Abstract
Complex morphological or functional traits are frequently considered evolutionarily unique and hence useful for taxonomic classification. Flea beetles (Alticinae) are characterized by an extraordinary jumping apparatus in the usually greatly expanded femur of their hind legs that separates them from the related Galerucinae. Here, we examine the evolution of this trait using phylogenetic analysis and a time-calibrated tree from mitochondrial (rrnL and cox1) and nuclear (small subunits and large subunits) genes, as well as morphometrics of femora using elliptic Fourier analysis. The phylogeny strongly supports multiple independent origins of the metafemoral spring and therefore rejects the monophyly of Alticinae, as defined by this trait. Geometric outline analysis of femora shows the great plasticity of this structure and its correlation with the type and diversity of the metafemoral springs. The recognition of convergence in jumping apparatus now resolves the long-standing difficulties of Galerucinae–Alticinae classification, and cautions against the value of trait complexity as a measure of taxonomic significance. The lineage also shows accelerated species diversification rates relative to other leaf beetles, which may be promoted by the same ecological factors that also favour the repeated evolution of jumping as an anti-predation mechanism.
Keywords: predator avoidance, Chrysomelidae, homoplasy, parallel evolution, species richness
1. Introduction
Predator–prey interactions are well established as a driving force in organismal evolution, and predator-related natural selection may bring about major changes in morphology and behaviour in evolutionary lineages [1]. Yet, the mechanisms determining the outcome of these evolutionary interactions are unclear: prey and predators may change in response to each other in a coevolutionary ‘arms race’ [2], while major predators may trigger the appearance of changes in design in the prey through episodic ‘escalation’ [3,4]. For beetles, varied strategies exist which function to avoid predation. Chemical defence is prevalent in various lineages and has long been recognized as prone to convergent evolution [5–7]. However, the role of anti-predator strategies in driving morphological adaptation is less clear. Leaf beetles (Chrysomelidae) offer a good system to study predator-induced morphological adaptation and the particular conditions under which complex defensive traits originate. Chrysomelids display a wide range of avoidance behaviours, including flying, running, dropping, feigning death, mimetic concealing and, largely unique to this group, jumping [8].
The ability to jump is a particularly interesting predator avoidance behaviour that has been shown to be very effective [9]. Take-off accelerations range from 15 to 270 times gravity, and some species are able to control jump direction and landing [10]. Within the Chrysomelidae, jumping ability is associated with dilated hind femora, which is largely confined to the subfamily Alticinae (‘flea beetles’), a group of more than 10 000 species. Jumping in flea beetles is enabled by an enlarged metafemoral endosclerite, known as ‘Maulik's organ’, ‘metafemoral spring’ or ‘Costa Lima's organ’ [9,11]. It is made of polysaccharide α-chitin together with protein, and generally consists of dorsal lobes attached to the metatibia and ventral lobes extended into a recurved flange [12,13]. The evolutionary history of this striking structure and its consequences for the species diversity of this group have not been studied.
Yet, the jumping mechanism has featured prominently in the classification of Chrysomelidae, as the presence of the metafemoral spring together with the dilation of hind femora are considered the chief characters distinguishing the Alticinae from the closely related Galerucinae [12,14–17]. In addition, variability in this trait has been considered informative for the classification of this group; e.g. taxa bearing ‘simple’ springs without or with weakly developed ventral lobes may represent a primitive state [15,16,18,19]. However, these and other attempts to clarify the basal relationships of the flea beetles using morphological traits, life-history traits of adults and larvae, and host plant associations have proven inconclusive, raising questions about the validity of the metafemoral spring for higher level classification [15,20]. Because of these inconsistencies of character variation, many genera have been referred to as ‘transitional’, ‘incertae sedis’, ‘members of the Galerucinae–Alticinae complex’ or simply ‘problematic’ [20–24].
Recent phylogenetic work on flea beetles (Alticinae) resulted in conflicting phylogenetic hypotheses regarding their relationship with the Galerucinae. Three alternatives have been proposed: (i) a sister group relationship of Alticinae and Galerucinae; this was supported by molecular analyses [25] combined with morphological data [26], and extended studies with additional taxa and genes showed similar results, but cautioned about insufficient sampling of the ‘problematic’ taxa [27,28]. (ii) A monophyletic Alticinae arising from within a paraphyletic Galerucinae; this was supported by morphological characters from 12 species of flea beetles and 10 species of Galerucinae [29]. (iii) The alternative paraphyletic scenario, a monophyletic Galerucinae within a grade of flea beetles; this was obtained by various morphological and molecular analyses [30–32], including recent phylogenetic studies of galerucines and the evolution of their chemical defence [33,34]. However, taxon sampling in each of these studies did not greatly overlap, and the conflicting results might reflect the differences in selection of exemplar taxa from greater than 15 000 species and greater than 1000 genera in both subfamilies combined. Existing molecular studies have at most included 28 flea beetle genera, with focus on Palaearctic and Nearctic taxa, leaving vast taxonomic and geographical gaps and a lack of comparability among studies.
In order to address the inter-related questions about classification and evolution of defensive strategies in the Galerucinae–Alticinae clade, gene sequences were generated for an expanded taxon set and for additional markers, with an emphasis on several genera considered ‘problematic’. The phylogenetic analysis reveals multiple origins of the metafemoral spring, demonstrating that an evolutionary classification based on this trait is not defensible, and disentangles ancestral and derived states in the morphological variation of this complex structure in the face of evolutionary convergence.
2. Material and methods
(a). Sampling and DNA sequencing
Taxon coverage had to be selective but attempted to (i) complement existing data mainly from the Holarctic, Oriental, Afrotropical regions and included several cosmopolitan genera, (ii) cover most suprageneric variation following the previous attempts of classification, and (iii) put particular emphasis on sampling of taxa whose status in the Galerucinae–Alticinae classification had been identified as ‘problematic’, following various literature sources that raise their inconclusive affinities or character combinations. Sampling included taxa from groups designated based on spring type by Furth and co-workers for the Palaearctic and Nearctic regions [18], the Neotropics [19] and the Oriental-Australian region [20] (electronic supplementary material, table S1). This selection also represents 60 per cent of the groups of genera (42 of 70) listed by Seeno & Wilcox [35] and has a high coverage of higher taxa established by previous studies including Horn [36], Leng [37] and Takizawa [38].
Most samples were preserved in 95 per cent ethanol after field collection but a few dry samples were also used in this study. DNA was extracted using a standard proteinase K digestion following guidelines and products from QIAGEN. Four genes were used for the PCR amplification, including the small (SSU) and large (LSU) subunit ribosomal RNA genes, and the mitochondrial 16S rRNA (rrnL) and cytochrome oxidase subunit 1 (cox1) genes. Primers are listed in the electronic supplementary material, table S2. Sequences were obtained with standard DNA sequencing and BigDye 3.1 technology (Applied Biosystems). New sequences were deposited in GenBank. Specimen voucher details and sequence accession numbers can be found in the electronic supplementary material, table S3.
(b). Phylogenetic analyses and divergence time estimate
Phylogenetic methodology largely followed Gómez-Zurita et al. [27], and involved extensive tests of the effect of length variation in rRNA sequences on the tree topology. The preferred alignment was obtained with Prank [39]. The resulting aligned data matrix of four markers was used for tree searches under maximum likelihood with RAxML [40]. Divergence times were estimated using the Bayesian relaxed molecular clock methods in BEAST 1.4.7 [41], again under six partitions and an uncorrelated lognormal prior model of rate change. The age of Chrysomelinae, Galerucinae and Alticinae was used as prior to estimate the divergence time of representative nodes, which was obtained from recent publications [27,28], and 95% CI based on the current data matrix. Two separate Bayesian analyses with 4 × 107 generation were performed and finally combined and further annotated with 10 per cent pre-burn-in to get the final dated tree and posterior probabilities in Tracer 1.4.1, TreeAnnotator 1.4.8, LogCombiner 1.4.8 (http://beast.bio.ed.ac.uk/) and displayed in FigTree 1.2.2 (http://tree.bio.ed.ac.uk/).
(c). Morphological analysis of metafemoral spring
A hind leg from DNA voucher specimens or other representative of the same species was incubated in cold 10 per cent KOH for 2 days. The metafemoral spring was removed from the femoral capsule with a fine needle and photographed under a microscope, then washed in distilled water and preserved in 75 per cent ethanol. The Shape package [42] was used for image analysis and visualization of femora outline shapes and elliptic Fourier analysis. The shape of femora was approximated by the first 20 harmonics, which related to 77 coefficients of normalized elliptic Fourier descriptors, using Principal Component Analysis (PCA) on a variance–covariance matrix of the coefficients. Correlation among the shape variation of the femora and the four states of the spring were calculated using normalized Mantel test in NTSYSpc with 999 permutations [43].
(d). Hypothesis testing
Spring types were categorized as (i) ventral lobe absent, (ii) the length of the ventral lobe reduced to at most half the length of the spring, or (iii) the ventral lobe extended beyond half the length of the spring and (iv) the recurved flange fully developed (figure 1). These states were mapped on the Bayesian trees pruned to include only the in-group and a single representative per genus in MacClade 4 [44]. The phylogenetic distribution of this trait was assessed against the number of changes for the available states reshuffled 1000 times on the original trees. Alternative hypotheses were tested by site bootstrapping using scores derived from individual columns in a multiple sequence alignment [45]. The top ranking topology was identified for each bootstrapped dataset under the likelihood criterion. For each hypothesis, a heuristic search of 5000 rearrangements was run under ML, with random stepwise sequence addition in PAUP* [46]. Individual site likelihoods were the output for bootstrap analysis and statistical testing in Consel [45]. This software assesses support for each topology as the length of the bootstrapped sequence changes and outputs p-values for an approximately unbiased test (AU), Bootstrap probability tests (NP, BP and PP), Shimodaira–Hasegawa test (SH), and weighted Shimodaira–Hasegawa test (WSH). We used the 10 default scaling factors of 0.5–1.4, with 10 000 pseudoreplicates for each.
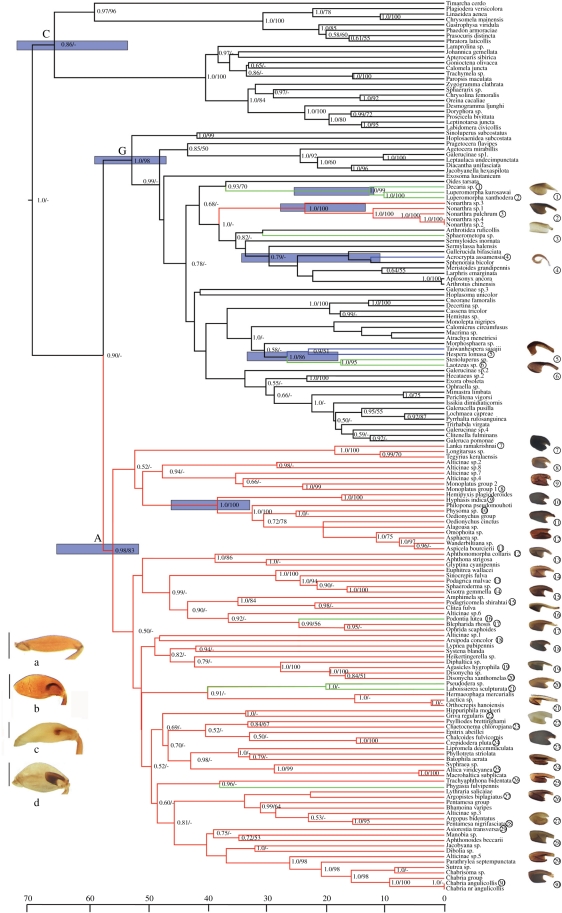
Figure 1.
Maximum clade credibility tree for flea beetles based on Bayesian analysis. Bayesian posterior probabilities (PP) ≥ 0.50 and maximum-likelihood bootstrap (BS) values ≥ 50% are shown on the branches (PP/BS). 95% CI for the ages of important nodes are indicated with blue shaded bars. Images in the bottom left show hind legs of four species to illustrate different states of metafemoral spring, illustrated by (a) Mimastra limbata, (b) Hespera sp., (c) Laboissierea sculpturata and (d) Clitea fulva (scale bar, 0.5 mm). Branches are coloured according to the parsimony optimization of these states. Numbers following the taxon names correspond to the metafemoral springs to the right. See electronic supplementary material, figure S1 for enlarged images. Large letters mark the major lineages of Chrysomelinae (C), Galerucinae (G) and Alticinae (A). Black lines, without spring; blue lines, spring simple, without ventral lobe; green lines, spring simple, ventral lobe no longer than half of spring; red lines, spring well developed, ventral lobe beyond half of spring.
(e). Diversification rate comparison
Measures of imbalance in species numbers for sister clades with and without metafemoral spring were performed with the Slowinsky–Guyer test [47] on a phylogenetic hypothesis for the Chrysomelidae [27,28] and an alternative hypothesis that reflects the findings from the current study. Statistical analysis of a stochastic birth–death process was performed using moment estimators [48,49]. The ages of subfamilies in Chrysomelidae estimated in the present study and elsewhere [27,28] were used to derive the expected diversification rates, calculating 95% CI for expected numbers across different ages with extremely low (ɛ = 0) or high (ɛ = 0.9) extinction rates.
3. Results
Phylogenetic analyses were based on nuclear ribosomal (SSU = 1351–1832 bp; LSU = 634–672 bp) and mitochondrial ribosomal (rrnL = 391–457 bp) and protein coding (cox1 = 629–782 bp) loci. Concatenation of the four loci resulted in an aligned data matrix of 4306 bp for 165 species, representing 83 species (81 genera) of Alticinae and 58 species (53 genera) of Galerucinae, in addition to 24 outgroup taxa. Bayesian phylogenetic analysis (figure 1) showed most species of Alticinae in a well-supported clade. Within this clade, we defined 16 groups of genera, each represented by between two and 10 genera, based on broadly supported lineages whose composition resembles that of suprageneric groups established in the earlier literature but whose precise composition differed in many cases [36,37,50]. Interestingly, support for these groups did not match well with the groups established based on metafemoral spring type [18–20] (electronic supplementary material, table S1), i.e. similarity in this trait does not reflect phylogenetic history.
The alticine clade was sister to all taxa classified as Galerucinae, but the latter group also included five separate lineages of spring-bearing beetles considered members of Alticinae based on this character. These included genera characterized by either a well developed or a simple metafemoral spring, including Hespera, Laotzeus, Stenoluperus, Luperomorpha, Decaria, Nonarthra, Acrocrypta, Taiwanhespera and Sphaerometopa. Among these, Hespera, Luperomorpha and Nonarthra exhibit elytron-to-body meshing structures of the Galerucinae type, which rendered their position within the ‘Galerucinae–Alticinae complex’ unclear [23], while for Stenoluperus affinities to the galerucine subtribe Luperina had already been recognized in morphological cladistic analysis [29]. Luperomorpha and Nonartha show similarity with galerucines in genitalic characters [21], and the metafemoral springs of Acrocrypta and Sphaerometopa were reported to constitute unique morphogroups [20]. In the DNA-based tree, the ‘problematic’ taxa were placed in the Luperini, Sermylini and as sister to the single representative of Oidini (Oides tarsata), in all cases with good support (figure 1). Among ‘problematic’ genera, some cases were also recovered with affinities to the main spring-bearing branch. For example, Phygasia, which was considered ‘problematic’ by Wilcox [24] and shows simplified springs but alticine type spermatheca and hind wing venation [21] were found included in the main branch of Alticinae. Likewise, Clitea, which exhibits spermatheca of the galerucine type [20] was found associated with the alticine Amphimela group.
Hypothesis testing on constrained tree topologies (table 1) strongly rejected the monophyly of all taxa with metafemoral spring (hypothesis 1.6 in table 1); the monophyly of the traditional Galerucinae excluding ‘problematic’ genera (hypothesis 1.4 in table 1); and the monophyly of all taxa classified as ‘problematic’ (hypothesis 1.5). In addition, we confirmed the monophyly of Alticinae + Galerucinae, by rejecting the close sister relationships of the Chrysomelinae either with Alticinae (in the new sense without the incertae sedis taxa; hypothesis 1.2) or Galerucinae (in the new sense with incertae sedis taxa included; hypothesis 1.3); or in the traditional sense, i.e. all taxa without spring) relative to Chrysomelinae (hypothesis 1.1), although the latter three hypotheses were not rejected in all of the tests (table 1).
Table 1.
Results of statistical tests on site likelihoods generated under constrained tree searches. The first column gives the topological constraint under which the site likelihoods were produced, where C, Chrysomelinae; A, Alticinae; G, Galerucinae and S, incertae sedis with spring present. Columns show the observed log-likelihood difference of trees (OBS), the results from approximately unbiased tests (AU); bootstrap probability of item/hypothesis (NP); non-scaled bootstrap probability (BP) and the Bayesian posterior probability (PP) calculated by the Bayesian Information Criterion (BIC) approximation, and the (weighted) Shimodaira–Hasegawa test (SH and WSH). See main text for a narrative of the tested hypotheses.
| hypothesis | OBS | AU | NP | BP | PP | SH | WSH | |
|---|---|---|---|---|---|---|---|---|
| 1.0 | no constraint | −56.3 | 0.969 | 0.866 | 0.874 | 1.000 | 0.996 | 0.998 |
| 1.1 | (((G,S),C),A) | 56.3 | 0.119 | 0.075 | 0.070 | 4e−25 | 0.416 | 0.267 |
| 1.2 | ((A,C),G,S) | 78.1 | 0.066 | 0.033 | 0.029 | 1e−34 | 0.250 | 0.146 |
| 1.3 | ((G,C),A,S) | 110.7 | 0.056 | 0.015 | 0.016 | 8e−49 | 0.076 | 0.093 |
| 1.4 | ((G),A,S,C) | 129.6 | 0.016 | 0.004 | 0.004 | 5e−57 | 0.032 | 0.042 |
| 1.5 | ((S),A,C,G) | 137.0 | 0.024 | 0.006 | 0.006 | 3e−60 | 0.033 | 0.045 |
| 1.6 | ((A,S),G,C) | 181.5 | 0.002 | 3e−04 | 0.001 | 1e−79 | 0.003 | 0.004 |
Morphological analysis of metafemoral springs revealed several types. For example, the springs of Decaria (labelled ‘1’ in the electronic supplementary material, figure S1) and Luperomorpha (‘2’ in the electronic supplementary material, figure S1) are spoon-like with the ventral lobe shorter or reduced to nearly half the length of the spring. In the closely related Podagrica (‘13’ in the electronic supplementary material, figure S1) and Nisotra (‘14’ in the electronic supplementary material, figure S1) the recurved flange is developed and the ventral lobe extended beyond half the length of the spring. Almost all taxa represented here bear ventral lobes, while Acrocrypta and Hespera exhibit extremely simplified springs (‘4’ and ‘5’ in the electronic supplementary material, figure S1), the former hook-shaped and the latter narrowly sheet-like. Plotting the shape of metafemoral springs on the tree using these three well-defined states and the absence of springs as a fourth state, clearly indicated the polyphyly of each of the spring types (and the polyphyly of the spring-bearing taxa themselves; figure 1 and electronic supplementary material, figure S1). The phylogenetic distribution of the spring was significantly non-random (p < 0.001), whether considered a binary (presence/absence) or multi-state (four states) character on the whole topology. Within the galerucines, five independent origins of the various spring types were inferred. Conversely, three groups with relatively simple springs (the Blepharida, Pseudodera and Phygasia groups of genera) were derived from within the Alticinae, indicating that this type of spring can also arise owing to reversals from the fully developed spring.
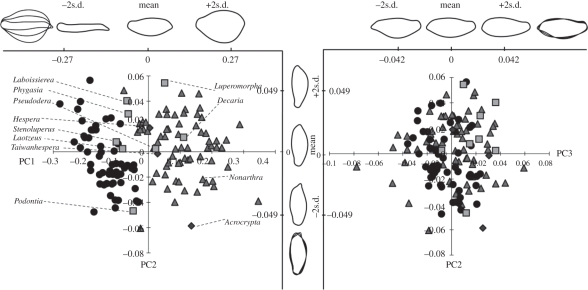
PCA on the shape variation of the femora showed that the first three principal components captured 96.4 per cent of the shape variation. The effect of the first three components on femoral shape variation was visualized through estimating elliptic Fourier descriptors inversely with mean and extreme values (±2° for each principal component [51]; figure 2). The presence of a metafemoral spring had a great impact on the shape of the femora and showed a largely non-overlapping distribution of taxa with and without spring along axis 1, which contributes 91.3 per cent of the total shape variation. Taxa with ‘simple’ springs generally assumed positions near the centre of the variation along axis 1 (intermediate between Alticinae and Galerucinae types). These include the secondarily simplified femora of the Blepharida, Pseudodera and Phygasia groups. Taxa with simple springs showed a great range of variation along axis 2 (although this is responsible for only 2.92% of the variation). The shape of femora in these taxa was not dependent of the phylogenetic associations, nor was it linked to the specific state of the extension lobe of the metafemoral spring (figure 2). The Mantel test also indicates significant correlation between shape variation of femora and metafemoral spring (r = 0.604, p = 0.002). This analysis indicates considerable plasticity of femur shape and shows that the variation is largely predictive of the metafemoral type (absent, simple, etc.).
Figure 2.
PCA based on elliptic Fourier analysis of hind femora. The inset shows the shape variation (mean and extreme values are figured) of the first three principal components. Taxa with simple springs are labelled and their names grouped according to the subclades in figure 1. Circles, metafemoral spring absent; diamonds, spring simple, without ventral lobes; squares, ventral lobes no longer than half of spring; triangles, spring well developed, ventral lobe beyond half of spring.
Dating the divergence time of Chrysomelinae, Alticinae and Galerucinae and several important nodes on the tree with dates selected from recent publications [27,28] as priors showed the age of Chrysomelinae at 61.9 Ma (95% CI range 52.6–71.0 Ma) versus 58.5 and 53.3 Ma for Alticinae and Galerucinae, respectively. Lineages with simple springs that had been thought to occupy early branches owing to their supposedly primitive metafemoral springs [15,16,18,19], showed a much more recent origin (from 18.2 Ma for the Luperomorpha group to 26.2 Ma for the Hespera group) when compared with the main branch of Alticinae (figure 1 and electronic supplementary material, table S4).
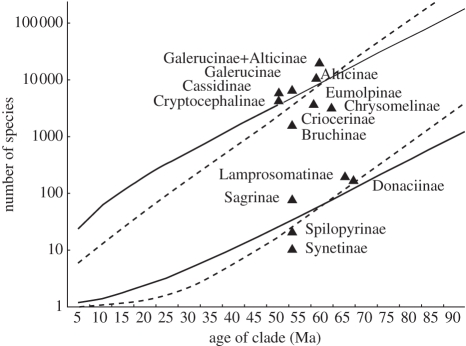
Numbers of species and genera for each subfamily were estimated based on various sources [24,35,52,53]. The Alticinae (570 genera, 10 000 species) are the most species-rich group in Chrysomelidae, and together with Galerucinae (488 genera, 6000 species), their diversity is much greater than observed in their sister clade, Chrysomelinae (150 genera, 3000 species). This difference was not significant in the Guyer–Slowinski test (electronic supplementary material, table S5), but the expected diversification rate of alticines and galerucines through time was above the confidence interval obtained from diversification rates estimated for major clades (subfamilies) of Chrysomelidae (figure 3 and electronic supplementary material, table S6, S7).
Figure 3.
Actual and expected species diversity for the subfamilies of Chrysomelidae. The x-axis gives the age of clades and y-axis gives the species numbers. The upper and lower 95% CI are shown for the expected species of a clade with either no extinction (dashed line) or a high (90%) extinction rate (solid line).
4. Discussion
Among the most important tasks in reconstructing phylogenetic relationships is to separate characters that have arisen through convergent evolution from those that share common ancestry, i.e. to distinguish homoplasy from homology [54]. The evolution of defensive traits has already been recognized to be more likely to result in trait diversity than selection related to capture in the predator, and instances of convergence and parallelism in prey evasion mechanisms are more common than generally realized [55]. Accordingly, based on phylogenetic evidence, we find clear support for multiple origins of the complex jumping mechanism in the galerucines–alticines. Metafemoral springs, albeit less highly developed (i.e. of the ‘simple’ type), exist in unrelated families of beetles including certain subfamilies of Curculionidae and Buprestidae [56], and therefore, the finding of multiple origins even within a particular family should be less surprising. The findings also explain the contradictory character states in some ‘problematic’ taxa. For example, Furth & Suzuki's [21] study of Oriental groups included three taxa used in our study. While they found Luperomorpha and Nonartha to show ‘galerucine tendencies’ in at least one of the four character systems investigated, all character systems were ‘typical Alticinae’ in Phygasia, strongly corroborating the phylogenetic position established here. The Alticinae–Galerucinae lineage is unique among leaf beetles in presenting this trait, and its recurrent appearance suggests an inherent propensity to acquire this specialized jumping apparatus, constituting an example of a parallelophyletic character as defined by Mayr & Ashlock [57].
The evolutionary scenario supported here suggests that the absence of the metafemoral spring is the ancestral state in the Galerucinae–Alticinae lineage (figure 1). A major early event gave rise to the great majority of jumping species and the most complex structural features in the Alticinae (sensu stricto), while later acquisitions from within the ‘galerucines’ generally show simplified features that were previously interpreted as an early evolutionary state [9] but here are established to be of fairly recent origin (figure 1). Likewise, on two occasions simplified spring types have arisen from the complex type; as those in the incertae sedis taxa, they also do not constitute a transitional state that reflects an ancestral condition of the Alticinae. Close relatives usually share similar types of spring, in accordance with the various studies by Furth (e.g. [9,16]) who established the metafemoral spring types to be conserved within genera. However, on a higher hierarchical level the various spring-bearing groups differ in the detailed design, again in accordance with earlier studies [16], and reveal a great deal of homoplasy (figure 2). The evolutionary plasticity of the metafemoral spring type was also evident from the lack of congruence of phylogenetic groups defined here (figure 1) with those groups established based on the similarity of metafemoral spring type (electronic supplementary material, table S1).
In addition, the morphometric analysis on the hind legs further supports the homoplasy of this trait. The four spring types were correlated with variation in the shape and proportions of the hind femora that harbour these springs, showing a good correlation of metafemoral shapes with the classification of presence or absence of the metafemoral spring and a clear trend from elongated femur shapes in taxa with simple springs to the highly extended shapes correlated with complex springs along axis 1 (figure 2). Yet, within these classes, various lineages greatly differ along axes 2 and 3, representing the great diversity in the morphology of the hind legs. This is also reflected in a Mantel test for the correlation of morphometric variation in the femur shape with the four discrete classes of springs, which was found to be significant. Yet, the correlation is incomplete (r = 0.604) suggesting that femur shape may vary independently of the metafemoral spring, while equally the variation within the four types is comparatively large and is not perfectly captured by discrete states (and the definition of the character states may be improved). Femur shape is probably a reflection of the musculature, which in turn is a reflection of the jumping ability, and function is also closely linked with the type of the metafemoral spring. Therefore, broadly the variation can be captured in the way performed here and tested for the fit with the phylogeny. The fact that the fit of femur shape and spring type is not close may be additional evidence for the high evolutionary plasticity of the jumping apparatus.
The difficulty of past workers to establish a satisfactory classification of Alticinae and Galerucinae may be explained by the focus on the metafemoral spring and the expansion of femora as key characteristics for separating the major lineages. Previous DNA-based studies were unable to resolve this problem because of limited taxon sampling, either owing to the omission of incertae sedis groups, biased taxon selection focused on galerucines, or minimal sampling of both galerucines and alticines that prevented the recognition of their non-monophyly (see §1). The current sampling scheme that included a great diversity of genera and focused on taxa with metafemoral springs of various kinds, revealed the true level of homoplasy in this character. While the shape of this structure is conserved within genera [16] and possibly related genera (figure 1), the plasticity of this structure and the general incongruence of spring-based groups with the phylogeny at higher hierarchical levels indicate that hind leg characters are not a reliable criterion for establishing a suprageneric classification, because they have been subjected to extensive natural selection leading to repeated functional diversification evident from the presence of spurs, grooves and swollen metatarsal segments.
It remains to be tested what drives the repeated origin of this trait and whether a well-developed metafemoral spring would be correlated with impaired alternative defence mechanisms (e.g. reduced toxicity) and ultimately how this trait might be coupled with the high diversification rate of this group. The number of species in Galerucinae and Alticinae by far exceeds that in other subfamilies of Chrysomelidae, in particular, considering their comparatively recent origin, while nearly two thirds of the species exhibit the metafemoral spring (electronic supplementary material, table S7). In this respect, the metafemoral spring could be considered an evolutionary ‘key innovation’ [58] that links the origin of a trait to high species diversity in a clade. However, imbalance of diversification rates associated with the appearance of this structure was not significant in the (highly conservative) Slowinsky–Guyer test, while high species richness is also evident in the sister clade that ancestrally lacks the metafemoral spring, i.e. the shift to higher diversification rates precedes the origin of the trait. This may suggest that the selective regime favouring the origin of this mechanism for predator avoidance is also associated with life traits that favour species diversification.
The great diversity of beetles has been ascribed to the greater niche diversity associated with the angiosperm diversification [25,59–61]. If true, the impact of these parameters would be greatest in the Galerucinae–Alticinae clade as the fastest-diversifying lineage of Chrysomelidae. Most species are root feeders in the larval stages, which are lifestyles that correlate with high species numbers in other groups of beetles [62]. In contrast, most larvae of the sister group, Chrysomelinae, are phyllophagous and in most species, the larvae feed together with the adults, relying on aggregation of chemically defended individuals of all life stages for predator avoidance. The trend to repeated acquisition of jumping abilities in this group may be related to decoupled larval and adult life histories in most flea beetles (although exceptions exist in the large-bodied species of Altica, Blepharida and others), as the concealed lifestyle of larvae that disassociates them ecologically from the adults may no longer prohibit the evolution of efficient escape mechanisms in the adults. In addition, the small body size common to most flea beetle genera may make the physics of jumping more effective. Hence, the link of escape mechanism and high species numbers may be more complex than suggested by a simple definition of ‘key innovation’, but an indirect link of the particular mode of larval plant utilization (root feeding and, to lesser degree, leaf mining) may favour both the high species richness and the evolution of adult escape mechanisms. Given the higher species richness in the jumping lineage compared with its sister (figure 3 and electronic supplementary material, table S7), the presence of the jumping mechanism may reinforce the greater diversification rate.
5. Conclusion
Ever since the discovery of metafemoral springs in Chrysomelidae, the phylogenetic relationship of galerucines and alticines has been controversial. We show here that this important structure related to anti-predation behaviour is in fact susceptible to rapid diversification and convergent evolution, rather than being an incrementally varying trait [20]. A diverse predator environment as experienced by exposed phyllophages is unlikely to result in mutual coevolutionary changes in prey and predators. Hence, a scenario of a major shift in design as postulated under the hypothesis of escalation [4] seems more plausible. This shift is associated directly or indirectly with the spectacular species richness of this group, perhaps confirming that species with well-developed defences are more prone to speciation [3,4]. Competition and predation have been suggested as the main mechanisms driving the diversification of lineages [63]. While the former received a lot of attention, the importance of predation has been underestimated. However, phenotypic diversity induced by predation is widespread in nature and includes changes in the prey's life history, variation in size and shape of particular organs, or phenological changes [63]. Unexpectedly, as shown in the present study, these morphological shifts can originate repeatedly in a lineage, and most of the trait diversity results from independent origins, rather than secondary refinement of an existing design. The structural and functional complexity of these traits has been considered as evidence against their convergent evolutionary history, but this assumption of the traditional classification is refuted here.
Acknowledgements
We acknowlegde Shuyong Wang, Hongbin Liang, Chaodong Zhu, Manfred Doberl, Alex Konstantinov, Maurizio Biondi, Jan Bezdek, K. D. Pranthapan, Sharon Shute, Max Barclay and particularly David Furth for help with obtaining and identifying samples and for constructive suggestions. Siqin Ge, Huaijun Xue, Ming Bai, Yong Zhang, Wenzhu Li and Junzhi Cui contributed to field collections. We thank Drs M. J. Sanderson and C. W. Wheat for their suggestions on diversification rate calculation. We are deeply indebted to Prof. Yapin Zhang for his help in the early stage of this project, as well as Norman Macleod, Jonathan Krieger and Hiroyoshi Iwata for their suggestions on the morphometrics analysis. We also appreciate comments by Prof. I. Cuthill and two anonymous reviewers. This project was partially supported by the Innovation Project of the Chinese Academy of Sciences (no. KSCX2-YW-Z-015), National Science Foundation of China (no. 30970393 and 31010103913), a grant from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences (no. O529YX5105) and The Leverhulme Trust (F/00 696/P). This project also received support from the Chinese state scholarship council. J.G.Z. received support from project no. CGL2008-0007 (Spanish Ministry of Science and Innovation). D.C. was funded by a NERC CASE studentship.
References
- 1.Vermeij G. J. 1994. The evolutionary interaction among species: selection, escalation, and coevolution. Ann. Rev. Ecol. Evol. Syst. 25, 219–236 [Google Scholar]
- 2.Ehrlich P. R., Raven P. H. 1964. Buttertlies and plants: a study in coevolution. Evolution 18, 586–608 10.2307/2406212 (doi:10.2307/2406212) [DOI] [Google Scholar]
- 3.Dietl G. P. 2003. The escalation hypothesis: one long argument. Palaios 18, 83–86 (doi:10.1669/0883-1351(2003)18<83:TEHOLA>2.0.CO;2) [DOI] [Google Scholar]
- 4.Vermeij G. J. 1987. Evolution and escalation: an ecological history of life. Princeton, NJ: Princeton University Press [Google Scholar]
- 5.Kopf A., Rank N. E., Roininen H., Julkunen-Tiitto R., Pasteels J. M., Tahvanainen J. 1998. The evolution of host-plant use and sequestration in the leaf beetle genus Phratora (Coleoptera: Chrysomelidae). Evolution 52, 517–528 10.2307/2411087 (doi:10.2307/2411087) [DOI] [PubMed] [Google Scholar]
- 6.Metcalf R. L., Lampman R. L. 1991. Evolution of Diabroticite rootworm beetle (Chrysomelidae) receptors for Cucurbita blossom volatiles. Proc. Natl Acad. Sci. USA 88, 1869–1872 10.1073/pnas.88.5.1869 (doi:10.1073/pnas.88.5.1869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogler A. P., Kelley K. C. 1998. Covariation of defensive traits in tiger beetles (genus Cicindela): a phylogenetic approach using mtDNA. Evolution 52, 529–538 10.2307/2411088 (doi:10.2307/2411088) [DOI] [PubMed] [Google Scholar]
- 8.Ohno T., Miyatake T. 2007. Drop or fly? Negative genetic correlation between death-feigning intensity and flying ability as alternative anti-predator strategies. Proc. R. Soc. B 274, 555–560 10.1098/rspb.2006.3750 (doi:10.1098/rspb.2006.3750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furth D. G. 1988. The jumping apparatus of flea beetles—the metafemoral spring. In Biology of Chrysomelidae (eds Jolivet P., Petitpierre E., Hsiao T.), pp. 285–297 Dordrecht, The Netherlands: Kluwer Academic Publishers [Google Scholar]
- 10.Brackenbury J., Wang R. 1995. Ballistics and visual targeting in flea-beetles (Alticinae). J. Exp. Biol. 198, 1931–1942 [DOI] [PubMed] [Google Scholar]
- 11.Furth D. G. 1982. The metafemoral spring of flea beetles (Chrysomelidae: Alticinae). Spixiana 7, 11–27 [Google Scholar]
- 12.Chen S. 1985. Phylogeny and classification of the Chrysomeloidea. Entomography 3, 465–475 [Google Scholar]
- 13.Furth D. G., Traub W., Harpaz I. 1983. What makes Blepharida jump? A structural study of the metafemoral spring of a flea beetle. J. Exp. Zool. 227, 43–47 10.1002/jez.1402270107 (doi:10.1002/jez.1402270107) [DOI] [Google Scholar]
- 14.Barth R. 1954. O aparelho saltatorio do Halticineo Homophoeta sexnotata Har. (Coleoptera). Mem. Inst. Osw. Cruz 52, 365–376 [Google Scholar]
- 15.Furth D. G., Suzuki K. 1990. The metatibial extensor and flexor tendons in Coleoptera. Syst. Entomol. 15, 443–448 10.1111/j.1365-3113.1990.tb00076.x (doi:10.1111/j.1365-3113.1990.tb00076.x) [DOI] [Google Scholar]
- 16.Furth D. G. 1980. Inter-generic differences in the metafemoral apodeme of flea beetles (Chrysomelidae: Alticinae). Syst. Entomol. 5, 263–271 10.1111/j.1365-3113.1980.tb00413.x (doi:10.1111/j.1365-3113.1980.tb00413.x) [DOI] [Google Scholar]
- 17.Maulik S. 1929. On the structure of the hind femur in halticine beetles. Proc. Zool. Soc. Lond. 2, 305–308 10.1111/j.1469-7998.1929.tb07744x (doi:10.1111/j.1469-7998.1929.tb07744x) [DOI] [Google Scholar]
- 18.Furth D. G. 1985. Relationships of Palearctic and Nearctic genera of Alticinae. Entomography 3, 375–392 [Google Scholar]
- 19.Furth D. G. 1989. Metafemoral spring studies of some Neotropical genera of Alticinae. Entomography 6, 497–510 [Google Scholar]
- 20.Furth D. G., Suzuki K. 1998. Studies on Oriental and Australian Alticinae genera, based on the comparative morphology of the metafemoral spring, genitalia, and hind wing venation. In Proc. 4th Int. Symp. on the Chrysomelidae (eds Biondi M., Daccordi M., Furth D. G.), pp. 91–124 Tornino, Italy: Atti Museo Regionale Scienze Naturali [Google Scholar]
- 21.Furth D. G., Suzuki K. 1994. Character correlation studies of problematic genera of Alticinae in relation to Galerucinae. (Coleoptera: Chrysomelidae). In Proc. 3rd Int. Symp. on the Chrysomelidae (ed. Furth D. G.), pp. 116–135 Beijing, China: Backhuys Publishers [Google Scholar]
- 22.Reid C. A. M. 1992. The leaf-beetle genus Microdonacia Blackburn (Coleoptera: Chrysomelidae: Galerucinae): revision and systematic placement. Syst. Entomol. 17, 359–387 10.1111/j.1365-3113.1992.tb00557.x (doi:10.1111/j.1365-3113.1992.tb00557.x) [DOI] [Google Scholar]
- 23.Samuelson G. A. 1996. Binding sites: elytron-to-body meshing structures of possible significance in the higher classification of Chrysomeloidea. In Chrysomelidae biology, the classification, phylogeny and genetics (eds Jolivet P. H. A., Cox M. L.), pp. 267–290 Amsterdam, The Netherlands: SPB Academic Publishing [Google Scholar]
- 24.Wilcox J. A. 1971–1975. Chrysomelidae: Galerucinae. In Coleopterum Catalogus (ed. Schenkling S.), (Pars 78 Fasc. 1-4) 1-770. Berlin: W. Junk. [Google Scholar]
- 25.Farrell B. D. 1998. ‘Inordinate fondness’ explained: why are there so many beetles? Science 281, 555–559 10.1126/science.281.5376.555 (doi:10.1126/science.281.5376.555) [DOI] [PubMed] [Google Scholar]
- 26.Reid C. A. M. 1995. A cladistic analysis of subfamilial relationships in the Chrysomelidae sensu lato (Chrysomeloidea). In Biology, phylogeny and classification of Coleoptera: papers celebrating the 80th birthday of Roy A. Crowson (eds Pakaluk J., Slipinski S.), pp. 1559–1631 Warzawa: Muzeum i Instytut Zoologii Pan [Google Scholar]
- 27.Gómez-Zurita J., Hunt T., Vogler A. P. 2008. Multilocus ribosomal RNA phylogeny of the leaf beetles (Chrysomelidae). Cladistics 24, 34–50 10.1111/j.1096-0031.2007.00167.x (doi:10.1111/j.1096-0031.2007.00167.x) [DOI] [Google Scholar]
- 28.Gómez-Zurita J., Hunt T., Kopliku F., Vogler A. P. 2007. Recalibrated tree of leaf beetles (Chrysomelidae) indicates independent diversification of angiosperms and their insect herbivores. PLoS ONE 2, e360. 10.1371/journal.pone.0000360 (doi:10.1371/journal.pone.0000360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingafelter S. W., Konstantinov A. S. 1999. The monophyly and relative rank of alticine and galerucine leaf beetles: a cladistic analysis using adult morphological characters (Coleoptera: Chrysomelidae). Entomol. Scand. 30, 397–416 [Google Scholar]
- 30.Crowson R. A., Crowson E. A. 1996. The phylogenetic relations of Galerucinae—Alticinae. In Chrysomelidae biology, vol. 1: The classification, phylogeny and genetics (eds Jolivet P. H., Cox M. L.), pp. 97–118 Amsterdam, The Netherlands: SPB Academic Publishing [Google Scholar]
- 31.Duckett C. N., Gillespie J. J., Kjer K. M. 2004. Relationships among the subfamilies of Chrysomelidae inferred from small subunit ribosomal DNA and morphology, with special emphasis on the relationship among the Flea Beetles and the Galerucinae. In New developments in the biology of Chrysomelidae (eds Jolivet P., Santiago-Blay J. A., Schmitt M.), pp. 803–818 The Hague, The Netherlands: SPB Academic Publishing [Google Scholar]
- 32.Kim S. J., Kjer K. M., Duckett C. N. 2003. The evolution of host-plant use and sequestration in the leaf beetle genus Phratora (Coleoptera: Chrysomelidae). Insect Syst. Evol. 34, 53–64 [Google Scholar]
- 33.Bünnige M., Hilker M., Dobler S. 2008. Convergent evolution of chemical defence in Galerucine larvae. Biol. J. Linnean. Soc. 93, 165–175 10.1111/j.1095-8312.2007.00912.x (doi:10.1111/j.1095-8312.2007.00912.x) [DOI] [Google Scholar]
- 34.Gillespie J. J., Tallamy D. W., Riley E. G., Cognato A. I. 2008. Molecular phylogeny of rootworms and related galerucine beetles (Coleoptera: Chrysomelidae). Zool. Scr. 37, 195–222 10.1111/j.1463-6409.2007.00320.x (doi:10.1111/j.1463-6409.2007.00320.x) [DOI] [Google Scholar]
- 35.Seeno T. N., Wilcox J. A. 1982. Leaf beetle genera (Coleoptera: Chrysomelidae). Entomography 1, 126–156 [Google Scholar]
- 36.Horn G. H. 1889. A synopsis of the Halticini of Boreal America. Trans. Am. Entomol. Soc. 16, 163–320 [Google Scholar]
- 37.Leng C. W. 1920. Catalogues of the Coleoptera of America, north of Mexico. Mount Vernon NY: John D. Sherman, Jr [Google Scholar]
- 38.Takizawa H. 2005. Supra-generic subdivisions of the subfamily Alticinae based on larval characters, with descriptions of larvae of Hispaniolan species (Coleoptera: Chrysomelidae). Insecta Matsumurana 62, 187–206 [Google Scholar]
- 39.Loytynoja A., Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA 102, 10 557–10 562 10.1073/pnas.0409137102 (doi:10.1073/pnas.0409137102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A., Hoover P., Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 10.1080/10635150802429642 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 41.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwata H., Ukai Y. 2002. SHAPE: a computer program packagefor quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J. Hered. 93, 384–385 10.1093/jhered/93.5.384 (doi:10.1093/jhered/93.5.384) [DOI] [PubMed] [Google Scholar]
- 43.Rohlf F. J. 1998. NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2.02. New York, NY: Exeter Publications Setauket [Google Scholar]
- 44.Maddison D. R., Maddison W. P. 2003. MacClade: analysis of phylogeny and character evolution, v. 4.08. Sunderland, MA: Sinauer; [DOI] [PubMed] [Google Scholar]
- 45.Shimodaira H. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247 10.1093/bioinformatics/17.12.1246 (doi:10.1093/bioinformatics/17.12.1246) [DOI] [PubMed] [Google Scholar]
- 46.Swofford D. L. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). V. 4. Sunderland, MA: Sinauer Associates [Google Scholar]
- 47.Slowinski J. B., Guyer C. 1989. Testing the stochasticity of patterns of organismal diversification: an improved null model. Am. Nat. 134, 907–921 10.1086/285021 (doi:10.1086/285021) [DOI] [Google Scholar]
- 48.Magallon S., Sanderson M. J. 2001. Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780 [DOI] [PubMed] [Google Scholar]
- 49.Wheat C. W., Vogel H., Wittstock U., Braby M. F., Uderwood D., Mitchell-Olds T. 2007. The genetic basis of a plant–insect coevolutionary key innovation. Proc. Natl Acad. Sci. USA 104, 20 427–20 431 10.1073/pnas.0706229104 (doi:10.1073/pnas.0706229104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chapuis F. 1875. Famille des Phytophages. In Histoire naturelle des insects. Genera des coleopteres, vol. 11 (eds Lacordaire J. T., Chapuis F.), pp. 124–134 Paris A la Libraire Encyclopédique de Roret [Google Scholar]
- 51.Ohsawa R., Tsutumi T., Uehara H., Namai H., Ninomiya S. 1998. Quantitative evaluation of common buckwheat (Fagopyrum esculentum Moench) kernel shape by elliptic Fourier descriptor. Euphytica 101, 175–183 10.1023/A:1018344707479 (doi:10.1023/A:1018344707479) [DOI] [Google Scholar]
- 52.Becerra J. X. 2004. Molecular systematics of Blepharida beetles (Chrysomelidae: Alticinae) and relatives. Mol. Phylogenet. Evol. 30, 107–117 10.1016/S1055-7903(03)00158-1 (doi:10.1016/S1055-7903(03)00158-1) [DOI] [PubMed] [Google Scholar]
- 53.Reid C. A. M., Jurado-Rivera J. A., Beatson M. 2009. A new genus of Chrysomelinae from Australia (Coleoptera: Chrysomelidae). Zootaxa 2207, 57–66 [Google Scholar]
- 54.Leander B. S. 2008. A hierarchical view of convergent evolution in microbial eukaryotes. J. Eukaryot. Microbiol. 55, 59–68 10.1111/j.1550-7408.2008.00308.x (doi:10.1111/j.1550-7408.2008.00308.x) [DOI] [PubMed] [Google Scholar]
- 55.Abrams P. A. 2000. The evolution of predator–prey interactions: theory and evidence. Ann. Rev. Ecol. Syst. 31, 79–105 10.1146/annurev.ecolsys.31.1.79 (doi:10.1146/annurev.ecolsys.31.1.79) [DOI] [Google Scholar]
- 56.Furth D. G., Suzuki K. 1992. The independent evolution of the metafemoral spring in Coleoptera. Syst. Entomol. 17, 341–349 10.1111/j.1365-3113.1992.tb00555.x (doi:10.1111/j.1365-3113.1992.tb00555.x) [DOI] [Google Scholar]
- 57.Mayr E., Ashlock P. D. 1991. Principles of systematic zoology. New York, NY: McGraw-Hill [Google Scholar]
- 58.Queiroz A. D. 2002. Contingent predictability in evolution: key traits and diversification. Syst. Biol. 51, 917–929 10.1080/10635150290102627 (doi:10.1080/10635150290102627) [DOI] [PubMed] [Google Scholar]
- 59.Farrell B. D., Sequeira A. S. 2004. Evolutionary rates in the adaptive radiation of beetles on plants. Evolution 58, 1984–2001 [DOI] [PubMed] [Google Scholar]
- 60.Marvaldi A. E., Sequeira A. S., O'Brien C. W., Farrell B. D. 2002. Molecular and morphological phylogenetics of weevils (Coleoptera, Curculionoidea): do niche shifts accompany diversification? Syst. Biol. 51, 761–785 10.1080/10635150290102465 (doi:10.1080/10635150290102465) [DOI] [PubMed] [Google Scholar]
- 61.Wilf P., Labandeira C. C., Kress W. J., Staines C. L., Windsor D. M., Allen A. L., Johnson K. R. 2000. Timing the radiations of leaf beetles: hispines on gingers from Latest Cretaceous to Recent. Science 289, 291–294 10.1126/science.289.5477.291 (doi:10.1126/science.289.5477.291) [DOI] [PubMed] [Google Scholar]
- 62.Ahrens D., Vogler A. P. 2008. Towards the phylogeny of chafers (Sericini): analysis of alignment-variable sequences and the evolution of segment numbers in the antennal club. Mol. Phylogenet. Evol. 47, 783–798 10.1016/j.ympev.2008.02.010 (doi:10.1016/j.ympev.2008.02.010) [DOI] [PubMed] [Google Scholar]
- 63.Benard M. F. 2004. Predator-induced phenotypic plasticity in organisms with complex life history. Ann. Rev. Ecol. Syst. 35, 651–673 10.1146/annurev.ecolsys.35.021004.112426 (doi:10.1146/annurev.ecolsys.35.021004.112426) [DOI] [Google Scholar]