Abstract
Conserved noncoding elements (CNEs) in vertebrate genomes often act as developmental enhancers, but a critical issue is how well orthologous CNE sequences retain the same activity in their respective species, a characteristic important for generalization of model organism studies. To quantify how well CNE enhancer activity has been preserved, we compared the anatomy-specific activities of 41 zebra fish CNEs in zebra fish embryos with the activities of orthologous human CNEs in mouse embryos. We found that 13/41 (∼30%) of the orthologous CNE pairs exhibit conserved positive activity in zebra fish and mouse. Conserved positive activity is only weakly associated with either sequence conservation or the absence of bases undergoing accelerated evolution. A stronger effect is that disparate activity is associated with transcription factor binding site divergence. To distinguish the contributions of cis- versus trans-regulatory changes, we analyzed 13 CNEs in a three-way experimental comparison: human CNE tested in zebra fish, human CNE tested in mouse, and orthologous zebra fish CNE tested in zebra fish. Both cis- and trans-changes affect a significant fraction of CNEs, although human and zebra fish sequences exhibit disparate activity in zebra fish (indicating cis regulatory changes) twice as often as human sequences show disparate activity when tested in mouse and zebra fish (indicating trans regulatory changes). In all four cases where the zebra fish and human CNE display a similar expression pattern in zebra fish, the human CNE also displays a similar expression pattern in mouse. This suggests that the endogenous enhancer activity of ∼30% of human CNEs can be determined from experiments in zebra fish alone, and to identify these CNEs, both the zebra fish and the human sequences should be tested.
Keywords: enhancer, conserved noncoding elements, evolution, development, transcription factor, expression, cis, trans
Introduction
Enhancer elements are a crucial component of the functional repertoire of vertebrate genomes (Woolfe et al. 2005; Pennacchio et al. 2006; Kikuta, Laplante et al. 2007; Visel, Bristow, et al. 2007; Visel, Minovitsky, et al. 2007; Woolfe et al. 2007; Antonellis et al. 2008; Engstrom et al. 2008; Kleinjan et al. 2008; Navratilova et al. 2009). Understanding of their biological importance has arisen from two intersecting approaches. The first is through genetic characterization of well-known developmental genes and the enhancers that control their expression (Antonellis et al. 2008; Kleinjan et al. 2008; Navratilova et al. 2009). The second is by identification of large numbers of highly conserved intergenic sequences (Bejerano et al. 2004; Ovcharenko et al. 2004; Visel, Minovitsky, et al. 2007; Engstrom et al. 2008; Persampieri et al. 2008; Stephen, Pheasant et al. 2008; Wang et al. 2008), large fractions of which can act as transcriptional enhancers in vertebrate genomes (Woolfe et al. 2005; Pennacchio et al. 2006; Visel et al. 2009). Together, these approaches have shown that many enhancer sequences are constrained by common functional pressures across species. However, a key shortcoming of this view is that it ignores species-specific changes in enhancer function. The extent to which enhancer functions are conserved across species is not well characterized.
Although a number of conserved noncoding sequences have been assayed within one host organism, the lineage-specific behaviors are less clear. Lineage-specific evolution in either the enhancer sequence itself (cis) or elsewhere in the host genome or cellular environment (trans) could affect the function of each enhancer. For example, both lineage-specific cis- and trans-effects are well known to cause differences in gene expression in closely related fly species (Wittkopp et al. 2004). These facts suggest that enhancers could have considerable lineage-specific activity too. As a comparison, orthologous genes can have strong sequence conservation across species even if their functions differ (Chan et al. 2009), and this may be true for enhancers as well.
Changes in cis-regulation associated with conserved noncoding elements (CNEs) have been investigated in a number of works (de la Calle-Mustienes et al. 2005; McEwen et al. 2009; Navratilova et al. 2010) because they require experiments in only one host organism but there are less data on the behaviors of CNEs in different hosts. A recent example of a two-host study is the work of Navratilova et al. (2009), who described five pairs of zebra fish–human CNE sequences near the SOX3 and PAX6 loci, each with homologous anatomical activity in zebra fish and mouse hosts, respectively. Strahle and Rastegar (2008) compared the functional behavior of six pairs of CNE sequences from zebra fish and mouse around the ngn1 and shh loci, finding that the activities of at least four differ in their native hosts. de la Calle-Mustienes et al. (2005) tested seven CNEs near the IRX locus in zebra fish and xenopus embryos, finding four cases of similar activity and three of disparate activity. Other works have also measured CNE activities in two hosts, for two to three CNEs at a time (Antonellis et al. 2008; Jarinova et al. 2008; Kleinjan et al. 2008). One concern is that the prior CNEs may not be representative, given their focus on individual loci. However, there now exist large data sets describing the activity of human CNE sequences within mouse (Pennacchio et al. 2006) as well as zebra fish sequences in zebra fish (ZZ) (Li et al. 2009), allowing a broader approach.
Here, we present a study of the functional evolution of orthologous CNE enhancers in distinct hosts. We consider data from 875 human CNEs whose enhancer activity has been tested in mouse and from 151 zebra fish CNEs, many of which are new, that have been tested by our laboratory in zebra fish. By identifying orthologous matches among these data sets, we were able to compare the enhancer activity of 41 ZZ host with the activities of their orthologous human sequences in a mouse (HM) host. Our results allow us to estimate the rate at which orthologous CNEs have conserved enhancer activity across species. This is of quantitative importance for determining the functions of human enhancer sequences because many thousands remain uncharacterized, and fish are a promising system for investigating their activity. This is because fish can be grown more quickly than mice, and fish experimental CNE techniques have shown notable recent advances (Bessa et al. 2009; Gehrig et al. 2009).
We analyze these 41 CNE sequences to determine how sequence conservation, lineage-specific accelerated sequence evolution, and divergence in transcription factor binding site motifs correlate with the preservation of activity across species. We also perform measurements of the enhancer activity of 13 human CNEs in zebra fish. For these 13 CNEs, we have a trio of corresponding experiments—ZZ, human sequence in zebra fish (HZ), and HM—allowing us to distinguish the relative importance of cis- and trans-regulatory effects.
Our results support the greater importance of cis-effects over trans-effects but indicate that both have played a significant role in enhancer evolution. Our studies also suggest that to characterize and identify those human sequences (∼30%) whose activity in zebra fish is the same as their activity in a mammalian host, both the zebra fish and the human sequences should be tested in zebra fish. Details on all CNEs are available in the supplementary data file, and we have built a Web site to make images of CNE activity publicly available (zebrafishcne.org).
Materials and Methods
Zebra Fish Experimental Data Set
Zebra fish sequences were chosen for verification based on >60% sequence identity in pairwise alignments between zebra fish and human and then measured for enhancer activity, as has been described elsewhere (Li et al. 2009). Note that to make the sequence conservation analysis parallel with the multispecies phyloP analysis, sequence identity values used for the sequence conservation analysis were based on the 44-species multiple alignments, which is why some CNEs have reported sequence identities <60%. Enhancer activity was measured in zebra fish using a minimal promoter/green fluorescent protein reporter system. Constructs of putative enhancer sequences located directly adjacent to the EB1 minimal promoter and GFP were integrated transgenically into the genome of zebra fish embryos using a Tol2 transposon system (Li et al. 2009). Robustness of enhancer activity was ascertained by performing injections in multiple embryos per CNE (n = 35 ± 12 among the 41 CNEs with mutual best hits), with most embryos (72 ± 20%) expressing in the assigned expression pattern. Sample images of replicates are shown in supplementary figure 1. Negative controls were also measured using GFP and a minimal promoter without a CNE. For the HZ experiments, the human sequence tested was the sequence listed for each element in the Enhancer database. Data on the activity of each CNE, including anatomical specificity and robustness, genomic location, predicted target genes, and so forth, are available in the downloadable supplementary data file. Images for each CNE are available at zebrafishcne.org.
Identification of Orthologous Enhancers
An initial set of 151 zebra fish CNE sequences whose enhancer activity was measured in zebra fish (from Li et al. 2009, as well as newly tested sequences) was Blasted versus 875 human sequences from the enhancer.lbl.gov database (Pennacchio et al. 2006) and three additional sequences tested by the Pennacchio laboratory. This yielded a data set of 41 mutual best hits (e-value cutoff of 0.05) that also aligned to 44-way University of California–Santa Cruz (UCSC) alignments, which were used for further analysis. The enhancer.lbl.gov database contains experiments of human sequences tested for enhancer activity in developing mouse embryos. Among these 41 matches, the length of human CNEs tested was ∼3.6 times longer than the zebra fish sequences (human: avg 1,573 bp, min 703 bp, max 2,200 bp; zebra fish: avg 432 bp, min 185 bp, max 790 bp).
Zebra Fish and Mouse Enhancer Activity Comparisons
Sequences tested in the zebra fish GFP enhancer assay were annotated with up to four expression anatomies at 24 h and up to four anatomies at 48 h. Both time points were used for comparison. For the LBL mouse enhancer assays (Pennacchio et al. 2006), up to five anatomy labels were assigned (single time point, 11.5 days). Sequences with more than four different expression tags were labeled as nonspecific. We manually compared the anatomical expression patterns of mutual best hit sequences in the zebra fish and mouse assays. At zebrafishcne.org, the zebra fish CNE image data can be accessed in conjunction with orthologous mouse data.
Analysis of Target Genes of CNEs
Akalin et al. (2009) predicted the gene targets of CNEs based on occurrence in genomic regulatory blocks (GRBs) having conserved synteny across vertebrate species (Akalin et al additional data file 1). We have listed the predicted gene target(s) for each CNE in the supplementary data file. In addition, the supplementary data file lists the flanking genes within 500 kb for each CNE along the human, zebra fish, and mouse genomes. Establishing correspondence between target gene expression and CNE enhancer activity depends on the availability of time-matched gene expression data, quality of the target gene prediction, and interpretation of the available gene expression images. We manually inspected the gene expression patterns for all predicted target genes for the 37 CNEs overlapping the Akalin et al. data. Zebra fish gene expression patterns were viewed using images curated by the ZFIN database (Sprague et al. 2006) at the 24-h time point. Annotations of the expression patterns of target genes are given in the supplementary data file, and notable cases of correspondence between CNE activity and target gene expression are given in supplementary figure 2.
Paralogous Zebra Fish CNEs
Paralogous CNEs were identified by Blasting zebra fish CNEs from the cneViewer database, a database of zebra fish sequences strongly conserved in the human genome (Persampieri et al. 2008) versus one another. All paralogous CNEs had a Blast e-value of 1e−14 or better with their paired sequence, although none of the four pairs were exactly identical. Each pair of CNEs was manually inspected along the zebra fish genome to rule out misassemblies. The pairs analyzed correspond to CNEs (6.03, 6.04), (6.05, 6.06), (6.07, 6.08), and (6.09, 6.10) in the experiment compendium at zebrafishcne.org.
Transcription Factor Binding Site Divergence
Binding sites were predicted on both strands of the human and zebra fish sequences using position-specific weight matrices derived from motifs of the 11 JASPAR FAM transcription factor families (Sandelin and Wasserman 2004; Wasserman and Sandelin 2004) using pseudocounts. For each family, significant binding affinity was determined using the cutoff balancing type 1 and type 2 error (Rahmann et al. 2003). Summing over strands, the combined number of predicted binding sites on both strands was determined separately for human (nh) and zebra fish (nz). For each sequence and family, these values were transformed into the fraction of diverged binding sites (fdivergence) by dividing the absolute value of the difference of counts in human versus zebra fish by the sum of the counts:
Sequence Conservation Analyses
We aligned 49 mutual best hits between human and zebra fish according to their Hg18 coordinates with the 44-way vertebrate alignments downloaded from the UCSC Browser. As the human coordinates are often larger than the zebra fish conserved sequences tested in experimental assays, we trimmed the overhanging human sequence and kept regions where zebra fish is aligned, although gapped, in the 44-way alignment. We discarded other zebra fish unaligned sequences. Doing this, we retained 41 enhancers from the 49 mutual best hits to the enhancer database. To get zebra fish–human percent sequence identity values for these sequences, we selected the zebra fish (danRer5) and human (Hg18) sequences from the 44-way MAF alignments in Galaxy and used the Emboss tool Needle to measure percent sequence identity in a global sequence alignment. Using this procedure, the average pairwise percent identity of the set of 41 enhancers was 65% (min 28.9%, max 84.5%, med 66.8%).
To quantify accelerated evolution, we defined an acceleration measure for each enhancer as the fraction of bases with a negative phyloP score in the mammalian (or other) lineage. PhyloP scores are −log base 10 P values from a likelihood ratio test for accelerated substitution rate (vs. the neutral rate estimated from 4-fold degenerate sites) at each base position in an alignment (Pollard et al. 2009).
Statistical Analysis
Receiver operating characteristic (ROC) curves were generated using ROC area under the curve (AUC) from the caTools package in R (R Development Core Team 2005), and statistical tests were generated using prop.test, wilcox.test, or t.test in R. For all ROC analyses, classes II and III (disparate positive and conserved positive, respectively) were compared. For percentage of accelerated bases (PAB) and pairwise percent identity, a stepwise increment of the minimum and maximum PAB or percent identity score for combined classes II and III was used as the classification threshold. For transcription factor binding site divergence (TFBD), for each sequence, the fraction of diverged transcription factor binding sites (fdivergence, see above) was averaged over families and used as the classification score.
Results
Comparison of Orthologous Fish and Mammalian Enhancers
We first compared the enhancer activity of ZZ embryos with the activity of orthologous HM embryos. Zebra fish sequences were chosen from zebra fish intergenic regions based on high sequence conservation with the human genome, a criterion that makes them strong candidates for enhancer activity (typically >60% identity; see Methods and Li et al. 2009). We then identified orthologous human sequences by comparing 151 zebra fish CNE sequences whose enhancer activity had been assayed to 875 human sequences in the Enhancer database (Pennacchio et al. 2006; Visel, Bristow, et al. 2007). We identified 41 zebra fish mutual best Blast hits to orthologous enhancer sequences in the human genome. Zebra fish sequences were tested for enhancer activity in a Tol2 transposon GFP assay as described in Li et al. (2009), whereas human sequences had been assayed for enhancer activity in mouse embryos as described in Visel, Minovitsky, et al. (2007).
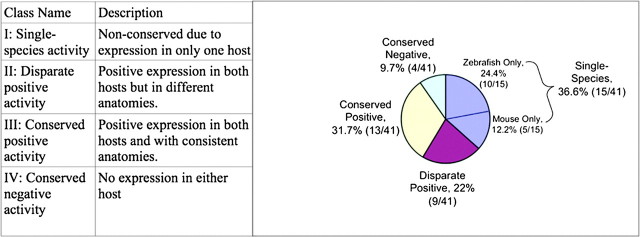
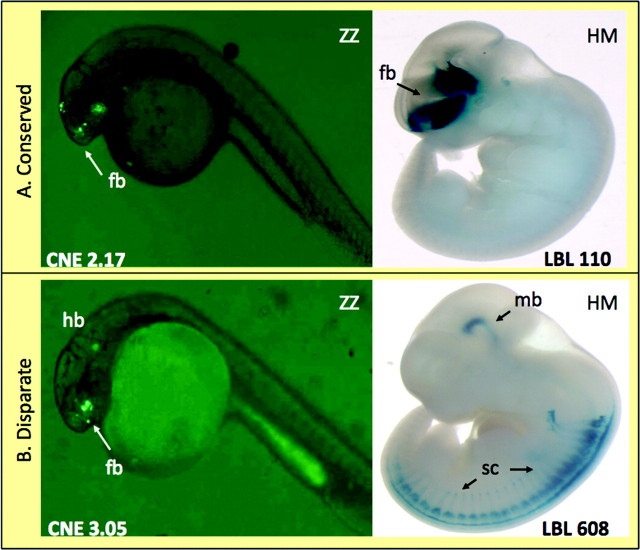
A minority of the enhancer pairs showed conserved anatomical activity (13/41, 31.7%). We classified the experiments into four classes that each contained substantial fractions of the data: 1) activity in only one host (36.6%), 2) positive expression in both hosts but different anatomies (22.0%), 3) positive and anatomically similar expression in both hosts (31.7%), and 4) no expression in either host (9.7%), as shown in figure 1. For those enhancer pairs driving gene expression in both hosts (categories II and III), the sequences exhibit conserved activity in 13 of 22 cases (59.1%). An example of conserved positive activity (class III) is shown in figure 2A in which the fish and mammalian experiments both display expression in the anterior brain. An example of disparate positive activity (class II) is shown in figure 2B in which the zebra fish sequence has weak activity in zebra fish forebrain, whereas the human sequence has activity in mouse midbrain and spinal cord.
FIG. 1.
Orthologous enhancer anatomical activity classes. Forty-one pairs of experiments comparing a zebra fish sequence tested in zebra fish with a human sequence tested in mouse. Each of the four classes contains a significant fraction of the data. I: single-species activity, II: disparate positive activity, III: conserved positive activity, and IV: conserved negative activity.
FIG. 2.
Activity of orthologous CNEs of zebra fish–zebra fish (ZZ) and human–mouse (HM). Examples show (A) conserved positive activity, and (B) disparate positive activity. fb, forebrain; hb, hindbrain; mb, midbrain; sc, spinal column.
The gene targets of CNE enhancers are generally not known, although Akalin et al. (2009) computationally predicted the targets of CNEs in GRBs based on conserved synteny across vertebrate species. Thirty-seven of the 41 CNEs occur in such blocks, so presence in a GRB is not a strong predictor of whether a CNE will have preserved activity across species. Correspondence between CNE activity patterns and target gene expression was irregular, with 14 zebra fish CNEs driving expression at least partially overlapping the expression of the predicted target gene. Notable examples of close correspondence include CNEs 1.01 and 2.01.2—both with FEZF2; CNE 12.03—with irx3 and sall1; CNE 2.04—with EMX2; and CNE 6.01—with SOX2 (see supplementary fig. 2). Details on predicted target genes and their expression patterns can be found in supplementary data file. A few CNEs have previously been tested in knockout experiments (Ahituv et al. 2007). For example, one CNE in our data overlaps a knockout by Ahituv et al. (CNE 2.19—uc248), but this did not reveal a target gene because knockout did not affect expression of genes nearby.
Sequence Conservation Is a Weak Predictor of Conserved Enhancer Activity
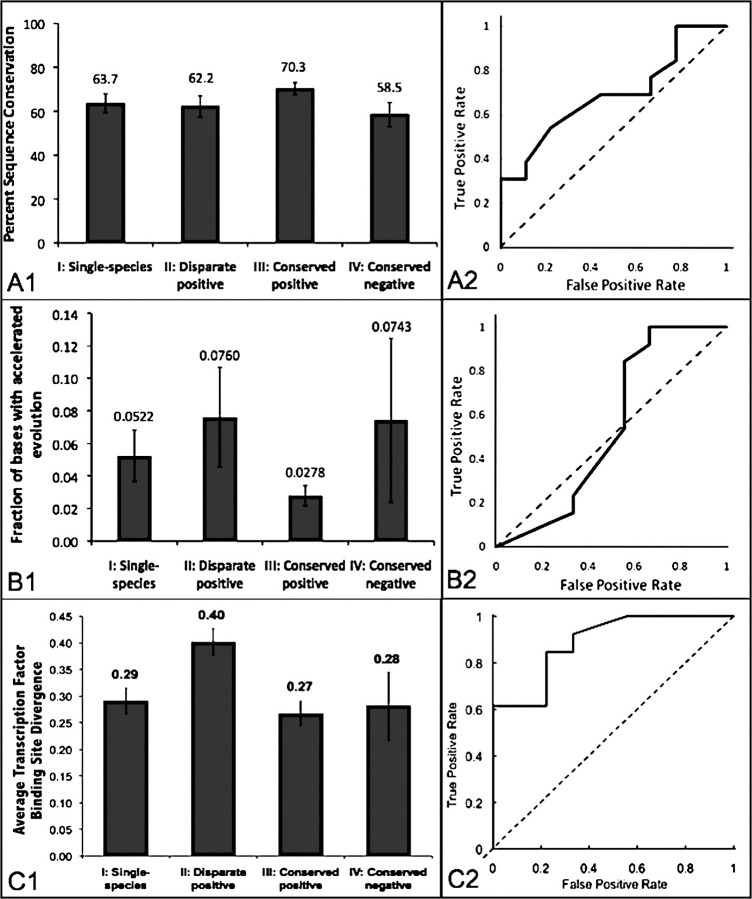
To explore why certain orthologous sequences exhibit conserved activity whereas others do not, we tested whether increased enhancer sequence conservation is associated with conservation of enhancer activity. For each CNE, we extracted the human and zebra fish sequences from UCSC 44-way vertebrate alignments and calculated pairwise percent identity. The average percent identities for CNE activity classes I–IV are shown in figure 3A1. All classes show high absolute sequence conservation, as expected. Sequences with conserved anatomical expression (class III) show greater sequence conservation than sequences in other classes, although these differences are small. P values for t-tests comparing mean expression of classes I, II, and IV with the most conserved class (III) are 0.20, 0.17, and 0.12, respectively.
FIG. 3.
(A1) Zebra fish–human sequence identity is marginally higher for sequences with conserved positive activity (class III) than those with other expression patterns. Error bars represent standard errors of the means. (A2) ROC curve to classify enhancers with conserved positive activity (class III) versus those with disparate positive activity (class II) based on sequence identity, marked by solid line. The AUC is 0.684, indicating marginal predictive power over random assignment (dashed line). (B1) Percentage of bases undergoing accelerated evolution in mammals (PAB). Enhancers with conserved positive activity exhibit a lower PAB than sequences in other classes. (B2) Mammalian PAB ROC curve. The area under the ROC curve is 0.56, slightly less than for the percent identity–based classifier. (C1) TFBD between human and zebra fish. The disparate positive category shows an increase in binding site divergence. (C2) Binding site divergence is superior at distinguishing classes II and III, with ROC area under curve equal to 0.88.
As a classification tool, sequence conservation has marginal power to predict expression class. We tested the effectiveness of using a sequence conservation threshold to distinguish between enhancers with disparate or conserved positive activity (classes II and III). By calculating the true-positive rate and false-positive rate at each threshold of sequence conservation, we computed an ROC curve (figure 3A2). The AUC is 0.684, indicating a small improvement over random guessing. This relatively weak predictive power is likely influenced by the fact that all the sequences in the data set were chosen based on high sequence conservation.
Percentage of Accelerated Bases Is a Comparable Predictor to Sequence Conservation
We next tested whether the presence of bases undergoing accelerated evolution (Prabhakar et al. 2006; Bird et al. 2007; Pollard et al. 2009) could discriminate enhancers with divergent activity because an enhancer with accelerated primate evolution has been shown to have lineage-specific human activity (Prabhakar et al. 2008). We used the program phyloP from the PHAST package (http://compgen.bscb.cornell.edu/phast/) applied to multiple sequence alignments of all mammals extracted from the UCSC 44-way alignments to assess whether bases in each CNE are rapidly evolving in the mammalian lineage (Pollard et al. 2009). We hypothesized that sequences with a larger PAB would be more likely to have divergent activity.
We found that mammalian PAB has similar discriminative power as human–zebra fish sequence percent identity (figure 3B1). For sequences with conserved positive activity, the average PAB is lower (0.0278) than the average PAB for sequences with disparate positive activity (0.0760), although this difference is not statistically significant (t-test: P = 0.16). Classes I and IV also have modest differences in PAB from class III (I vs. III: P = 0.16, IV vs. III: P = 0.43).
To determine if PAB score could be a useful discriminator for identifying enhancers with conserved cross-species expression, we calculated an ROC curve (figure 3B2). The area under the ROC curve is 0.560, lower than that for the human–zebra fish percent identity classifier. However, no enhancers with conserved positive activity have PAB > 0.07, whereas three of the nine enhancers with disparate positive activity have unusually large PAB values (0.13, 0.22, and 0.23). This suggests a rule of thumb that enhancers with high outlier PAB values are unlikely to have conserved positive activity. Because extreme PAB values are more predictive than a larger range of values, we considered whether a more stringent phyloP cutoff (considering only bases with PhyloP scores less than −3) would improve our classification. However, this stringency level reduced the data size too much to make any general conclusions.
Applying phyloP to detect accelerated evolution in other clades besides mammals gives similar results. For both the full vertebrate phylogeny and the primate phylogeny, the conserved positive activity class has the lowest PAB value, whereas the conserved negative activity has the highest PAB value. In each case, the tests distinguishing the classes have only marginal P values.
TFBD Is a Better Predictor of Conserved Enhancer Activity
Because enhancer activity is expected to act via transcription factor binding, we tested whether transcription factor binding site gain or loss (Pennacchio et al. 2007; Antonellis et al. 2008; Hare et al. 2008) might better predict conserved enhancer activity. Using 11 JASPAR FAM transcription factor family motifs (Sandelin and Wasserman 2004; Wasserman and Sandelin 2004), for each sequence and family, we calculated the TFBD as the absolute value of the difference of transcription factor binding site counts in human and zebra fish divided by the sum of the counts in both species. Figure 3C1 shows that increased TFBD is associated with enhancer class II, disparate positive activity. The ROC curve comparing the average of TFBD of all transcription factor families found in classes II and III (figure 3C2) has an AUC of 0.88, reflecting far superior separation compared with sequence conservation and PAB. Thus, by using known transcription factor binding profiles, it is possible to more accurately identify orthologous CNE sequences that conserve anatomical enhancer activity.
Cis-Effects Are More Important but Trans-Changes Also Influence Enhancer Activity
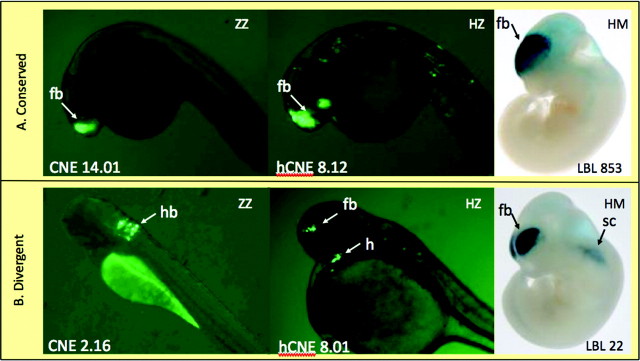
Although the sequence analysis above demonstrates that cis-evolution is important to changes in enhancer activity, trans-effects also play a role. Trans-effects include substitutions elsewhere in the genome as well as differences in the cellular environment of fish versus mammals. To distinguish the relative importance of cis- and trans-effects, we chose 13 sequences with robust anatomy-specific expression in zebra fish embryos from the above set of 41 enhancer sequences and assayed the corresponding HZ. This approach created 13 orthologous trios with a zebra fish sequence tested in zebra fish and a human sequence tested in both mouse and zebra fish. For these 13 CNE trios, we were then able to compare 1) the activity of the ZZ, 2) the activity of the HZ, and 3) the activity of the HM. Examples of these data are shown in figure 4 and summarized in figure 5.
FIG. 4.
Examples of comparisons of enhancer activity. (A) Conserved activity: the ZZ, HZ, and HM experiments all show enhancer activity in forebrain. (B) Divergent activity: the ZZ experiment shows expression in hindbrain, whereas the HZ and HM experiments show expression in forebrain. In addition, the HZ experiment shows expression in heart, and the HM experiment shows expression in the spinal column. This is an example of divergent cis-effects and partially divergent trans-effects fb, forebrain; h, heart; sc, spinal column.
FIG. 5.
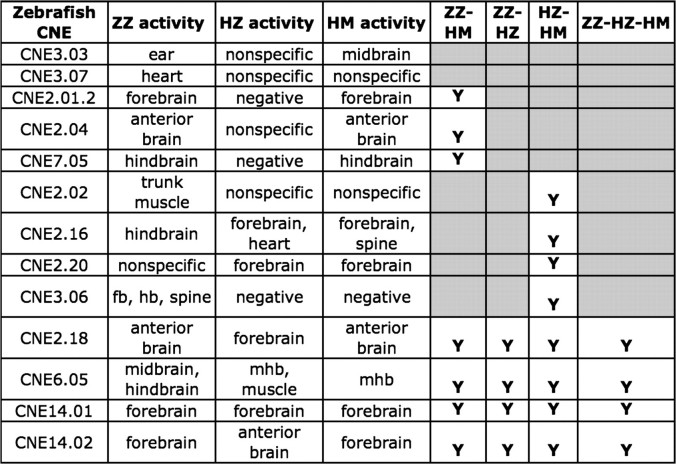
Comparison of cis- and trans-effects in the anatomical activity of orthologous enhancers. For 13 enhancers, we measured the activity of ZZ, the orthologous HZ, and the HM. We then compared whether the enhancer activity of orthologous experiments was similar (Y) or dissimilar (gray). A value of “Y” was assigned when at least one anatomy showed similar positive expression. Cis-changes cause differences in the orthologous activity 80% more often than trans-changes, as can be seen by the larger number of experiments yielding dissimilar expression (9) in the ZZ–HZ column than in the HZ–HM column (5). Furthermore, in all cases where there are no cis-effects (ZZ–HZ = Y), there are no trans-effects either (ZZ–HZ–HM = Y), suggesting that a ZZ and HZ experiment can substitute for an HM experiment for 4/13∼30% of enhancers mhb, midbrain–hindbrain boundary.
Comparisons of HZ and HM activities control for cis-effects because they make use of the same human CNE sequence so that differences in the HZ and HM enhancer activities should be mainly due to trans-changes. We found that 5/13 (39%) of the HZ–HM comparisons showed different activity patterns, indicating that trans-changes affect a substantial minority of CNE enhancers. Comparison of ZZ and HZ enhancer activities controls for trans-changes by using a consistent host species, and therefore, differences in activity should be due to differences in the zebra fish and human CNE sequences. We observed that 9/13 (69%) of ZZ–HZ experiment pairs display disparate anatomical expression. This result indicates that almost twice as many CNEs have expression domains affected by cis-changes compared with trans-changes.
Next, we compared the ZZ, HZ, and HM activity patterns simultaneously. In 4/13 (31%) of the cases, all three experiments drive similar anatomical expression in homologous tissues. This is a higher fraction than expected if cis- and trans-evolution were independent ([1 − 0.39] × [1 − 0.69] = 19%), suggesting concerted cis- and trans-evolution to maintain function in each species. In other words, positive selection has likely acted on ∼10% of our tested enhancer sequences to optimize them for species-specific trans-regulatory factors.
Intriguingly, in the four cases where the ZZ and HZ data agree, HM activity also agrees with ZZ and HZ. That is, in this data set, CNEs without cis-changes in enhancer activity also never show trans-changes. This result suggests that the anatomy-specific activity of many human enhancers can be determined by measuring the enhancer activity of each of the zebra fish and HZ. If the observed HZ and ZZ expression domains agree, then this activity is likely to be the same as the activity of the HM model. Our study suggests that ∼30% of zebra fish enhancers will have activity patterns that can be extrapolated to human in a mammalian host following this protocol. In contrast, when there are no trans-changes (8/13 experiments show HZ and HM having similar expression), the fact that activity is conserved is not sufficient to imply a lack of cis-changes. This reiterates the greater importance of cis-information over trans-information for determining changes in enhancer activity.
Sequence Predictors of Conserved Activity for Trios
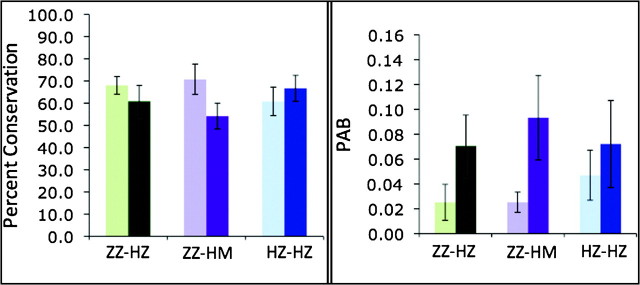
Do predictors of conserved activity perform better when cis- and trans-effects can be isolated? To answer this, we classified the sequences into six nonexclusive expression classes: ZZ–HZ Agree (n = 4) and ZZ–HZ Dissimilar (n = 9); ZZ–HM Agree (n = 7) and ZZ–HM Dissimilar (n = 6); HZ–HM Agree (n = 8) and HZ–HM Dissimilar (n = 5). Figure 6 (left) shows the average pairwise zebra fish–human sequence percent identity for enhancers in these classes.
FIG. 6.
Left: Sequence conservation for enhancer activity comparisons involving zebra fish sequences tested in zebra fish (ZZ), human sequences tested in zebra fish (HZ), and human sequences tested in mouse (HM). Bars show mean values ± the standard error of the mean. Sequence conservation is lower when enhancer activities disagree (dark bar) across experiments. This behavior is apparent for the cases involving cis-effects (ZZ–HZ and ZZ–HM) but not for the case where trans-effects have been isolated (HZ–HM). Right: The fraction of bases undergoing accelerated evolution (PAB) is higher when enhancer activities disagree (dark bar) across experiments. This behavior is stronger for the cases involving cis-effects (ZZ–HZ and ZZ–HM) than for the case where trans-effects have been isolated (HZ–HM).
Again, we found that sequence conservation has a weak positive association with conserved enhancer activity, but this association is stronger for the comparisons that include cis-effects. For example, the ZZ–HZ comparison is a test of cis-changes, and we found that the ZZ–HZ Agree set (light green) has a slightly higher sequence conservation than the ZZ–HZ Dissimilar set (dark green) (68% vs. 61%, t-test P = 0.45). Likewise, the ZZ–HM Agree set (light purple) has higher sequence conservation than the ZZ–HM Dissimilar set (dark purple) (71% vs. 54%, P = 0.08). However, the HZ–HM Agree class (light blue) actually exhibits a slightly lower sequence conservation than the HZ–HM Dissimilar class (dark blue) (61% vs. 67%, P = 0.51). This discrepancy can be explained by the fact that the HZ–HM comparison measures trans-changes, which are not expected to be related to sequence conservation.
The percentage of bases undergoing accelerated evolution (PAB) provides similar results (figure 6, right). For the comparisons involving cis-changes, ZZ–HZ and ZZ–HM, the enhancers with similar anatomical activity, have a lower PAB than those where the activities differ (P = 0.14 and 0.10, respectively). For the comparison where trans-effects are isolated, HZ–HM, the enhancers with conserved activity, have a smaller average PAB than those with differing activity, but this effect is even less significant than for comparisons involving cis-changes (P = 0.57).
Discussion
Zebra fish is a valuable model system for studying enhancer activity in vivo. While enhancer activity has been previously compared in mammalian and fish hosts for various developmental loci, the general prevalence of conserved activity has not been well quantified. By comparing a set of 41 zebra fish CNE sequences to their orthologous mammalian counterparts, we have shown that 13/41 (∼30%) of these predicted enhancers exhibit conserved positive activity. Moreover, when one restricts analysis to the cases where zebra fish shows positive activity, 13/32 (∼40%) display conserved anatomical activity with mammals. This finding quantifies the substantial fraction of zebra fish enhancer results that can be generalized to human.
Importantly, enhancers with conserved activity can be more exactly identified by simply testing the HZ. In the 4/13 three-way experiments (∼30%) where the zebra fish sequence and its orthologous human sequence display the same positive activity in zebra fish, similar activity is also found for the HM. This result shows that experiments in just zebra fish can identify native mammalian enhancer activities. It also provides a reference value from which to judge the cost and efficiency advantages of enhancer studies in fish over studies in a mammalian host, although because of the cis- and trans-changes only some human enhancers will be characterizable in this way. Note also that simply testing the HZ is not sufficient to extrapolate mammalian activity because trans-changes affect 5/13 (38%) cases. To determine human enhancer activity, both the HZ and ZZ experiments should be performed. When these are positive and agree, our data suggest that the activity of the HM can be reliably predicted.
One caveat is that a human in mouse measurement is only an approximation to the human in human activity, and we do not have direct data on the endogenous activity of the human sequence. This is a consequence of our use of HM data to compare with ZZ and HZ. This choice was made to take advantage of the Lawrence Berkeley National Lab Enhancer database of HM enhancer experiments, which provides the only large set of mammalian enhancer activity data available. Endogenous mammalian enhancer data (e.g., mouse in mouse) would be preferable if they were available. The human sequences we tested do have high sequence identities with their orthologous mouse sequences (avg 77% for those in the three-way experiments), and this is higher than the typical zebra fish–human identities (avg 65%).
Why do some enhancers exhibit conserved activity whereas others do not? Cis-changes influence approximately twice as many CNEs as trans-changes. This is consistent with the common argument that cis-regulation can change more easily because it acts locally, whereas trans-changes may influence a large number of target genes. Cis-effects have been reported to be more important than trans-effects in studies of transcription factor binding within the mammalian phylogeny (Odom et al. 2007; Wilson et al. 2008). Because the rate of trans-evolution is not fully understood, it is not clear at what divergence distance one should expect cis-effects to lose this primacy. For the CNEs we have studied, we saw that cis-effects continue to overshadow trans-effects even after the ∼450 million years since humans and fish diverged (Blair and Hedges 2005).
Our finding that a sizable minority of CNEs have been subject to trans-regulatory changes (5/13 trios) is different from previous studies, which have reported on CNEs that apparently do not have trans-changes (de la Calle-Mustienes et al. 2005; Navratilova et al. 2009) or for which trans-changes have been of minor effect (Jarinova et al. 2008). It is possible that the rate of trans-evolution we have observed is an overestimate because it is difficult to tell if the mouse and zebra fish developmental time points in the HZ and HM comparisons are perfectly matched. However, it is unlikely that this explains all the trans-changes. For example, in CNEs 2.01.2, 2.04, and 7.05, the ZZ and HM experiments show similar activity, whereas the HZ behavior is different (figure 5; images at zebrafishcne.org). Because the ZZ and HM behavior are similar, it is likely that the correct developmental time points are being considered. Moreover, the sequences in the HZ experiments each include the complete human regions orthologous to the sequences in the ZZ experiment, plus additional flanking sequence (supplementary data file). Therefore, the HZ sequences should be long enough to drive the enhancer activity unless there have been trans-changes. We also see in our data set that CNEs only have trans-changes when cis-changes occur as well, consistent with cis- and trans-behaviors reported from Drosophila gene expression studies (Wittkopp et al. 2004).
Previous studies have indicated that trans-evolution via changes in the DNA-binding domain of a protein is very slow—duplicate yeast transcription factors have maintained similar binding profiles despite divergence 100 million years ago (Wolfe and Shields 1997; Badis et al. 2008), and DNA-binding specificities of some homeodomain proteins appear to have been preserved since the vertebrate–invertebrate divergence (Berger et al. 2008). This suggests that trans-evolution is more likely to be occurring at the level of protein–protein interactions or protein expression. Another possibility is that these trans-effects are related to acetylation of the CNEs (Akalin et al. 2009).
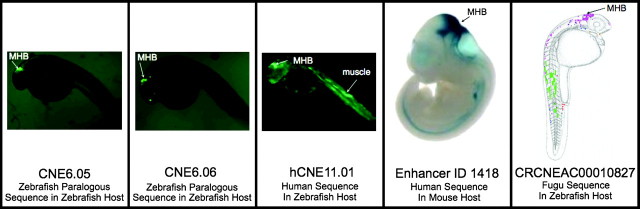
Despite the greater importance of cis-effects, the encoding of cis-regulatory changes in enhancers is subtle. We found that measures of sequence conservation are only weakly predictive of conserved activity among the CNEs we tested, with the best characterizations involving the use of transcription factor binding profiles. Simpler characterizations, such as sequence conservation and the absence of bases undergoing accelerated evolution (PAB), are only weakly associated with conserved enhancer activity. This result is consistent with findings that enhancers need not have high sequence conservation to be functional (Fisher et al. 2006; Hare et al. 2008; McGaughey et al. 2008). p300 ChIP-seq studies have shown recent promise for identifying tissue-specific mammalian enhancer activity (Visel et al. 2009), although p300-binding and cross-species activity conservation are not related to one another in our CNE data set. Mouse sequences for which p300 binding has been observed are not more likely to exhibit conserved cross-species activity (conserved positive vs. disparate positive; binomial test: P = 0.16, n=10). Paralogous CNEs in the zebra fish genome also show little association between sequence identity and activity conservation. We assayed four paralogous zebra fish enhancer pairs in zebra fish embryos and found no correlation of conserved activity with sequence identity. Despite the subtleties in enhancer regulatory signals, their functional robustness can be remarkable. Figure 7 shows consistent midbrain enhancer activity for a CNE in zebra fish, its paralogous CNE in zebra fish, its human ortholog in zebra fish, its human ortholog in mouse, and its fugu ortholog in zebra fish (Woolfe et al. 2007). Cases such as these illustrate the incredible fidelity with which evolution has conserved the activity of many extremely distantly related orthologous enhancers.
FIG. 7.
High fidelity conservation of activity of homologs of CNE6.05. This sequence shows midbrain expression for paralogous ZZ, HM, HM, and fugu sequences in zebra fish.
Supplementary Material
Supplementary data and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Axel Visel and Len Pennacchio for assistance with the mouse enhancer experiments and Jessica Lehoczky for assistance with the zebra fish paralog analysis. This research was supported by the National Institute of General Medical Sciences (grant GM82901), an early career award from the Alfred P. Sloan Foundation, and National Institutes of Health (grants NS042626 and HD051835). J.H.C. was also supported by a PhRMA Foundation Informatics Research Starter Grant.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio L.A, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Fredman D, Arner E, Dong X, Bryne J, Suzuki H, Daub C, Hayashizaki Y, Lenhard B. Transcriptional features of genomic regulatory blocks. Genome Biol. 2009;10:R38. doi: 10.1186/gb-2009-10-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Huynh JL, Lee-Lin S-Q, et al. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, et al. A Library of Yeast Transcription Factor Motifs Reveals a Widespread Function for Rsc3 in Targeting Nucleosome Exclusion at Promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, et al. Variation in Homeodomain DNA Binding Revealed by High-Resolution Analysis of Sequence Preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tenna JJ, de la Calle-Mustienes E, et al. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- Bird C, Stranger B, Liu M, Thomas D, Ingle C, Beazley C, Miller W, Hurles M, Dermitzakis E. Fast-evolving noncoding sequences in the human genome. Genome Biol. 2007;8:R118. doi: 10.1186/gb-2007-8-6-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Chan E, Quon G, Chua G, et al. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P, Fredman D, Lenhard B. Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome Biol. 2008;9:R34. doi: 10.1186/gb-2008-9-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Gehrig J, Reischl M, Kalmar E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Muller F. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat Methods. 2009;6:911–916. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarinova O, Hatch G, Poitras L, Prudhomme C, Grzyb M, Aubin J, Bérubé-Simard FA, Jeannotte L, Ekker M. Functional resolution of duplicated hoxb5 genes in teleosts. Development. 2008;135(21):3543–3553. doi: 10.1242/dev.025817. [DOI] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, Bancewicz RM, Gautier P, et al. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 2008;4:e29. doi: 10.1371/journal.pgen.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ritter D, Yang N, Dong Z, Li H, Chuang JH, Guo S. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Developmental Biol. 2009;337:484–495. doi: 10.1016/j.ydbio.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen GK, Goode DK, Parker HJ, Woolfe A, Callaway H, Elgar G. Early evolution of conserved regulatory sequences associated with development in vertebrates. PLoS Genet. 2009;59:e1000762. doi: 10.1371/journal.pgen.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, McCallion AS. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, Becker TS. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Developmental Biol. 2009;327:526–540. doi: 10.1016/j.ydbio.2008.10.044. [DOI] [PubMed] [Google Scholar]
- Navratilova P, Fredman D, Lenhard B, Becker T. Regulatory divergence of the duplicated chromosomal loci sox11a/b by subpartitioning and sequence evolution of enhancers in zebrafish. Mol Genet Genomics. 2010;283:171. doi: 10.1007/s00438-009-0503-1. [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucl Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persampieri J, Ritter DI, Lees D, Lehoczky J, Li Q, Guo S, Chuang JH. cneViewer: a database of conserved non-coding elements for studies of tissue-specific gene regulation. Bioinformatics. 2008;24:2418–2419. doi: 10.1093/bioinformatics/btn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of non-neutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: 2005. ISBN 3-900051-07-0, Available from http://www.R-project.org. [Google Scholar]
- Rahmann S, Muller T, Vingron M. On the power of profiles for transcription factor binding site detection. Stat Appl Genet Mol Biol. 2003;2 doi: 10.2202/1544-6115.1032. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J Mol Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, et al. The zebrafish information network: the zebrafish model organism database. Nucl Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S, Pheasant M, Makunin IV, Mattick JS. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the Molecular Clock. Mol Biol Evol. 2008;25:402–408. doi: 10.1093/molbev/msm268. [DOI] [PubMed] [Google Scholar]
- Strähle U, Rastegar S. Conserved non-coding sequences and transcriptional regulation. Brain Res Bull. 2008;75:225. doi: 10.1016/j.brainresbull.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, et al. ChiP-Seq accurately predicts tissue specificity of enhancers. Nature. 2009;454:854. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Bristow J, Pennacchio LA. Enhancer identification through comparative genomics. Semin Cell Dev Biol. 2007;18:140. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio L. VISTA enhancer browser—a database of tissue-specific human enhancers. Nucl Acids Res. 2007;35(Suppl 1):D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lee AP, Kodzius R, Brenner S, Venkatesh B. Large number of ultraconserved elements were already present in the jawed vertebrate ancestor. Mol Biol Evol. 2009;26:487–490. doi: 10.1093/molbev/msn278. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VLJ, Fisher EMC, Tavare S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goode D, Cooke J, Callaway H, Smith S, Snell P, McEwen G, Elgar G. CONDOR: a database resource of developmentally associated conserved non-coding elements. BMC Dev Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.