Abstract
Alternative reproductive tactics are ubiquitous in many species. Tactic expression often depends on whether an individual's condition surpasses thresholds that are responsible for activating particular developmental pathways. Two central goals in understanding the evolution of reproductive tactics are quantifying the extent to which thresholds are explained by additive genetic effects, and describing their covariation with condition-related traits. We monitored the development of early sexual maturation that leads to the sneaker reproductive tactic in Atlantic salmon (Salmo salar L.). We found evidence for additive genetic variance in the timing of sexual maturity (which is a measure of the surpassing of threshold values) and body-size traits. This suggests that selection can affect the patterns of sexual development by changing the timing of this event and/or body size. Significant levels of covariation between these traits also occurred, implying a potential for correlated responses to selection. Closer examination of genetic covariances suggests that the detected genetic variation is distributed along at least five directions of phenotypic variation. Our results show that the potential for evolution of the life-history traits constituting this reproductive phenotype is greatly influenced by their patterns of genetic covariance.
Keywords: alternative sexual tactics, G-matrix, salmon, heritability
1. Introduction
Alternative reproductive phenotypes are generally viewed as different sexual strategies or tactics that ensure some level of reproductive success under strong competition for reproductive resources [1,2]. Alternative reproductive tactics are widespread throughout the animal kingdom and are characterized by extreme discontinuity in the variation of reproductive traits within members of the same sex. This is observed, for example, in the horn or wing length in many species of insects [3,4], or body size of numerous fish species [1,5].
Many alternative phenotypes depend on switch mechanisms that are sensitive to internal and external aspects of an individual's environment and that mediate development towards the particular phenotypes [6]. The activation of these switch mechanisms depends on an individual's threshold of sensitivity to its somatic state and is usually quantified with measures of body size or energy storage (but is also affected by a combination of physiological factors and social interactions) [4,6,7].
Within a population, body size and surrogate measurements of these threshold values (such as hormone concentrations) may show important levels of phenotypic variation and covariation. For example, the threshold value that switches on a particular developmental pathway may vary with size (or growth) during sensitive ontogenetic stages. The potential for evolutionary change, however, depends on the extent of the additive genetic variance and covariance in these traits (as measured by heritability and genetic correlation, respectively) [8]. Providing that size and the surrogate measure of the thresholds have a heritable component, a strong genetic correlation may be expected to occur [8] and consequently affect how each trait responds to selection [4,9,10].
However, our current understanding of how alternative reproductive tactics evolve is limited by our knowledge about the patterns of genetic variance and covariance between the thresholds and the traits responsible for the surpassing of these values. Furthermore, consideration of the additive genetic covariances among all the measured traits in a phenotype (as described by the G-matrix, or G) may result in a biased distribution of additive genetic variance so that it is mostly summarized by a few trait combinations, or dimensions [11–13]. Thus, a form of constraint in the capacity for evolution emerges if some phenotypic aspects lack additive genetic variance [11,12]. Quantifying the multivariate nature of inheritance of such key life-history traits is therefore a major goal in evolutionary ecology because it allows us to understand how the genetic relationships among traits facilitate or impose constraints to phenotypic change [9,14].
The development of alternative reproductive pathways results in extreme divergence in virtually every aspect of the life cycle of almost all salmonid fish species (as well as many other vertebrates). The Atlantic salmon (Salmo salar L.) is remarkable among salmonids because alternative tactics are almost exclusively found in males and are characterized by extreme size dimorphism (approx. 10× order of magnitude). Individuals that mature early in life divert energetic resources to the production of gonad tissue at the expense of somatic growth. This occurs at young ages, while still living in natal freshwater habitats, during a life stage referred to as parr [15]. In contrast, other males mature later in life after having completed a marine feeding migration phase and returning to freshwater breeding habitats at much larger sizes (the anadromous or fighter tactic). During the spawning activity of anadromous fish, precociously mature males behave as opportunistic sneakers, darting into nests to fertilize the eggs deposited by females [5,15].
Great variation exists within and among salmon populations in the incidence of sneaker males, the age and size at which maturation occurs, and the developmental trajectories that follow [16,17]. Although inter-population variation in patterns of early maturity is explained partially by variation in growth opportunity [16,18], empirical evidence exists for differences in sensitive threshold values to body condition (which is usually described using body-size measures) [19–21]. Although some evidence also suggests genetically based spatial variation in thresholds for early sexual maturity and other life-cycle characteristics [21,22], we lack a detailed characterization of the evolvability of alternative developmental trajectories in this species.
The genetic basis of such alternative reproductive tactics is traditionally modelled as a threshold trait with an underlying liability that is normally distributed [8]. The liability is hypothesized to represent a compound trait (e.g. including body size, energetic state and social hierarchy) that, when taken in reference to a fixed threshold value, results in the expression of the dichotomous phenotypes [8]. The quantification of the threshold value in this approach is necessarily made at the population level (commonly using logistic regression techniques), rather than at the individual level. However, inherent in the prediction that individuals develop a reproductive tactic depending on aspects of their condition is the notion that each individual must have a threshold value sensitive to their somatic state [7,23]. A difficulty with this lies thus in the quantification of the threshold value, as it is likely to represent a composite energetic or physiological state of the individual that is affected by a variety of traits. In this study, we quantify the point in time when the onset of sexual maturation has a visible effect on individual growth (i.e. the timing of early sexual maturity). We use this time as a surrogate measure of the surpassing of threshold values activating early maturity, and we estimate this variable by examining the entire developmental trajectory of individual sneaker males.
We then assess the potential for phenotypic change by natural selection of this sexual tactic. Specifically, we first estimate the additive genetic relationships between the timing of early sexual maturity and body-size traits (measured before and after sexual maturation). With this, we test the hypothesis of no genetic (co)variance between the measured traits, allowing us to assess whether the correlated potential for phenotypic change varies across the ontogeny of sneaker males. We then use multivariate techniques to summarize the major directions of additive genetic variation from G [9,24,25]. This allows us to discuss the distribution of genetic variance across the measured phenotypes, as well as the likelihood of genetic constraints.
2. Material and methods
(a). Mating design and rearing conditions
In autumn 2004, four anadromous females, six anadromous males and eight mature male parr were captured in the northeastern branch of the Sainte-Marguerite River (Québec, Canada). They were used to produce a pedigree with both a maternal and paternal half-sib structure (table 1). Details on the early-life rearing conditions are found in [26]. In May 2005, once active swimming and feeding behaviour was observed in recently emerged fish, each of the 25 families was transferred from the incubation rearing environment to 200 l radial tanks. Individuals were reared in these tanks until January 2006. Over this time, mean water temperature was 11.8 ± 1.7°C (±1 s.d.), and mean oxygen concentration was 10.4 ± 0.6 mg l−1. In addition, at this point, all fish (n = 2683) were sufficiently large to be surgically tagged with a passive integrated transponder device, allowing us to monitor individual body size from then on.
Table 1.
Pedigree of the 574 mature parr. The average maternal and paternal number of offspring was equal to 143.5 and 41 individuals, respectively. The mean pairwise relatedness among individuals = 0.3 [54]. A, anadromous sires; P, mature parr sires; F, anadromous dams. The dashes represent families for which fertilization was unsuccessful, whereas the cross represents the family where all individuals died during the experiment.
| upstream |
downstream |
|||||
|---|---|---|---|---|---|---|
| dam |
dam |
|||||
| F1 | F2 | F3 | F4 | |||
| sire | A1 | 66 | 31 | A3 | 20 | 49 |
| A2 | 14 | — | A4 | — | 23 | |
| P1 | 20 | 7 | A5 | 2 | 32 | |
| P2 | 23 | 15 | A6 | 60 | 16 | |
| P3 | — | 35 | P5 | 5 | 8 | |
| P4 | × | 19 | P6 | 10 | 39 | |
| P7 | 29 | 11 | ||||
| P8 | 4 | 36 | ||||
| total = 574 | ||||||
Once tagged, individuals within families were split into two 3 m3 tanks where they were reared until early November 2006, terminating the experiment. Mean water temperature during this latter part of the experiment was 10.11 ± 0.8°C and the mean oxygen concentration was 10.4 ± 0.81 mg l−1. Throughout the experiment, fish were fed 4 per cent of their body weight with commercial salmonid food pellets containing 20 per cent lipids and 50 per cent protein. Photoperiod was adjusted with a SunMatch controller (AquabioLab, Québec, Canada), which was set to follow the natural photoperiod regime in Québec at 70 per cent light intensity. From July to November 2005, mortality was high, resulting in the loss of one full-sib family. From December 2005 to late October 2006, the median mortality was 19.2 ± 27.9 per cent. A small (but significant) bias in mortality was observed between January 2006 and July 2006, when heavier individuals were more likely to survive (results not shown).
To reduce tank densities and for the purposes of other experiments, randomly selected individuals from large families were sacrificed in the samplings conducted in April and July. The body length (BL) between sacrificed and live individuals was not significantly different in April 2006 (linear mixed model (LMM): F1,2406 = 0.23, p = 0.63). However, individuals sampled in July 2006 were, on average, 1.02 cm smaller (LMM: F1,1661 = 78.4, p < 0.001). All the points discussed above lead to variation in sample sizes throughout the experiment. However, sampling variation was mostly restricted to changes in the number of individuals within families rather than to changes in the number of families measured (except for the one family lost in August 2005). Overall, at least one phenotypic record was obtained for 2683 individuals, of which 574 were mature male parr. The statistical properties of the pedigree constituted by the mature parr are described in table 1. Note that we were logistically constrained to a small number of progenitors. However, genetic variation should be sufficiently represented in our design as only the variance arising from the effects of rare alleles is likely to be excluded [27]. The additional loss of genetic variation owing to the asymmetry in the number of sires and dams is also expected to be small [28].
(b). Measurements
Body length up to the fork of the caudal fin, body mass (BM) and body depth (BD; the distance from the origin of the dorsal fin to the most ventral point of the body) were measured at hatching (April) and emergence (July) in 2005, and in January, April, July and early November 2006. The sampling protocol conducted at hatching and emergence is described by Páez et al. [26]. For the other sampling dates, all individuals were anaesthetized with 10 per cent eugenol, photographed against a millimetric ruler and then allowed to recover before being returned to the corresponding tank.
The software ImageJ [29] was used to calibrate images to the nearest millimetre and to measure BL and BD. BM measurements to the nearest gram were taken at sampling. Alternative male reproductive tactics were first observed in April 2006 and were readily distinguishable by early November 2006. Mature male parr conserved the cryptic juvenile coloration, produced milt and reached a growth plateau. In contrast, male smolts (i.e. the migratory, immature phenotype) had developed a silver coloration, were more streamlined and were growing exponentially by the last sampling date. Thus, we used these characteristics to class individuals as mature parr or as immature smolts. Smolts were then classed into males and females by examining gonads after dissection.
(c). Estimation of the timing of sexual maturity
In Atlantic salmon, the mechanism signalling early maturity results in investment in reproductive tissue at a cost to future somatic growth [17,30]. This finding was corroborated by comparisons between several growth models based on Akaike information criteria (AIC). This suggested that sigmoidal growth models, having a parameter representing a point of inflection of the growth trajectory, fitted the data much better than either linear or exponential models. Among the models tested, we found that the logistic growth model had the best fit, with the highest squared correlation between observed and predicted values (r2 = 0.97), as well as the lowest AIC score (such that differences in AIC scores between the linear, exponential, von Bertalanffy, and Gompertz models and the logistic growth model were 109.5, 855.5, 827.9 and 46.0, respectively).
Therefore, for each individual mature parr for which all six BL measurements were available (that is, 216 individuals), the logistic growth model
was fitted to the data. Here, BL is a function (∼) of the parameters a, k and b, which describe, respectively, the asymptote, rate of increase and inflection point of the curve. ‘date’ is the number of days after hatching when the measurements were made. The individual estimates of inflection points obtained represent the specific date during the growth trajectory at which males divert energetic resources to gonad tissue [17,31,32].
(d). Quantitative genetic analyses
Here, we briefly describe the main steps in our procedure to estimate genetic (co)variance components (§2e–h) using animal models. We have assumed that the measurements made at each date are age-specific traits, for a total of 18 traits. Such an assumption is realistic, given the impressive morphological and physiological changes between juvenile and precocious sexual maturation states that occur over a short period (e.g. [33]).
(e). Univariate animal models: choice of fixed effects
The effects of six variables were evaluated for all traits to determine whether the estimation of quantitative genetic parameters should be conditioned on potential sources of variation owing to the experimental set-up and rearing conditions [34]. A description of these variables is found in the electronic supplementary material, table S1. The effects of these variables were evaluated by adding them as fixed effects in an LMM, which also included, as random effects, the sire and dam identity, the early-life common environment and the common environment experienced after tagging. The significance of the fixed effects was evaluated using F-tests [35] (and are also presented in electronic supplementary material, table S1). Only those variables found to have a significant effect (at α < 0.05) on the trait were retained for the subsequent analyses concerning the partitioning of variance components.
In a previous study, we documented an important contribution of the maternal environment on the traits measured at emergence [26]. However, because of the smaller number of dams used in this study and because maternal effects are known to decline shortly after emergence in the Atlantic salmon [36], we fitted maternal identity as a fixed effect only for these traits.
(f). Univariate animal models: choosing random effects and estimating heritability
Similarly, the effects of common environmental sources of variation induced by the rearing environment can potentially inflate additive genetic variance estimates if unaccounted for [37]. The initial structure of each animal model therefore consisted of the fixed effect(s), as found above, and the random effects, as described by the animal term and the tank environment both before and after tagging. Therefore, for each trait, the full animal model
was fitted, where y is a vector of phenotypic observations of all individuals and μ is a vector of fitted fixed effects related to phenotypic observations through the incidence matrix X. The vector a represents the additive genetic variance to be estimated, whereas the vectors EC1 and EC2 represent effects from common rearing conditions, characteristic of the tank environment before and after tagging, respectively. These random effects are related to the phenotypic observations through the incidence matrices Z1, Z2 and Z3. Finally, the vector e contains the residual variation.
The significance of the common environmental effects was then assessed with log-likelihood ratio tests (LRTs) between the full model and reduced models, omitting the term. We interpreted a significant common environmental effect as a potential biasing factor in the estimation of heritability, therefore justifying its inclusion in the final model. However, we excluded the effect from subsequent analyses if it did not significantly increase deviance of the model [37].
Significant effects of the tank environment after tagging were found for the BD measured in November (LRT = 5.03, d.f. = 1, p = 0.03) and the organ weight (LRT = 5.55, d.f. = 1, p = 0.02). The inclusion of the common environmental effects before tagging, however, never improved the log likelihood of the models, indicating that the effects of the common environment are adequately partitioned out through the fixed effects. Once the random effects structure of the animal models was established, we tested the significance of the additive genetic variance, using LRT. Narrow-sense heritability (h2) was then calculated as the ratio of additive variance (VA) to total phenotypic variance (VP).
(g). Bivariate animal models: estimating genetic correlations
All possible genetic correlations between the 18 traits were then estimated, using bivariate animal models. The significance of genetic covariances was also evaluated, using the LRT score obtained from comparing a full model with a reduced model, where genetic covariance is set at 0. The genetic correlation (rg) was then calculated as
where COVtrait1,trait2 is the additive genetic covariance between the traits, and VAtrait1 and VAtrait2 are the additive genetic variance for trait 1 and trait 2, respectively. All genetic, phenotypic and residual pairwise estimates are presented in the electronic supplementary material, tables S2 and S3.
(h). Multivariate animal model: determining dimensionality and major patterns of additive genetic variation
We used the ‘subset’ and ‘itsum’ functions implemented in WOMBAT on the estimates of the bivariate analyses to construct a covariance matrix of starting values with non-negative eigenvalues suitable for multivariate modelling [38]. The number of dimensions or principal components of the G-matrix summarizing genetic variation was determined using factor-analytic modelling [39]. In this method, the G-matrix was constrained at 18 to 1 dimensions and a series of nested LRTs were used to determine whether removal of a dimension resulted in a significant worsening of the model fit [39,40]. For the statistically supported dimensions, we identified significant trait loadings (greater than +0.1245 or lower than −0.1245), using Fisher's inverse hyperbolic tangent transformation [41] with a Bonferroni correction to control the type I error rate. Finally, we used AIC as a concomitant measure to determine the number of dimensions required to adequately represent the patterns of additive genetic covariance between the traits [42].
3. Results
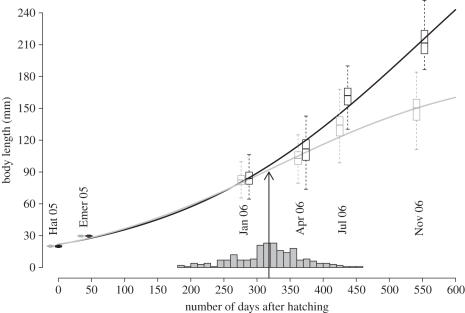
The development of alternative life histories in the Atlantic salmon was characterized by divergent growth trajectories (figure 1). The activation of early sexual maturity, as estimated from the point of inflection in the logistic growth curves, occurs on average at 317.8 ± 59.9 days after hatching (figure 1). Individual body sizes at these days correspond on average to a BL of 93.3 ± 15.2 mm and a BM of 8.34 ± 3.03 g. These values agree closely to those reported in other studies [19,21]. In addition, as expected, higher values in the timing of sexual maturity are positively correlated with bigger body sizes of sneaker males by the end of the experiment (phenotypic Pearson's correlation coefficient with BL in November = 0.66 ± 0.037, p < 0.001; phenotypic Pearson's correlation with BM in November = 0.63 ± 0.041, p < 0.001; table 2).
Figure 1.
Growth patterns observed for sneaker (grey) and faster-growing (black) males throughout the experiment. Boxes encompass the interquartile range of the observed data, with the medians represented by the dark lines drawn midpoint in the box. The whiskers extend to approximately 2 s.d. of the data. The curves represent the growth trajectories of each life history based on a logistic function. Labels within the figure indicate the month when sampling was conducted. From left to right: hatching (Hat 05) and emergence (Emer 05) in 2005, and January (Jan 06), April (Apr 06), July (Jul 06) and November (Nov 06) in 2006. The grey bars represent the distribution of the timing of early sexual maturation for 216 sneaker males. After removing two outliers, this distribution does not differ from that expected under normality (Shapiro–Wilk normality test: W = 0.989, p = 0.1001), with a skew = −0.18±1.048, and a kurtosis = 0.048 ± 0.443. The mean of this distribution occurs at 318.9 days after hatching and is represented by the black arrow.
Table 2.
Variance components estimates of the measured traits. All values in parentheses are s.e. of the estimates. Values in bold represent significant estimates at α < 0.01. Values in bold and italic represent significant estimates with α between 0.01 and 0.05. p-values were adjusted using the Benjamini–Hochberg method to control for false discovery rates [55].
| traita |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| parameterb | EmL | EmB | EmM | JaL | JaB | JaM | ApL | ApB | ApM | JuL | JuB | JuM | NoL | NoB | NoM | Gon | Org | Time |
| n | 430 | 430 | 430 | 536 | 536 | 574 | 461 | 461 | 495 | 433 | 433 | 433 | 281 | 281 | 283 | 284 | 282 | 216 |
| mean | 29.486 | 4.187 | 0.177 | 82.58 | 15.202 | 6.203 | 102.918 | 21.894 | 11.809 | 133.065 | 28.817 | 30.306 | 149.119 | 30.972 | 38.669 | 3.932 | 6.083 | 318.854 |
| VP | 0.590 (0.0651) | 0.0380 (0.00618) | 0.25 × 10−3 (0.38×10−4) | 40.791 (5.478) | 3.118 (0.355) | 4.284 (0.309) | 80.421 (6.419) | 6.988 (0.589) | 12.271 (0.905) | 176.506 (14.872) | 9.624 (0.808) | 82.822 (6.805) | 287.224 (28.153) | 17.566 (2.132) | 162.301 (17.986) | 5.492 (0.578) | 7.781 (0.932) | 2949.66 (359.854) |
| VA | 0.171 (0.113) | 0.023 (0.012) | 0.16 × 10−3(0.73 × 10−4) | 12.858 (10.274) | 1.089 (0.646) | 0.632 (0.421) | 14.801 (8.989) | 1.789 (0.893) | 2.204 (1.171) | 41.932 (22.012) | 2.299 (1.187) | 18.431 (9.703) | 64.422 (39.830) | 6.832 (3.375) | 55.748 (29.477) | 1.540 (0.876) | 2.867 (1.432) | 911.002 (572) |
| h2 | 0.290 (0.165) | 0.605 (0.222) | 0.629 (0.205) | 0.315 (0.214) | 0.349 (0.172) | 0.148 (0.092) | 0.184 (0.103) | 0.256 (0.113) | 0.180 (0.088) | 0.238 (0.112) | 0.239 (0.110) | 0.223 (0.106) | 0.224 (0.126) | 0.389 (0.160) | 0.343 (0.155) | 0.280 (0.140) | 0.368 (0.155) | 0.309 (0.169) |
| COVTime | −3.549 (6.673) | 1.352 (2.340) | 0.202 (0.182) | 70.945 (71.193) | 7.869 (15.262) | 11.184 (13.619) | 77.638 (59.960) | 26.215 (18.326) | 10.382 (16.437) | 184.930 (91.405) | 45.613 (22.091) | 108.501 (54.897) | 184.781 (109.506) | 66.492 (36.006) | 192.141 (98.957) | 37.171 (18.956) | 45.694 (24.194) | — |
| rgTime | −0.286 (0.515) | 0.250 (0.404) | 0.485 (0.346) | 0.454 (0.404) | 0.241 (0.463) | 0.446 (0.481) | 0.591 (0.321) | 0.610 (0.304) | 0.280 (0.419) | 1.000 (0.182) | 1.000 (0.092) | 1.000 (0.143) | 0.987 (0.087) | 0.888 (0.129) | 1.000 (0.064) | 0.960 (0.131) | 0.853 (0.154) | — |
| rpTime | −0.085 (0.159) | 0.122 (0.207) | 0.233 (0.197) | −0.181 (0.114) | −0.237 (0.097) | −0.05 (0.08) | −0.012 (0.084) | −0.04 (0.086) | −0.042 (0.075) | 0.389 (0.065) | 0.445 (0.060) | 0.402 (0.060) | 0.633 (0.043) | 0.576 (0.053) | 0.568 (0.051) | 0.261 (0.078) | 0.367 (0.072) | — |
aTrait codes: EmL, emergence length; EmB, emergence body depth; EmM, emergence body mass; JaL, January length; JaB, January body depth; JaM, January body mass; ApL, April length; ApB, April body depth; ApM, April body mass; JuL, July length; JuB, July body depth; JuM, July body mass; NoL, November length; NoB, November body depth; NoM, November body mass; Gon, gonad mass; Org, organ mass; Time, timing of early sexual maturity.
bParameter codes: n, the number of individuals phenotyped for a given trait; VP, phenotypic variance of unscaled traits; VA, additive genetic variance of unscaled traits; h2, narrow-sense heritability; COVTime, additive genetic covariance between the threshold and each trait; rgTime, genetic correlation between the timing of sexual maturity and each trait; rpTime, phenotypic correlation between the timing of sexual maturity and each trait.
Our study reveals a significant amount of additive genetic variance characterizing the variation in the timing of early maturity (h2 = 0.31 ± 0.17, LRT = 12.27, d.f. = 1, p < 0.001) and in body-size traits (mean h2 over BL, BM and BD = 0.31 ± 0.14; table 2).
Based on our bivariate comparisons, we found strong and significant genetic correlations between these traits after the activation of sexual maturation. In contrast, the genetic correlations with traits measured at early life stages are smaller and always non-significant (table 2). Furthermore, positive genetic correlations between traits measured at the different sampling dates were also detected (particularly after January 2006), implying a largely shared genetic basis for some traits across ontogeny (electronic supplementary material, table S2; figure 2).
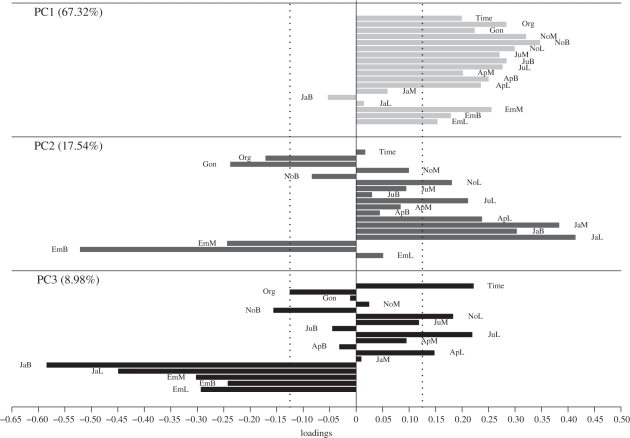
Figure 2.
Trait loadings on the first three leading principal components (depicted with different grey shadings) extracted from the G-matrix. A loading is considered to significantly contribute to the ordination if it extends over ±0.1245, marked by the black dotted lines (see table 1 for an explanation of trait codes).
We detected genetic variance on five principal components of G (as constraining G from five to four PCs results in a worsening of the model fit: χ2 = 56.02, d.f. = 14, p < 0.01; electronic supplementary material, table S4). PC1 summarized 67.32 per cent of the additive genetic variance, with decreasing (but significant) amounts found from PC2 to PC5 (summarizing 17.54, 8.98, 4.03 and 2.13 per cent of the variance in G). Different trait loadings, and hence a diverse pattern of trait covariation, characterized these PCs, as depicted in figure 2 (electronic supplementary material, tables S4–S7).
4. Discussion
A major component in our understanding of the consequences of natural selection and other evolutionary processes lies in knowing the extent of the genetic variation characterizing phenotypes. Particularly relevant to condition-dependent phenotypes, quantifying the genetic basis of the key life-history traits associated with the available developmental pathways allows us to better comprehend the mechanisms that maintain this variation and facilitate or impose constraints to their evolution.
Our experiment reveals two growth trajectories among juvenile salmon, the fastest being associated with migration and the slowest (male-biased) being associated with either delayed migration or precocious maturity. Note that several studies suggest that mature parr display the fastest growth when compared with immature, juvenile fish. This result generally occurs when mature parr growth is compared with the growth of delayed migrants, rather than with that of the earliest migrants, thus confusing the relationship between growth rate and tactic choice (J. J. Dodson, N. Aubin-Horth, V. Thériault & D. J. Páez 2010, unpublished data).
Overall, our results show that the variation in the development of early sexual maturation and the sneaker reproductive tactic in male Atlantic salmon is characterized by genetic variation. Selection can therefore modify the patterns of sexual development through its effects on the timing of early sexual maturity and/or body size. As a result, the capacity to respond to selection at these life-history traits may contribute to the striking differences in the incidence of early maturation, as well as in other life-history characteristics, such as size and age at maturity, observed across the distribution of this species [17,20,21].
However, selection acting only on the timing of maturation will cause a correlated response on body size (and vice versa) because these traits are genetically correlated. The specific pattern of covariation observed suggests that while small pleiotropic interactions between genes influence the covariation of these traits in the early life stages, much stronger interactions characterize the stages later in life. Therefore, selection occurring on traits after the developmental threshold is surpassed is likely to generate a stronger correlated response than selection occurring earlier in life, when the genetic correlations between these traits are not as strong.
The possibility that directional selection may act on the timing of sexual maturity while taking a stabilizing form on body size may potentially occur (e.g. when delayed sexual maturity is selected for in the commercial production of this species). In this case, selection could favour changes in the timing of maturation without causing a correlated response on size [43]. However, for the timing of this event to evolve independently from other traits, part of the additive genetic variance must be unique to this trait alone [43]. Although we have not estimated such trait-specific genetic variances, we have shown that the timing of early sexual maturity has a heritable basis that covaries to different degrees across ontogeny with body size. Therefore, further partitioning of the additive genetic variance into trait-specific and shared components is necessary to understand the capacity of independent evolution of these traits [43,44].
We also found positive genetic correlations between the very early- and late-life body sizes, suggesting that the effects of selection at one age are likely to have correlated effects at other ages, even in the case where a developmental switch mechanism is activated. A more holistic examination of G, based on its decomposition into all possible patterns of trait covariation, confirmed that most of the genetic variance is described by positive covariation between traits. This pattern is the expected outcome of a principal component describing the size variation particular to each trait [45–47]. Therefore, because PC1 summarizes most of the observed additive genetic variance, phenotypic aspects related to size are expected to be the least resistant to selection [25,45].
Smaller but significant amounts of additive genetic variance were also represented by at least four other directions of trait covariation, suggesting that phenotypes with such characteristics may also evolve under specific selective regimes [48]. Given the high heterogeneity of habitat features, variable selection regimes are plausible across the distribution of the Atlantic salmon [49–51]. Therefore, providing that selection acts on the phenotypic combinations containing additive genetic variance, contrasting patterns of trait variation across the distribution of this species may occur.
Our results show that the genetic variance of the measured traits lies along five directions of phenotypic variation. Although the lack of genetic variance in the remaining 13 dimensions is affected by the small sampling design [39] and reflects either the covariation between traits through time or among the three body measures within sampling dates, it may also reflect the presence of genetic constraints to the response to selection [40]. However, caution is required when interpreting our results in this context because the high genetic correlations between the repeated measures obtained for each trait from April 2006 onwards suggest that the measures represent largely the same biological trait. This pattern of covariation coincides with the approach of an asymptotic rate of change in trait development. Thus, as this asymptote is reached, it is possible that the development of these traits does not draw on additional sources of genetic variation, causing the genetic correlations to approach a value of 1.
Another form of constraint to the evolution of alternative life histories may occur if shared traits across the sexual tactics are positively genetically correlated, but optimal trait values are different (or alternatively, if the genetic correlation is negative but selection occurs in the same direction for both sexual tactics). Such conflict would resemble the intra-locus conflict observed between the sexes of many species and may similarly influence the evolution of alternative life histories [52,53].
This study is a first attempt at a comprehensive understanding of the genetic relationships between traits involved in the development of the sneaker male phenotype in the Atlantic salmon. We show that high additive genetic variation, and hence a high capacity to respond to selection, is found in the timing of sexual maturation and size-related traits. Although selection can thus influence early sexual development, a correlated response between these traits is also expected to occur, with the rate of this response changing throughout ontogeny. Our results also provide preliminary evidence about the distribution of genetic variance in the mature parr phenotype, thus setting a framework for the study of the genetic constraints that arise from trait correlations.
Acknowledgements
This study contributes towards the research programmes of CIRSA and Quebec Ocean. Funding was provided by an NSERC strategic grant awarded to L.B., J.J.D. and Helga Guderley, and by an NSERC Discovery grant awarded to J.J.D. We thank LARSA and O. Rossignol for logistical help with this project. We also thank E. L. McAdam, G. Daigle, N. Aubin-Horth, D. Garant, K. McGuigan, N. Bennett, F. Allendorf, M. Taborsky and three anonymous reviewers for insightful comments that improved the quality of this manuscript.
References
- 1.Oliveira R. F., Taborsky M., Brockmann H. J. 2008. Alternative reproductive tactics. In An integrative approach, 1st edn. New York, NY: Cambridge University Press [Google Scholar]
- 2.Shuster S. M., Wade M. J. 2003. Mating systems and strategies, 1st edn. Princeton, NJ: Princeton University Press [Google Scholar]
- 3.Emlen D. J., Corley Lavine L., Ewen-Campen B. 2007. On the origin and evolutionary diversification of beetle horns. Proc. Natl Acad. Sci. USA 104(Suppl. 1), 8661–8668 10.1073/pnas.0701209104 (doi:10.1073/pnas.0701209104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roff D. A. 1996. The evolution of threshold traits in animals. Q. Rev. Biol. 71, 3–35 10.1086/419266 (doi:10.1086/419266) [DOI] [Google Scholar]
- 5.Taborsky M. 1998. Sperm competition in fish: ‘bourgeois’ males and parasitic spawning. Trends Ecol. Evol. 13, 222–227 10.1016/S0169-5347(97)01318-9 (doi:10.1016/S0169-5347(97)01318-9) [DOI] [PubMed] [Google Scholar]
- 6.West-Eberhard M. J. 2003. Developmental plasticity and evolution, 1st edn. New York, NY: Oxford University Press [Google Scholar]
- 7.Gross M. R. 1996. Alternative reproductive strategies and tactics: diversity within sexes. Trends Ecol. Evol. 11, 92–98 10.1016/0169-5347(96)81050-0 (doi:10.1016/0169-5347(96)81050-0) [DOI] [PubMed] [Google Scholar]
- 8.Falconer D. S., Mackay T. F. C. 1996. Introduction to quantitative genetics, 4th edn. Harlow, UK: Pearson Education Limited [Google Scholar]
- 9.Blows M. W. 2007. A tale of two matrices: multivariate approaches in evolutionary biology. J. Evol. Biol. 20, 1–8 10.1111/j.1420-9101.2006.01164.x (doi:10.1111/j.1420-9101.2006.01164.x) [DOI] [PubMed] [Google Scholar]
- 10.Lande R. 1979. Quantitative genetic analysis of multivariate evolution, applied to brain : body size allometry. Evolution 33, 402–416 10.2307/2407630 (doi:10.2307/2407630) [DOI] [PubMed] [Google Scholar]
- 11.Arnold S. J. 1992. Constraints on phenotypic evolution. Am. Nat. 140, S85–S107 10.1086/285398 (doi:10.1086/285398) [DOI] [PubMed] [Google Scholar]
- 12.McGuigan K. 2006. Studying phenotypic evolution using multivariate quantitative genetics. Mol. Ecol. 15, 883–896 10.1111/j.1365-294X.2006.02809.x (doi:10.1111/j.1365-294X.2006.02809.x) [DOI] [PubMed] [Google Scholar]
- 13.Mezey J. G., Houle D. 2005. The dimensionality of genetic variation for wing shape in Drosophila melanogaster. Evolution 59, 1027–1038 [PubMed] [Google Scholar]
- 14.Walsh B., Blows M. W. 2009. Abundant genetic variation + strong selection = multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Syst. 40, 41–59 10.1146/annurev.ecolsys.110308.120232 (doi:10.1146/annurev.ecolsys.110308.120232) [DOI] [Google Scholar]
- 15.Fleming I. A. 1996. Reproductive strategies of Atlantic salmon: ecology and evolution. Rev. Fish. Biol. Fish. 6, 379–416 10.1007/BF00164323 (doi:10.1007/BF00164323) [DOI] [Google Scholar]
- 16.Letcher B. H., Gries G. 2003. Effects of life history variation on size and growth in stream-dwelling Atlantic salmon. J. Fish Biol. 62, 97–114 10.1046/j.1095-8649.2003.00009.x (doi:10.1046/j.1095-8649.2003.00009.x) [DOI] [Google Scholar]
- 17.Myers R. A., Hutchings J. A., Gibson R. J. 1986. Variation in male parr maturation within and among populations of Atlantic salmon, Salmo salar. Can. J. Fish. Aquat. Sci. 43, 1242–1248 10.1139/f86-154 (doi:10.1139/f86-154) [DOI] [Google Scholar]
- 18.Metcalfe N. B. 1998. The interaction between behavior and physiology in determining life history patterns in Atlantic salmon (Salmo salar). Can. J. Fish. Aquat. Sci. Suppl. 1, 93–103 10.1139/cjfas-55-S1-93 (doi:10.1139/cjfas-55-S1-93) [DOI] [Google Scholar]
- 19.Aubin-Horth N., Bourque J.-F., Daigle G., Hedger R. D., Dodson J. J. 2006. Longitudinal gradients in threshold sizes for alternative male life history tactics in a population of Atlantic salmon (Salmo salar). Can. J. Fish. Aquat. Sci. 63, 1–9 10.1139/F06-103 (doi:10.1139/F06-103) [DOI] [Google Scholar]
- 20.Baum D., Laughton R., Armstrong J. D., Metcalfe N. B. 2004. Altitudinal variation in the relationship between growth and maturation rate in salmon parr. J. Anim. Ecol. 73, 253–260 10.1111/j.0021-8790.2004.00803.x (doi:10.1111/j.0021-8790.2004.00803.x) [DOI] [Google Scholar]
- 21.Piché J., Hutchings J. A., Blanchard W. 2008. Genetic variation in threshold reaction norms for alternative reproductive tactics in male Atlantic salmon, Salmo salar. Proc. R. Soc. B 275, 1571–1575 10.1098/rspb.2008.0251 (doi:10.1098/rspb.2008.0251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicieza A. G., Reyes-Gavilán F. G., Braña F. 1994. Differentiation in juvenile growth and bimodality patterns between northern and southern populations of Atlantic salmon (Salmo salar L.). Can J. Zool. 72, 1603–1610 10.1139/z94-213 (doi:10.1139/z94-213) [DOI] [Google Scholar]
- 23.Hazel W. N., Smock R., Johnson M. D. 1990. A polygenic model for the evolution and maintenance of conditional strategies. Proc. R. Soc. Lond. B 242, 181–187 10.1098/rspb.1990.0122 (doi:10.1098/rspb.1990.0122) [DOI] [PubMed] [Google Scholar]
- 24.Arnold S. J. 1981. Behavioral variation in natural populations. I. Phenotypic, genetic and environmental correlations between chemoreceptive responses to prey in the garter snake, Thamnophis elegans. Evolution 35, 489–509 [DOI] [PubMed] [Google Scholar]
- 25.Schluter D. 1996. Adaptive radiation along genetic lines of least resistance. Evolution 50, 1766–1774 10.2307/2410734 (doi:10.2307/2410734) [DOI] [PubMed] [Google Scholar]
- 26.Páez D. J., Morrissey M., Bernatchez L., Dodson J. J. 2010. The genetic basis of early-life morphological traits and their relation to alternative male reproductive tactics in Atlantic salmon. J. Evol. Biol. 23, 757–768 10.1111/j.1420-9101.2010.01941.x (doi:10.1111/j.1420-9101.2010.01941.x) [DOI] [PubMed] [Google Scholar]
- 27.Barton N. H., Charlesworth B. 1984. Genetic revolutions, founder effects, and speciation. Annu. Rev. Ecol. Evol. Syst. 15, 133–164 10.1146/annurev.es.15.110184.001025 (doi:10.1146/annurev.es.15.110184.001025) [DOI] [Google Scholar]
- 28.Waples R. S. 1987. Sperm storage, multiple insemination, and genetic variability in mosquitofish: a reassessment. Copeia 4, 1068–1072 10.2307/1445580 (doi:10.2307/1445580) [DOI] [Google Scholar]
- 29.Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophoton. Int. 11, 36–42 [Google Scholar]
- 30.Stearns S. C. 1992. The evolution of life histories, 1st edn. New York, NY: Oxford University Press [Google Scholar]
- 31.Stamps J. A., Mangel M., Phillips J. A. 1998. A new look at relationships between size at maturity and asymptotic size. Am. Nat. 152, 470–479 10.1086/286183 (doi:10.1086/286183) [DOI] [PubMed] [Google Scholar]
- 32.Wootton R. J. 1990. Ecology of teleost fishes, 1st edn. London, UK: Chapman & Hall [Google Scholar]
- 33.Aubin-Horth N., Landry C., Letcher B. H., Hofmann H. A. 2005. Alternative life histories shape brain gene expression profiles in males of the same population. Proc. R. Soc. B 272, 1655–1662 10.1098/rspb.2005.3125 (doi:10.1098/rspb.2005.3125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson A. J. 2008. Why h2 does not always equal VA/VP. J. Evol. Biol. 21, 647–650 10.1111/j.1420-9101.2008.01500.x (doi:10.1111/j.1420-9101.2008.01500.x) [DOI] [PubMed] [Google Scholar]
- 35.Baayen R. H. 2008. LanguageR: data sets and functions with ‘analyzing linguistic data: a practical introduction to statistics’, R Package v. 0.953. See http://cran.r-project.org/package=languageR [Google Scholar]
- 36.Garant D., Dodson J. J., Bernatchez L. 2003. Differential reproductive success and heritability of alternative reproductive tactics in wild Atlantic salmon (Salmo salar L.). Evolution 57, 1133–1141 [DOI] [PubMed] [Google Scholar]
- 37.Kruuk L. E. B. 2004. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. Lond. B 359, 873–890 10.1098/rstb.2003.1437 (doi:10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer K. 2007. WOMBAT—a tool for mixed model analyses in quantitative genetics by REML. J. Zhejiang Univ. Sci. B. 8, 815–821 10.1631/jzus.2007.B0815 (doi:10.1631/jzus.2007.B0815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hine E., Blows M. W. 2006. Determining the effective dimensionality of the genetic variance–covariance matrix. Genetics 173, 1135–1144 10.1534/genetics.105.054627 (doi:10.1534/genetics.105.054627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGuigan K., Blows M. W. 2007. The phenotypic and genetic covariance structure of drosophilid wings. Evolution 61, 902–911 10.1111/j.1558-5646.2007.00078.x (doi:10.1111/j.1558-5646.2007.00078.x) [DOI] [PubMed] [Google Scholar]
- 41.Sokal R. R., Rohlf F. J. 1995. In Biometry: the principles and practice of statistics in biological research. New York, NY: W. H. Freeman and Co [Google Scholar]
- 42.Meyer K. 2005. Genetic principal components for live ultrasound scan traits of Angus cattle. Anim. Sci. 81, 337–345 10.1079/ASC50850337 (doi:10.1079/ASC50850337) [DOI] [Google Scholar]
- 43.Hansen T., Armbruster W. S., Carlson M., Pélabon C. 2003. Evolvability and genetic constraint in Delechampia blossoms: genetic correlations and conditional evolvability. J. Exp. Zool. (Mol. Dev. Evol.) 296B, 23–39 10.1002/jez.b.14 (doi:10.1002/jez.b.14) [DOI] [PubMed] [Google Scholar]
- 44.McGuigan K., Blows M. W. 2010. Evolvability of individual traits in a multivariate context: partitioning the additive genetic variance into common and specific components. Evolution 64, 1899–1911 10.1111/j.1558-5646.2010.00968.x (doi:10.1111/j.1558-5646.2010.00968.x) [DOI] [PubMed] [Google Scholar]
- 45.McGuigan K., Chenoweth S. F., Blows M. W. 2005. Phenotypic divergence along lines of genetic variance. Am. Nat. 165, 32–43 10.1086/426600 (doi:10.1086/426600) [DOI] [PubMed] [Google Scholar]
- 46.Ragland G. J., Carter P. A. 2004. Genetic covariance structure of growth in the salamander Ambystoma macrodactylum. Heredity 92, 569–578 10.1038/sj.hdy.6800462 (doi:10.1038/sj.hdy.6800462) [DOI] [PubMed] [Google Scholar]
- 47.Wilson A. J., Kruuk L. E. B., Coltman D. W. 2005. Ontogenetic patterns in heritable variation for body size: using random regression models in a wild ungulate population. Am. Nat. 166, E177–E192 10.1086/497441 (doi:10.1086/497441) [DOI] [PubMed] [Google Scholar]
- 48.Kirkpatrick M., Lofsvold D. 1992. Measuring selection and constraint in the evolution of growth. Evolution 46, 954–971 10.2307/2409749 (doi:10.2307/2409749) [DOI] [PubMed] [Google Scholar]
- 49.Koskinen M. T., Haugen T. O., Primmer C. R. 2002. Contemporary fisherian life-history evolution in small salmonid populations. Nature 419, 826–830 10.1038/nature01029 (doi:10.1038/nature01029) [DOI] [PubMed] [Google Scholar]
- 50.Metcalfe N. B., Thorpe J. E. 1990. Determinants of geographical variation in the age if seaward-migrating salmon, Salmo salar. J. Anim. Ecol. 59, 135–145 10.2307/5163 (doi:10.2307/5163) [DOI] [Google Scholar]
- 51.O'Malley K. G., Ford M. J., Hard J. J. 2010. Clock polymorphism in Pacific salmon: evidence for variable selection along a latitudinal gradient. Proc. R. Soc. B 277, 3703–3714 10.1098/rspb.2010.0762 (doi:10.1098/rspb.2010.0762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rowe L., Day T. 2006. Detecting sexual conflict and sexually antagonistic coevolution. Phil. Trans. R. Soc. B 361, 277–285 10.1098/rstb.2005.1788 (doi:10.1098/rstb.2005.1788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poissant J., Wilson A. J., Festa-Bianchet M., Hogg J. T., Coltman D. W. 2008. Quantitative genetics and sex-specific selection on sexually dimorphic traits in bighorn sheep. Proc. R. Soc. B 275, 623–628 10.1098/rspb.2007.1361 (doi:10.1098/rspb.2007.1361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrissey M. B., Wilson A. J. 2009. PEDANTICS: an R package for pedigree-based genetic simulation and pedigree manipulation, characterization and viewing. Mol. Ecol. Res. 10, 711–719 10.1111/j.1755-0998.2009.02817.x (doi:10.1111/j.1755-0998.2009.02817.x) [DOI] [PubMed] [Google Scholar]
- 55.R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]