoa. We infer that the TSP C-terminal region domain architecture originated in the last common ancestor of the metazoa. Of the different domains included in the TSP C-terminal region, L-type lectin domains are present in plants, animals, and other eukaryotes (Dodd and Drickarmer 2001), the TSP type 3 repeats are present only in TSPs within the metazoa but have also been identified in a few bacterial proteins (e.g., Rigden and Galperin 2004; Akimana and Lafontaine 2007), and EGF domains are present in viruses and bacteria as well as in eukaryotes (SMART database entry SM00181). In T. adhearens and the majority of protostome genomes examined, one TSP-encoding sequence per genome was identified. In the Cnidaria, N. vectensis encoded multiple TSPs (fig. 1). No TSP-encoding sequences were identified in nematode (C. elegans and C. briggsae) or planarian (S. mediterranea) worms.
FIG. 1.
Evolution of thrombospondins in the metazoa. Accession numbers are from GenBank (+), JGI (*) or HGSC (^). In the domain models, red lines indicate a coiled coil; short vertical lines indicate positions of paired cysteine residues; dotted lines incomplete sequence regions. coil, coiled coil; CX2X, CX2C domain; EGF, EGF domain; LG, laminin G–like domain; L-lectin, L-type lectin-like domain; vWF_C, von Willebrand factor type C domain.
Many protostome TSPs have different numbers of EGF-like domains to vertebrate TSPs, but in other regards, the domain organization that is characteristic of mammalian TSP-3 and TSP-4 has been remarkably conserved between Cnidaria, annelids, molluscs, arthropods, and deuterostomes (fig. 1). Furthermore, the coiled-coil domains of representative Cnidarian, arthropod, and mollusc TSPs have sequence characteristics equivalent to the coiled-coil domains of Drosophila TSP, TSP-3, TSP-4, and TSP-5/COMP, which have all been established experimentally to assemble as pentamers. In particular, these pentamerizing coiled coils of TSPs are defined by two closely spaced cysteine residues positioned at the C-terminal end of the heptad repeats (marked by asterisk in supplementary fig. S1, Supplementary Material online) (Efimov et al. 1994; Adams et al. 2003). This clearly distinguishes the pentamerizing coiled coils of TSPs from the coiled coils of trimerizing TSPs that are defined by cysteine residues adjacent to the N-terminal end of the heptad repeats (Adams et al. 2003; McKenzie et al. 2006, and see examples in supplementary fig. S4, Supplementary Material online). We infer that pentameric assembly of TSPs evolved early in the metazoa. However, Cnidarian N. vectensis TSP22035 contains only a short coiled coil that lacks the characteristic C-terminal cysteine residues (supplementary fig. S1, Supplementary Material online).
An interesting exception to the dominance of pentamerizing TSPs in protostomes is provided by TSPs identified from cDNA in prawn and shrimp species, Marsupenaeus japonicus and Fenneropenaeus chinensis. These lack a coiled coil and contain in their amino-terminal regions three tandem type 2 chitin-binding domains (type 2 CB domain; entry SM00494 in the SMART database) (supplementary fig. S1, Supplementary Material online). This domain organization (designated here TSP-CB, for chitin binding) appears specific to this crustacean family because TSPs from other crustacea (D. pulex, P. humanus corporis) have the same domain organization as other protostome TSPs (fig. 1).
Expansion of the TSP Gene Family in Basal Deuterostomes Preceded 2R and Includes Noncanonical TSPs
Multiple TSPs were identified in all the deuterostome genomes examined and verified from ESTs. We previously identified two TSPs in the urochordate C. intestinalis corresponding to subgroups A and B domain architectures (Adams et al. 2003). The availability of complete genome sequences for both C. intestinalis and its close relative, C. savigyni, and the very large database of ESTs for C. intestinalis enabled identification of two other TSPs. The TSPs of C. intestinalis thus include one with the major domain architecture of a subgroup A TSP, designated TSP-A (Adams et al. 2003); two with the domain architectures of subgroup B TSPs, corresponding to our original TSP-B (Adams et al. 2003), here renamed TSP-Bi; the gene product of Gene Cluster 31959, here designated TSP-Bii; and the gene product of gene cluster 35494, here designated TSP-DD, that has a novel domain architecture including an amino-terminal F5/F8 or discoidin domain (DD) and an unusual cysteine-rich domain. No coiled-coil domain is present in TSP-DD (fig. 1). The four predicted protein sequences were confirmed by DNA sequencing of corresponding cDNAs. Each of the encoding genes has a distinct location in the C. intestinalis genome, and four orthologous TSPs are encoded in C. savigyni (table 1).
Table 1.
TSPs of Ciona Species and Their Genomic Locations.
|
Ciona intestinalisa |
Ciona savignyib | ||
| TSP | Genomic Scaffold Number | Chromosome | Paired Genomic Scaffold Number |
| A | 182 | 4 | 431 and 437 |
| Bi | 626 and 998 | Not mapped | 66 |
| Bii | 186 | 11 | 4 |
| DD | 22 and 2901 | 3/Not mapped | 360 |
Data on C. intestinalis from the Ghost database (http://ghost.zool.kyoto-u.ac.jp/).
Data on C. savignyi from the Broad Institute (http://www.broadinstitute.org/annotation/ciona/).
TSPs with equivalent domain architectures including one or more TSP-DD paralogues are encoded in the genome of the basal deuterostomes S. purpuratus and S. kowalevskii and the cephalochordate B. floridae (fig. 1) (S. purpuratus TSP-A and TSP-B were also identified in Whittaker et al. 2006). Whereas the DD is widely present in eukaryotes and prokaryotes (Kiedzierska et al. 2007), the novel cysteine-rich domain could be identified only in TSP-DD of echinoderms and basal chordates. The newly identified, 180 aa domain contains 24 conserved cysteine residues including multiple CX2C motifs (supplementary fig. S2, Supplementary Material online); we therefore designated it the CX2C domain.
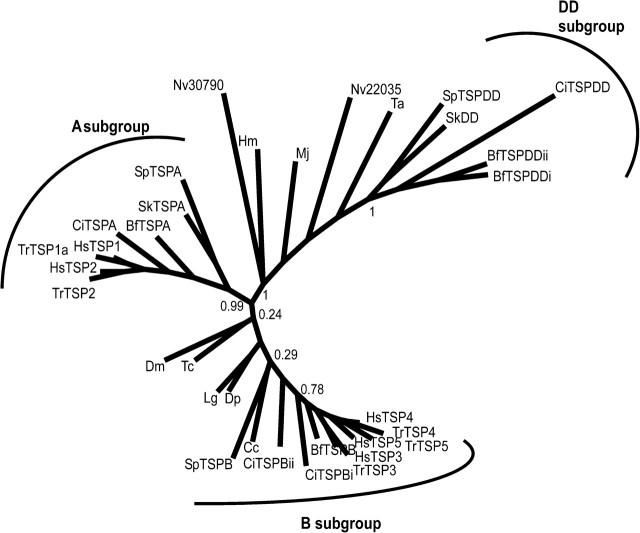
Although the general TSP-A domain architecture has been conserved in vertebrate TSP-1 and TSP-2, TSP-DD is specific to basal deuterostomes and basal chordates and was not identified in vertebrates. This gene loss likely occurred early in the vertebrate lineage because TSP-DD was not identified in the P. marinus (lamprey) preliminary genome assembly or ESTs, or the genome of a cartilaginous fish, C. milii. Five distinct TSP-encoding sequences, corresponding to TSP-1, TSP-2, TSP-3, TSP-4, and the fish equivalent of TSP-5/COMP, are present in the C. milii genome (table 2). A phylogenetic analysis based on C-terminal sequences of 33 TSPs from basal metazoa, protostomes, and deuterostomes clearly supported the three subgroups of deuterostome TSPs (fig. 2). The analysis also highlighted high sequence diversity within the basal and protostome TSPs, with some individual sequences appearing more closely related to the A or DD subgroup sequences (fig. 2) despite their overall subgroup B domain architecture (see fig. 1).
Table 2.
TSPs of the Elephant Shark, Callorhinchus milii.
| TSP | Genomic Scaffold Number |
| TSP-1 | AAVX01400762.1, AAVX01126766.1, AAVX01133838.1, AAVX01228990.1, AAVX01088989.1 |
| TSP-2 | AAVX01056006.1, AAVX01074659.1, AAVX01015865.1, AAVX01479228.1, AAVX01363948.1, AAVX01009577.1 |
| TSP-3 | AAVX01143361.1, AAVX01522440.1, AAVX01136414.1 |
| TSP-4 | AAVX01409157.1 |
| TSP-5 | AAVX01228699.1, AAVX01202029.1, AAVX01326016.1, AAVX01255302.1 |
NOTE.—Genomic scaffolds containing partial sequences for TSP-1 to TSP-5 were identified by TblastN searches of the 1.4× genome assembly v1 at http://esharkgenome.imcb.a-star.edu.sg. No TSP-DD sequence was identified.
FIG. 2.
Unrooted phylogenetic tree of the TSPs. Sequences of 450 residues including the type 3 repeats and L-lectin–like domain from 33 TSPs representing basal metazoa, protostomes, and the deuterostome TSP family were aligned in MUSCLE and analyzed in PHYML with 100 bootstrap cycles. Bootstrap values are given for major internal nodes. Branch length is proportional to the number of substitutions/site. The three subgroups of deuterostome TSPs are well supported, and the basal and protostome TSPs exhibit high sequence diversity with some individual sequences appearing most closely related to the A or DD subgroups. Bf, Branchiostoma floridae; Cc, Capitella capitella; Ci, Ciona intestinalis; Dp, Daphnia pulex; Dm, Drosophila melanogaster; Hm, Hydra magnipapillata; Hs, Homo sapiens; Lg, Lottia gigantea; Mj, Marsupenaeous japonicus; Nv, Nematostella vectensis; Sk, Saccoglossus kowalevskii; Sp, Strongylocentrotus purpuratus; Ta; Trichoplax adhaerens; Tc, Tribolium castena; Tr, Takifugu rubripes.
Analysis of Oligomerization Capacity of Ciona TSP-A and TSP-DD
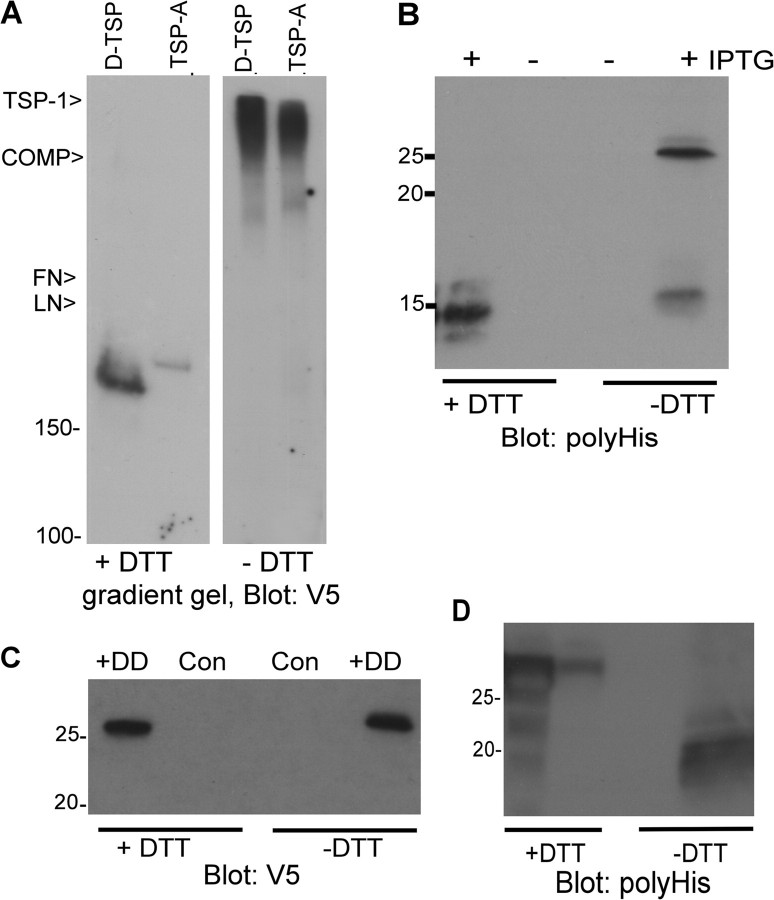
The TSP-A proteins of sea urchin and Ciona species contain the major domains characteristic of vertebrate TSP-1 and TSP-2, yet lack a coiled-coil domain, as does TSP-DD (fig. 1). Thus, the oligomerization capacity of these TSPs could not be ascertained by sequence analysis. This point is important for understanding both the evolution of TSPs within the deuterostomes and the evolution of TSP subgroup A. To determine if TSP-A or TSP-DD are secreted as monomers or oligomers, we analyzed their oligomerization capacity experimentally. Full-length C. intestinalis TSP-A secreted from COS-7 cells had an apparent molecular mass of 180 kDa under reducing conditions and showed a major increase in molecular mass under nonreducing conditions, as did the known oligomer, D-TSP (fig. 3A). Because this approach does not adequately resolve different states of oligomerization (TSP-A, TSP-1, D-TSP, and TSP-5/COMP all have similar migration characteristics under the nonreducing conditions; fig. 3A), we turned to experiments with single domains to identify the oligomerization domain. The N-terminal laminin G–like (LG) domain and the C-terminal region of TSPs do not have oligomerization activity, and indeed, secreted Ciona TSP-A LG domain (aa 1–250) had the same molecular mass under both reducing and nonreducing conditions (data not shown). Thus, the intervening region (IVR) (aa 251–315) was the prime candidate for an oligomerization domain and indeed a protein containing the LG domain plus IVR showed increased molecular mass under nonreducing conditions (data not shown). The IVR does not have coiled-coil character because it contains no heptad repeats (supplementary fig. S3, Supplementary Material online). The IVR sequence region is poorly conserved in S. purpuratus and S. kowalevskii compared with Ciona species. However, two closely spaced cysteine residues are conserved at the C-terminal end of all these IVR, suggesting that oligomerization might be mediated by disulfide bonds alone (supplementary fig. S3, Supplementary Material online). In contrast, B. floridae TSP-A contains a clear coiled-coil domain with paired cysteine residues adjacent to its N-terminal end that is closely homologous to the trimerizing coiled-coil domain of vertebrate TSP-1 and TSP-2 (supplementary fig. S4, Supplementary Material online). Recombinantly expressed C. intestinalis IVR had an apparent molecular mass of 14 kDa when reduced and 25 and 14 kDa under nonreducing conditions, indicative of dimerization capacity (fig. 3B, Supplementary Material online). Under the conditions of this assay, dimerization may not proceed to completion.
FIG. 3.
Oligomerization properties of Ciona TSP-A and TSP-DD. (A) Oligomerization of Ciona intestinalis TSP-A. TSP-A and Drosophila TSP were collected from conditioned media of transiently transfected COS-7 cells onto heparin–Sepharose, resolved on 4–10% polyacrylamide gradient gels under reducing or nonreducing conditions, and analyzed by immunoblotting. Migration positions of purified ECM glycoproteins used nonreduced as additional markers are shown to the left of the panel. (B) The IVR has independent dimerization activity. The 6His-tagged IVR of TSP-A was inducibly expressed in Escherichia coli, purified on metal affinity beads, resolved on 15% polyacrylamide gels under reducing or nonreducing conditions, and analyzed by immunoblotting. (C,D) The DD and CX2C domain of C. intestinalis TSP-DD lack oligomerization activity. Tagged versions of the discoidin domain (C) or CX2C domain (D) were collected from conditioned media of transiently transfected COS-7 cells (C) or bacterial lysates (D) onto metal affinity beads, resolved on 12.5% polyacrylamide gels under reducing or nonreducing conditions, and analyzed by immunoblotting.
In TSP-DD, the DD and CX2C domain were the candidates for possible oligomerization domains. The secreted DD (aa 1–146) was monomeric (fig. 3C) and a protein containing both the DD and CX2C domains (aa 1–336) could not be analyzed due to poor expression and secretion (data not shown). The CX2C domain (aa 191–336) purified from bacteria had an apparent molecular mass of 30 kDa under reducing conditions and migrated as a broad band of lower apparent molecular mass under nonreducing conditions (fig. 3D). Thus, the CX2C domain likely forms intradomain disulfide bonds. These properties identify TSP-DD as an unusual monomeric TSP. These data on Ciona TSP-A and TSP-DD can be applied to TSP-A and TSP-DD of other basal deuterostomes.
Evolution of Major TSP Ligands
Mammalian TSPs are multifunctional glycoproteins that bind to many cell-surface proteins, ECM components, and growth factors (Adams 2001; Bornstein et al. 2004; Zhang and Lawler 2007). These interactions have been most intensively studied for TSP-1, TSP-2, and TSP-5/COMP. Ligand-binding peptide motifs have been mapped, and crystal structures of the relevant TSP domains have identified the peptide motifs that are appropriately surface exposed for ligand binding (Tan et al. 2002, 2006, 2009; Kvansakul et al. 2004; Carlson et al. 2005). With this structural criterion as a basis, we made a phylogenomic search for the representation of the most validated ligand-binding motifs of TSPs and their respective ligands.
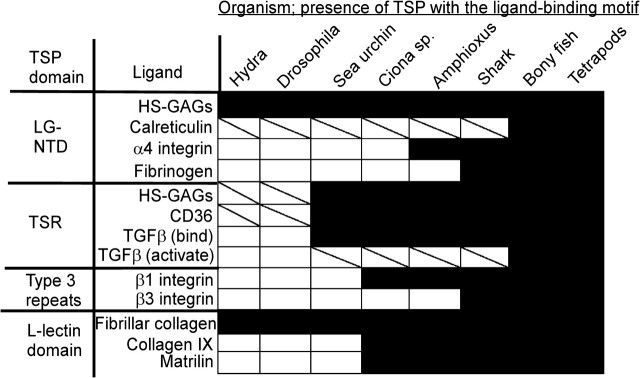
Figure 4 summarizes our findings. Within the N-terminal LG domain, a polybasic patch on one face binds heparin or heparan sulfate (Tan et al. 2006). This patch is conserved (Tan et al. 2006) and heparan glycosaminoglycans (GAGs) are also present throughout the metazoa (Cássaro and Dietrich 1977). This interaction is thus a strong candidate for an ancestral activity of TSPs. In contrast, although the ligand calreticulin is present in metazoa, yeast, and higher plants (Christensen et al. 2008; Madeo et al. 2009), the specific calreticulin-binding motif is only present in vertebrate TSP-1 (Goicoechea et al. 2000). This identifies calreticulin binding as a neofunction of TSP-1 (fig. 4). The TSP-1 LG domain also contains binding sites for α4β1 integrin and fibrinogen (Voland et al. 2000; Calzada et al. 2004). These ligands are chordate and vertebrate innovations, respectively (Huhtala et al. 2005; Doolittle 2009), and their binding motifs also represent neofunctions specific to TSP-1 (fig. 4).
FIG. 4.
Representation of TSP ligand-binding motifs and their cognate ligands across the metazoa. Results from Hydra and Drosophila are representative of other basal metazoa and protostomes. White boxes, ligand not encoded within the indicated species genome; diagonal line, ligand encoded; black box, the ligand and a TSP with the appropriate binding motif are both encoded.
The TSR domains are deuterostome innovations specific to TSP-A, TSP-1, and TSP-2, most likely as a result of exon shuffling events. Binding motifs for CD36 and GAGs are present in the TSR of TSP-A, TSP-1, and TSP-2 (Asch et al. 1992; Guo et al. 1992), and both these ligands are present throughout the metazoa (Cássaro and Dietrich 1977; Müller et al. 2004). Although TSP-A, TSP-1, and TSP-2 contain motifs for binding TGFβ1 in their TSR, TSP-1 alone contains the TGFβ1 activation motif, RFK (Schultz-Cherry et al. 1995; Ribeiro et al. 1999; Young and Murphy-Ullrich 2004). The TGFβ superfamily is conserved throughout the metazoa, yet TGFβ senso stricto is present only in deuterostomes (Lapraz et al. 2006). Thus, the binding interactions of the TSR represent both neofunctions of evolutionarily ancient molecules (e.g., CD36) and coevolutionary innovations (the TSR/TGFβ binding system), all of which evolved within the deuterostome lineage (fig. 4).
Within the C-terminal region, the RGD motif in the seventh type 3 repeat of TSP-1 binds αvβ3 and αIIbβ3 integrins and β1 integrins including α5β1 (Lawler and Hynes 1989, reviewed by Adams 2004). The activity of this RGD motif for cell attachment is regulated by the extent of calcium ion binding (Kvansakul et al. 2004) and an RGD motif at this precise position is conserved only in TSP-1, TSP-2, and TSP-A. The integrin β3 subunit is a vertebrate innovation (Huhtala et al. 2005), thus interactions with β3 integrins are specific neofunctions of the A subgroup trimers in vertebrates (fig. 4). However, β1 integrins are present in all chordates, integrins in general are conserved throughout the metazoa, and RGD or KGD motifs are present at various positions within the type 3 repeats of most TSPs throughout the metazoa (Adams 2004 and this analysis). Some of these motifs are of known functionality: the RGD motif of TSP-5 binds α5β1 integrin (Chen et al. 2005) and the KGD motif of Drosophila TSP is implicated in PS2 integrin-dependent attachment (Chanana et al. 2007; Subramanian et al. 2007). Vertebrate αv and α5 subunits both belong to the PS2 clade of integrins (Huhtala et al. 2005); thus, in view of the universality of integrins in metazoa, it is plausible that labile RGD-dependent integrin binding could be an ancestral function of TSPs.
The L-lectin–like domain of TSP-5/COMP interacts with fibrillar collagens, collagen IX, and matrilin-3, and these interactions are important for correct cartilage ECM assembly (Merritt et al. 2007). Collagen IX binding involves the motif GVDFEGTFHVNTVTDDD (Holden et al. 2001). The DDD motif is important for ECM incorporation of TSP-1 (Adams et al. 2008), and our results in this study identify that the DDD motif is conserved throughout all known TSPs except TSP-DD and T. adhaerens TSP. Fibrillar collagens are universal throughout the metazoa (Exposito et al. 2008) and the interaction with fibrillar collagen is a possible ancestral function of the L-lectin domain (fig. 4). In contrast, collagen IX is a member of the chordate-specific fibril-associated collagens, and matrilins are also only present in chordates (fig. 4). Thus, even for the most highly conserved domain of TSPs, neofunctions have been acquired in the chordate lineage.
We also examined the evolutionary record of CD47, a transmembrane immunoglobulin superfamily member reported to bind to TSP-1 L-lectin–like domain (Gao et al. 1996). The identified binding motifs, RFYVVMWK and IRVVM, are conserved but not surface exposed in the L-lectin domain structures of TSP-1, TSP-2, or TSP-5/COMP (Kvansakul et al. 2004; Carlson et al. 2005; Tan et al. 2009) and likely serve a structural role within the L-lectin domain fold. However, similar phenotypic traits in the vascular system have been reported in TSP-1–null and CD47-null mice (Isenberg et al. 2006). The searches revealed that CD47 is encoded only in the genomes of amniotes. No CD47-encoding sequences were identified in basal chordates, lamprey, shark, bony fish, or amphibia nor were any ESTs identified in these species. This corresponds with the evolutionary range of the CD47 counterreceptor, SIRPα, that functions in immune self-recognition in mammals (reviewed by Barclay and Brown 2006). In conclusion, CD47 cannot be an evolutionarily ancient ligand of TSPs: most likely, CD47 evolved primarily as a specialized component of the self-recognition and immune regulation systems of amniotes.
Discussion
Through comparative genomics, we have identified that the TSPs, a family of glycoproteins that have been studied principally as accessory components of mammalian ECM, in fact have an extended evolutionary history throughout the metazoa. These data, in combination with an analysis of the species representation of the most well-validated ligands of TSPs, demonstrate a broad relevance of TSPs in animal ECM biology. With the exceptions of nematode (C. elegans and C. briggsae) or planarian (S. mediterranea) worms, TSPs were identified in species from all major metazoan phyla. Nematode and planarian worms are both unusual in terms of ECM. Nematodes have a vastly expanded repertoire of collagen genes that are major components of the protective cuticle (Page and Johnstone 2007). Planarian worms have proteoglycan-rich pericellular ECM with little obvious structure (Hori 1991). Although encoding sequences for integrin β subunit, syndecan, fibrillar collagen, and fibrillin-like sequences can be recognized from the S. mediterranea genome or ESTs, encoding sequences for laminin subunits or nidogen are not identifiable (our observations from Blast searches of SmedDB). The absence of TSP from these worms likely reflects these specialized ECM characteristics.
Many protostome TSPs have different numbers of EGF-like domains to vertebrate TSPs, but overall the domain organization characteristic of mammalian TSP-3 and TSP-4 has been remarkably conserved between basal metazoa, protostomes, and deuterostomes. A notable exception is TSP-CB of prawns and shrimps, in which three tandem type 2 chitin-binding domains replace the LG domain and coiled coil. In other insect proteins that contain these domains, the peritrophins, the tandem type 2 chitin-binding domains act to cross-link chitin polymer fibrils (Hegedus et al. 2009). Thus, although its domain organization and absence of a coiled coil indicate TSP-CB to be a monomeric TSP, it is plausible that TSP-CB forms dense extracellular aggregates that allow for “polymeric” ligand binding by its C-terminal region. The prawn TSPs are highly expressed in the ovary as components of the cortical rods and in the lymphoid organ upon bacterial challenge and are postulated to serve specialized adhesive functions at these sites (Yamano et al. 2004; Pongsomboon et al. 2008). Knowledge of a prawn or shrimp genome sequence will be needed to distinguish whether TSP-CB represents a lineage-specific evolutionary variant or if more than one TSP gene per genome is encoded in this crustacean family.
In contrast to the encoding of a single TSP gene in most protostomes, all deuterostome genomes examined encode multiple TSP genes with diverse domain architectures. Thus, expansion of the TSP gene family to include the A and DD domain architectures must have been a property of the last common deuterostome ancestor. This defines that TSP gene family expansion must have occurred before the two rounds of genome-wide large-scale duplications (2R) early in vertebrate evolution (Dehal and Boore 2005). This contrasts with other ECM protein families such as tenascins or syndecans, in which gene family expansion is clearly specific to the vertebrate lineage (Chakravarti and Adams 2006; Tucker and Chiquet Ehrismann 2009).
In conjunction with the experimental analyses of the noncanonical TSP-A and TSP-DD that lack coiled coils, our data establish clearly that the trimer and pentamer subgroups of vertebrate TSPs represent the outcomes of very different evolutionary events. A coiled-coil domain was acquired not once but, at minimum, twice during the evolution of TSPs. As a conservative estimate, in relation to the Pre-Cambrian origin of Cnidaria in the fossil record (Chen et al. 2002), the pentamerizing coiled coil has been conserved over at least 580 My, since the divergence of Hydra and human. In contrast, the trimerizing coiled coil of TSP-1 and TSP-2 is a chordate innovation that evolved in place of the dimerizing IVR of TSP-A. Although recent genomic studies place urochordates, not cephalochordates, as the closest relatives of vertebrates (Delsuc et al. 2006), the presence of a coiled coil in the single TSP-A of B. floridae that is homologous to the trimeric coiled coil of TSP-1 and TSP-2 (supplementary fig. S4, Supplementary Material online) puts this TSP as the most closely related to vertebrate TSP-1 and TSP-2. Either this represents a closer evolutionary relationship of cephalochordates with vertebrates as formerly considered, or else a trimerizing coiled coil evolved independently in both the Amphioxus and chordate lineages. Several activities of TSP-1 and -2 depend on oligomerization (Anilkumar et al. 2002; Adams et al. 2008), and it can be speculated that the evolution of stable oligomerization by a coiled-coil domain in combination with the increased ligand-binding avidity provided by a trimer was favored strongly under natural selection.
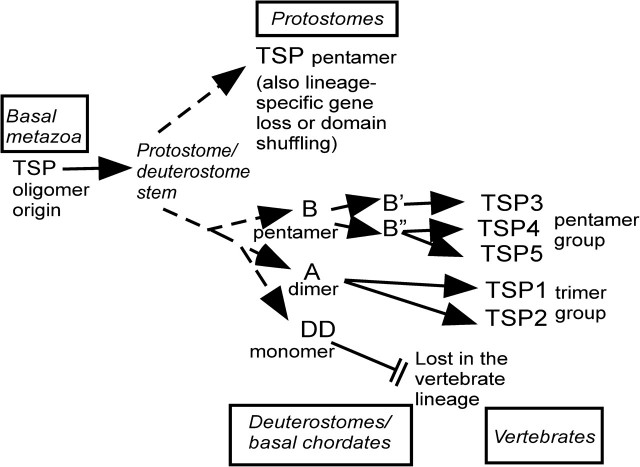
Together, the phylogenomic and experimental analyses identify three major transitions in the evolution of TSPs (fig. 5). First, the hallmark domain organization of TSPs originated in conjunction with the evolution of the metazoa and has been completely conserved subsequently. Changes in domain architecture have been restricted to domains at the N-terminal end (see fig. 1). Trichoplax adhaerens and most protostomes encode a single TSP; however, species-specific gene family expansion took place in Cnidaria and the TSP gene has been lost in planarian and nematode worms. Despite the conservation of overall domain architecture, basal metazoan and protostomal TSPs exhibit high sequence diversity relative to the deuterostome subgroups (fig. 2). Second, TSP gene duplication and domain shuffling events occurred in connection with the origin of deuterostomes and resulted in the evolution of TSPs with novel domain architectures and distinct oligomerization properties: TSP-A and TSP-DD. Third, TSP-DD was lost early in the vertebrate lineage and additional duplications of the TSP-A and TSP-B genes took place, leading to the existence of multiple trimeric and pentameric TSPs in modern vertebrates (fig. 5 and McKenzie et al. 2006). It is clear that a significant component of TSP evolution has been a considerable diversification of oligomerization properties. TSP pentamers have been particularly robust under evolution, yet monomer, dimmer, and trimer forms evolved through specific loss or gain of domains or intersubunit disulfide bonds or replacement of the IVR of TSP-A by a coiled coil. The relative transience of TSP-DD under evolution implies that the increased ligand-binding avidity provided by multivalency is biologically important for the binding interactions of the TSP C-terminal region. In other DD-containing extracellular proteins, such as clotting factors V and VIII, the DD mediates binding to negatively charged membrane phospholipids (Fuentes-Prior et al. 2002); it will be of interest in future to examine whether this is also a property of the discoidin domain TSP-DD.
FIG. 5.
Model of TSP evolution in the metazoa. See text for details. Details of TSP family evolution in vertebrates can be found in McKenzie et al. 2006).
The combined analysis of TSPs and their major ligands (fig. 4) reveals that the repertoire of binding interactions of mammalian TSPs represents, for the most part, innovations within the deuterostome lineage. These involve either deuterostome- or chordate-specific ligands or novel binding activities of ancient molecules (e.g., CD36, calreticulin). These de novo interactions likely contribute to specific features of deuterostome tissue organization and function, a concept supported by the nonlethal, tissue-specific, yet highly complex phenotypes of TSP gene knockouts in mice. The many activities of TSP-1 and TSP-2 that are mediated by their TSR domains in vivo (e.g., TGFβ binding/activation, CD36-dependent antiangiogenesis and CD36-dependent regulation of nitric oxide-regulated vascular responses [Ribeiro et al. 1999; Isenberg et al. 2007; Zhang and Lawler 2007]) demonstrate the extent to which the functional properties of TSPs were impacted by the evolution of TSP subgroup A (Zhang and Lawler 2007). Overall, it can be considered that interactions that are most suitable as therapeutic targets should be included within the deuterostome innovations because these interactions will be amenable to manipulation without causing complete disruption of the ECM. Examples of such interactions would include fibrinogen or calreticulin binding by the LG-amino-terminal domain or CD36 binding by the TSR. Reciprocally, investigations of TSP activities in engineered or synthetic ECM would most appropriately focus on apparent ancestral activities of TSPs in binding to ECM components or PS2 clade integrins.
Both the LG domain and C-terminal region of TSPs are palimpsests of ancient and chordate-specific binding capacities. Candidate ancestral ligands include heparan glycosaminoglycans, RGD- and KGD-dependent integrins, and fibrillar collagen. On this basis, we propose that the TSPs of basal metazoa and protostomes will be found to have significant roles in ECM organizational processes. Major structural components of ECM, collagens, fibrillin, and laminin are also conserved throughout the metazoa and the role of TSP in basal metazoa is now a question of high interest.
Supplementary Material
Supplementary table S1 and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Stephanie Haase and David Loftis for experimental assistance; Noriyaki Satoh and the Center for Genetic Resource Information, Japan, for providing C. intestinalis cDNAs; CCF Genomics core for DNA sequencing; and the Acorn Worm Genome Sequencing Consortium and BCM-HGSC for public availability of Skow1.0 draft assembly. The DNA sequence of the acorn worm, S. kowalevskii, was supported by a grant from the National Human Genome Research Institute of the National Institutes of Health to Richard Gibbs at Baylor College of Medicine-HGSC. Research in this article was supported by the National Institutes of Health, National Heart, Lung and Blood Institute (HL077107), the National Institute of General Medical Sciences (GM068073), and the University of Bristol, UK. Contributions: J.C.A. designed research; J.C.A. and A.A.B. performed research and analyzed data; and J.C.A. wrote the article.
References
- Adams JC. Thrombospondins: multifunctional regulators of cell interactions. Annu Rev Cell Dev Biol. 2001;17:25–51. doi: 10.1146/annurev.cellbio.17.1.25. [DOI] [PubMed] [Google Scholar]
- Adams JC. Functions of the conserved thrombospondin carboxy-terminal cassette in cell-extracellular matrix interactions and signaling. Int J Biochem Cell Biol. 2004;36:1102–1114. doi: 10.1016/j.biocel.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Adams JC, Bentley AA, Kvansakul M, Hatherley D, Hohenester E. Extracellular matrix retention of thrombospondin 1 is controlled by its conserved C-terminal region. J Cell Sci. 2008;121:784–795. doi: 10.1242/jcs.021006. [DOI] [PubMed] [Google Scholar]
- Adams JC, Monk R, Taylor AL, Ozbek S, Fascetti N, Baumgartner S, Engel J. Characterisation of Drosophila thrombospondin defines an early origin of pentameric thrombospondins. J Mol Biol. 2003;328:479–494. doi: 10.1016/s0022-2836(03)00248-1. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, et al. (195 co-authors) The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Akimana C, Lafontaine ER. The Moraxella catarrhalis outer membrane protein CD contains two distinct domains specifying adherence to human lung cells. FEMS Microbiol Lett. 2007;271:12–19. doi: 10.1111/j.1574-6968.2007.00693.x. [DOI] [PubMed] [Google Scholar]
- Anilkumar N, Annis DS, Mosher DF, Adams JC. Trimeric assembly of the C-terminal region of thrombospondin-1 or thrombospondin-2 is necessary for cell spreading and fascin spike organisation. J Cell Sci. 2002;115:2357–2366. doi: 10.1242/jcs.115.11.2357. [DOI] [PubMed] [Google Scholar]
- Asch AS, Silbiger S, Heimer E, Nachman RL. Thrombospondin sequence motif (CSVTCG) is responsible for CD36 binding. Biochem Biophys Res Commun. 1992;182:1208–1217. doi: 10.1016/0006-291x(92)91860-s. [DOI] [PubMed] [Google Scholar]
- Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6:457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- Bauer DJ. The Daphnia Genomics Consortium Meeting: the genome biology of the model crustacean Daphnia pulex. Expert Rev Proteomics. 2007;4:601–602. doi: 10.1586/14789450.4.5.601. [DOI] [PubMed] [Google Scholar]
- Bornstein P, Agah A, Kyriakides TR. The role of thrombospondins 1 and 2 in the regulation of cell-matrix interactions, collagen fibril formation, and the response to injury. Int J Biochem Cell Biol. 2004;36:1115–1125. doi: 10.1016/j.biocel.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Calzada MJ, Zhou L, Sipes JM, Zhang J, Krutzsch HC, Iruela-Arispe ML, Annis DS, Mosher DF, Roberts DD. α4β1 integrin mediates selective endothelial cell responses to thrombospondins 1 and 2 in vitro and modulates angiogenesis in vivo. Circ Res. 2004;94:462–470. doi: 10.1161/01.RES.0000115555.05668.93. [DOI] [PubMed] [Google Scholar]
- Carlson CB, Bernstein DA, Annis DS, Misenheimer TM, Hannah BL, Mosher DF, Keck JL. Structure of the calcium-rich signature domain of human thrombospondin-2. Nat Struct Mol Biol. 2005;12:910–914. doi: 10.1038/nsmb997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cássaro CM, Dietrich CP. Distribution of sulfated mucopolysaccharides in invertebrates. J Biol Chem. 1977;252:2254–2261. [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chakravarti R, Adams JC. Comparative genomics of the syndecans defines an ancestral genomic context associated with matrilins in vertebrates. BMC Genomics. 2006;7:83. doi: 10.1186/1471-2164-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanana B, Graf R, Koledachkina T, Pflanz R, Vorbrüggen G. AlphaPS2 integrin-mediated muscle attachment in Drosophila requires the ECM protein thrombospondin. Mech Dev. 2007;124:463–475. doi: 10.1016/j.mod.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Chen FH, Thomas AO, Hecht JT, Goldring MB, Lawler J. Cartilage oligomeric matrix protein/thrombospondin 5 supports chondrocyte attachment through interaction with integrins. J Biol Chem. 2005;280:32655–32661. doi: 10.1074/jbc.M504778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Oliveri P, Gao F, Dornbos SQ, Li CW, Bottjer DJ, Davidson EH. Precambrian animal life: probable developmental and adult cnidarian forms from Southwest China. Dev Biol. 2002;248:182–196. doi: 10.1006/dbio.2002.0714. [DOI] [PubMed] [Google Scholar]
- Christensen A, Svensson K, Persson S, Jung J, Michalak M, Widell S, Sommarin M. Functional characterization of Arabidopsis calreticulin1a: a key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 2008;49:912–924. doi: 10.1093/pcp/pcn065. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, et al. (87 co-authors) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. (12 co-authors) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd RB, Drickamer K. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology. 2001;11:71R–9R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. Step-by-step evolution of vertebrate blood coagulation. Cold Spring Harb Symp Quant Biol. 2009 doi: 10.1101/sqb.2009.74.001. Advance Access published August 10, 2010, doi:10.1101/sqb.2009.74.001. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Lustig A, Engel J. The thrombospondin-like chains of cartilage oligomeric matrix protein are assembled by a fivestranded alpha-helical bundle between residues 20 and 83. FEBS Lett. 1994;341:54–58. doi: 10.1016/0014-5793(94)80239-4. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glöckner G, et al. (97 co-authors) The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito JY, Larroux C, Cluzel C, Valcourt U, Lethias C, Degnan BM. Demosponge and sea anemone fibrillar collagen diversity reveals the early emergence of A/C clades and the maintenance of the modular structure of type V/XI collagens from sponge to human. J Biol Chem. 2008;283:28226–28235. doi: 10.1074/jbc.M804573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Fujikawa K, Pratt KP. New insights into binding interfaces of coagulation factors V and VIII and their homologues lessons from high-resolution crystal structures. Curr Protein Pept Sci. 2002;3:313–339. doi: 10.2174/1389203023380639. [DOI] [PubMed] [Google Scholar]
- Hegedus D, Erlandson M, Gillott C, Toprak U. New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol. 2009;54:285–302. doi: 10.1146/annurev.ento.54.110807.090559. [DOI] [PubMed] [Google Scholar]
- Holden P, Meadows RS, Chapman KL, Grant ME, Kadler KE, Briggs MD. Cartilage oligomeric matrix protein interacts with type IX collagen, and disruptions to these interactions identify a pathogenetic mechanism in a bone dysplasia family. J Biol Chem. 2001;276:6046–6055. doi: 10.1074/jbc.M009507200. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori I. Role of fixed parenchyma cells in blastema formation of the planarian Dugesia japonica. Int J Dev Biol. 1991;35:101–118. [PubMed] [Google Scholar]
- Huhtala M, Heino J, Casciari D, de Luise A, Johnson MS. Integrin evolution: insights from ascidian and teleost fish genomes. Matrix Biol. 2005;24:83–95. doi: 10.1016/j.matbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Gao AG, Lindberg FP, Dimitry JM, Brown EJ, Frazier WA. Integrin-associated protein is a receptor for the C-terminal domain of thrombospondin. J Biol Chem. 1996;271:21–24. doi: 10.1074/jbc.271.1.21. [DOI] [PubMed] [Google Scholar]
- Goicoechea S, Orr AW, Pallero MA, Eggleton P, Murphy-Ullrich JE. Thrombospondin mediates focal adhesion disassembly through interactions with cell surface calreticulin. J Biol Chem. 2000;275:36358–36368. doi: 10.1074/jbc.M005951200. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guo NH, Krutzsch HC, Nègre E, Vogel T, Blake DA, Roberts DD. Heparin- and sulfatide-binding peptides from the type I repeats of human thrombospondin promote melanoma cell adhesion. Proc Natl Acad Sci U S A. 1992;89:3040–3044. doi: 10.1073/pnas.89.7.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg JS, Jia Y, Fukuyama J, Switzer CH, Wink DA, Roberts DD. Thrombospondin-1 inhibits nitric oxide signaling via CD36 by inhibiting myristic acid uptake. J Biol Chem. 2007;282:15404–15415. doi: 10.1074/jbc.M701638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg JS, Ridnour LA, Dimitry J, Frazier WA, Wink DA, Roberts DD. CD47 is necessary for inhibition of nitric oxide-stimulated vascular cell responses by thrombospondin-1. J Biol Chem. 2006;281:26069–26080. doi: 10.1074/jbc.M605040200. [DOI] [PubMed] [Google Scholar]
- Kiedzierska A, Smietana K, Czepczynska H, Otlewski J. Structural similarities and functional diversity of eukaryotic discoidin-like domains. Biochim Biophys Acta. 2007;1774:1069–1078. doi: 10.1016/j.bbapap.2007.07.007. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, et al. (36 co-authors) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoa. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvansakul M, Adams JC, Hohenester E. Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats. EMBO J. 2004;23:1223–1233. doi: 10.1038/sj.emboj.7600166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakides TR, Zhu YH, Smith LT, et al. (12 co-authors) Mice that lack thrombospondin 2 display connective tissue abnormalities that are associated with disordered collagen fibrillogenesis, an increased vascular density, and a bleeding diathesis. J Cell Biol. 1998;140:419–430. doi: 10.1083/jcb.140.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapraz F, Röttinger E, Duboc V, et al. (16 co-authors) RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev Biol. 2006;300:132–152. doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PubMed] [Google Scholar]
- Lawler J, Hynes RO. An integrin receptor on normal and thrombasthenic platelets that binds thrombospondin. Blood. 1989;74:2022–2027. [PubMed] [Google Scholar]
- Lawler J, Sunday M, Thibert V, Duquette M, George EL, Rayburn H, Hynes RO. Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J Clin Invest. 1998;101:982–992. doi: 10.1172/JCI1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Durchschlag M, Kepp O, Panaretakis T, Zitvogel L, Fröhlich KU, Kroemer G. Phylogenetic conservation of the preapoptotic calreticulin exposure pathway from yeast to mammals. Cell Cycle. 2009;8:639–642. doi: 10.4161/cc.8.4.7794. [DOI] [PubMed] [Google Scholar]
- McKenzie P, Chadalavada SC, Bohrer J, Adams JC. Phylogenomic analysis of vertebrate thrombospondins reveals fish-specific paralogues, ancestral gene relationships and a tetrapod innovation. BMC Evol Biol. 2006;6:33. doi: 10.1186/1471-2148-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt TM, Bick R, Poindexter BJ, Alcorn JL, Hecht JT. Unique matrix structure in the rough endoplasmic reticulum cisternae of pseudoachondroplasia chondrocytes. Am J Pathol. 2007;170:293–300. doi: 10.2353/ajpath.2007.060530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller WE, Thakur NL, Ushijima H, et al. (11 co-authors) Matrix-mediated canal formation in primmorphs from the sponge Suberites domuncula involves the expression of a CD36 receptor-ligand system. J Cell Sci. 2004;117:2579–2590. doi: 10.1242/jcs.01083. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Page AP, Johnstone IL. The cuticle. WormBook. 2007;19:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongsomboon S, Wongpanya R, Tang S, Chalorsrikul A, Tassanakajon A. Abundantly expressed transcripts in the lymphoid organ of the black tiger shrimp, Penaeus monodon, and their implication in immune function. Fish Shellfish Immunol. 2008;25:485–493. doi: 10.1016/j.fsi.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Posey KL, Hankenson K, Veerisetty AC, Bornstein P, Lawler J, Hecht JT. Skeletal abnormalities in mice lacking extracellular matrix proteins, thrombospondin-1, thrombospondin-3, thrombospondin-5, and type IX collagen. Am J Pathol. 2008;172:1664–1674. doi: 10.2353/ajpath.2008.071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, et al. (37 co-authors) The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, et al. (19 co-authors) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Ribeiro SM, Poczatek M, Schultz-Cherry S, Villain M, Murphy-Ullrich JE. The activation sequence of thrombospondin-1 interacts with the latency-associated peptide to regulate activation of latent transforming growth factor-beta. J Biol Chem. 1999;274:13586–13593. doi: 10.1074/jbc.274.19.13586. [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Galperin MY. The DxDxDG motif for calcium binding: multiple structural contexts and implications for evolution. J Mol Biol. 2004;343:971–84. doi: 10.1016/j.jmb.2004.08.077. [DOI] [PubMed] [Google Scholar]
- Robb SM, Ross E, Sánchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36:D599–D606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. The molecular origins of multicellular transitions. Curr Opin Genet Dev. 2008;18:472–478. doi: 10.1016/j.gde.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, et al. (14 co-authors) A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Schéele S, Nyström A, Durbeej M, Talts JF, Ekblom M, Ekblom P. Laminin isoforms in development and disease. J Mol Med. 2007;85:825–836. doi: 10.1007/s00109-007-0182-5. [DOI] [PubMed] [Google Scholar]
- Schultz-Cherry S, Chen H, Mosher DF, Misenheimer TM, Krutzsch HC, Roberts DD, Murphy-Ullrich JE. Regulation of transforming growth factor-beta activation by discrete sequences of thrombospondin 1. J Biol Chem. 1995;1270:7304–7310. doi: 10.1074/jbc.270.13.7304. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KS, Brudno M, Hill MM, Sidow A. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007;8:R41. doi: 10.1186/gb-2007-8-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, et al. (21 co-authors) The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, et al. (36 co-authors) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Wayburn B, Bunch T, Volk T. Thrombospondin-mediated adhesion is essential for the formation of the myotendinous junction in Drosophila. Development. 2007;134:1269–1278. doi: 10.1242/dev.000406. [DOI] [PubMed] [Google Scholar]
- Sun YD, Zhao XF, Kang CJ, Wang JX. Molecular cloning and characterization of Fc-TSP from the Chinese shrimp Fennerpenaeus chinensis. Mol. Immunol. 2006;43:1202–1210. doi: 10.1016/j.molimm.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Tan K, Duquette M, Joachimiak A, Lawler J. The crystal structure of the signature domain of cartilage oligomeric matrix protein: implications for collagen, glycosaminoglycan and integrin binding. FASEB J. 2009;23:2490–2501. doi: 10.1096/fj.08-128090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Duquette M, Liu JH, Dong Y, Zhang R, Joachimiak A, Lawler J, Wang JH. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol. 2002;159:373–382. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Duquette M, Liu JH, Zhang R, Joachimiak A, Wang JH, Lawler J. The structures of the thrombospondin-1 N-terminal domain and its complex with a synthetic pentameric heparin. Structure. 2006;14:33–42. doi: 10.1016/j.str.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;453:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R. Evidence for the evolution of tenascin and fibronectin early in the chordate lineage. Int J Biochem Cell Biol. 2009;41:424–434. doi: 10.1016/j.biocel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Vakonakis I, Campbell ID. Extracellular matrix: from atomic resolution to ultrastructure. Curr Opin Cell Biol. 2007;19:578–583. doi: 10.1016/j.ceb.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, et al. (12 co-authors) Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voland C, Serre CM, Delmas P, Clezardin P. Platelet-osteosarcoma cell interaction is mediated through a specific fibrinogen-binding sequence located within the N-terminal domain of thrombospondin 1. J Bone Miner Res. 2000;15:361–368. doi: 10.1359/jbmr.2000.15.2.361. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Bergeron KF, Whittle J, Brandhorst BP, Burke RD, Hynes RO. The echinoderm adhesome. Dev Biol. 2006;300:252–266. doi: 10.1016/j.ydbio.2006.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Qiu GF, Unuma T. Molecular cloning and ovarian expression profiles of thrombospondin, a major component of cortical rods in mature oocytes of penaeid shrimp, Marsupenaeus japonicus. Biol Reprod. 2004;70:1670–1678. doi: 10.1095/biolreprod.103.025379. [DOI] [PubMed] [Google Scholar]
- Young GD, Murphy-Ullrich JE. The tryptophan-rich motifs of the thrombospondin type 1 repeats bind VLAL motifs in the latent transforming growth factor-beta complex. J Biol Chem. 2004;279:47633–47642. doi: 10.1074/jbc.M404918200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lawler J. Thrombospondin-based antiangiogenic therapy. Microvasc Res. 2007;74:90–99. doi: 10.1016/j.mvr.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.