Abstract
A new method for DNA diagnostics based on template-directed primer extension and detection by fluorescence polarization is described. In this method, amplified genomic DNA containing a polymorphic locus is incubated with oligonucleotide primers (designed to hybridize to the DNA template adjacent to the polymorphic site) in the presence of allele-specific dye-labeled dideoxyribonucleoside triphosphates and a commercially available modified Taq DNA polymerase. The primer is extended by the dye-terminator specific for the allele present on the template, increasing ∼10-fold the molecular weight of the fluorophore. At the end of the reaction, the fluorescence polarization of the two dye-terminators in the reaction mixture are analyzed directly without separation or purification. This homogeneous DNA diagnostic method is shown to be highly sensitive and specific and is suitable for automated genotyping of large number of samples.
[The data shown in Figure 3 are available as an online supplement at http://www.genome.org.]
DNA analysis is becoming increasingly important in the diagnosis of hereditary diseases, detection of infectious agents, tissue typing for histocompatability, identification of individuals in forensic and paternity testing, and monitoring the genetic makeup of plants and animals in agricultural research (Alford and Caskey 1994). In addition, DNA analysis is crucial in large-scale genetic studies to identify susceptibility alleles associated with common diseases involving multiples genetic and environmental factors (Risch and Merikangas 1996). Recently, attention was focused on single nucleotide polymorphisms (SNPs), the most common DNA sequence variation found in mammalian genomes (Cooper et al. 1985). Although most of the SNPs do not give rise to detectable phenotypes, a fraction of them are mutations responsible for genetic diseases. As the DNA sequence of the human genome is completely elucidated, large-scale DNA analysis will play a crucial role in determining the relationship between genotype (DNA sequence) and phenotype (disease and health) (Cooper and Clayton 1988). Although they have considerable promise for high throughput genetic analysis, the recently developed DNA diagnostic methods, including the high-density chip arrays for allele-specific hybridization analysis (Yershov et al. 1996; Wang et al. 1998), the homogeneous 5′ nuclease (TaqMan) assay (Livak et al. 1995), the homogeneous template-directed dye-terminator incorporation (TDI) assay (Chen and Kwok 1997; Chen et al. 1997), and the homogeneous molecular beacon allele-specific oligonucleotide (ASO) assay (Tyagi et al. 1998), all require specialty probes and expensive detection instrumentation.
Recently, we explored a new detection strategy for the TDI assay using an unmodified oligonucleotide probe, eliminating the need for specialty probes. Our approach combines the specificity of enzymatic discrimination between the two alleles of an SNP in a template-directed primer extension reaction and the sensitivity of fluorescence polarization.
Template-directed primer extension is a dideoxy chain-terminating DNA-sequencing protocol designed to ascertain the nature of the one base immediately 3′ to the sequencing primer that is annealed to the target DNA immediately upstream from the polymorphic site. In the presence of DNA polymerase and the appropriate dideoxyribonucleoside triphosphate (ddNTP), the primer is extended specifically by one base as dictated by the target DNA sequence at the polymorphic site. By determining which ddNTP is incorporated, the alleles present in the target DNA can be inferred. This genotyping method has been widely used in different formats and proved to be highly sensitive and specific (Nikiforov et al. 1994; Syvanen 1994; Pastinen et al. 1997).
Fluorescence polarization (FP) is based on the observation that when a fluorescent molecule is excited by plane-polarized light, it emits polarized fluorescent light into a fixed plane if the molecules remain stationary between excitation and emission (Perrin 1926). Because the molecule rotates and tumbles in space, however, FP is not observed fully by an external detector. The FP of a molecule is proportional to the molecule’s rotational relaxation time (the time it takes to rotate through an angle of 68.5°), which is related to the viscosity of the solvent, absolute temperature, molecular volume, and the gas constant. Therefore, if the viscosity and temperature are held constant, FP is directly proportional to the molecular volume, which is directly proportional to the molecular weight. If the fluorescent molecule is large (with high molecular weight), it rotates and tumbles more slowly in space and FP is preserved. If the molecule is small (with low molecular weight), it rotates and tumbles faster and FP is largely lost (depolarized) (Fig. 1). The FP phenomenon has been used to study protein–DNA and protein–protein interactions (Checovich et al. 1995; Heyduk et al. 1996), DNA detection by strand displacement amplification (Walker et al. 1996), and in genotyping by hybridization (Gibson et al. 1997). Currently, >50 fluorescence polarization immunoassays (FPIA) are commercially available, many of which are routinely used in clinical laboratories for the measurement of therapeutics, metabolites, and drugs of abuse in biological fluids (Checovich et al. 1995).
Figure 1.

Fluorescence polarization.
FP is expressed as the ratio of fluorescence detected in the vertical and horizontal axes and, therefore, is independent of the fluorescence intensity. This is a clear advantage over other fluorescence detection methods in that as long as the fluorescence is above detection limits of the instrument used, FP is a reliable measure. The degree of FP increases more or less linearly up to 10,000 Daltons in molecular mass before it levels off. Because a nucleotide bearing a fluorescent molecule has a molecular mass of ∼1000 Daltons and a fluorescent 25- to 30-mer is ∼10,000 Daltons, FP is well suited as a detection method for the primer extension reaction.
The total polarization reflects the sum of FP from all species in solution emitting at that wavelength. For a system in which the fluorophore is attached to a low molecular weight nucleotide producing a low polarization and is then incorporated into the probe oligomer at the allelic site, the polarization observed is described by the equation
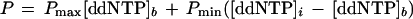 |
where Pmax is the polarization for dye-labeled ddNTP incorporated onto the TDI probe, Pmin is the polarization of the unincorporated dye-labeled ddNTP, [ddNTP]i is the initial concentration of dye-labeled ddNTP, and [ddNTP]b is the concentration of incorporated dye-labeled ddNTP. The maximum change in signal occurs with 100% incorporation of the ddNTP. Therefore, an important aspect in experimental design is to ensure that the initial concentration of dye-labeled ddNTP used in the reaction is kept at a minimum.
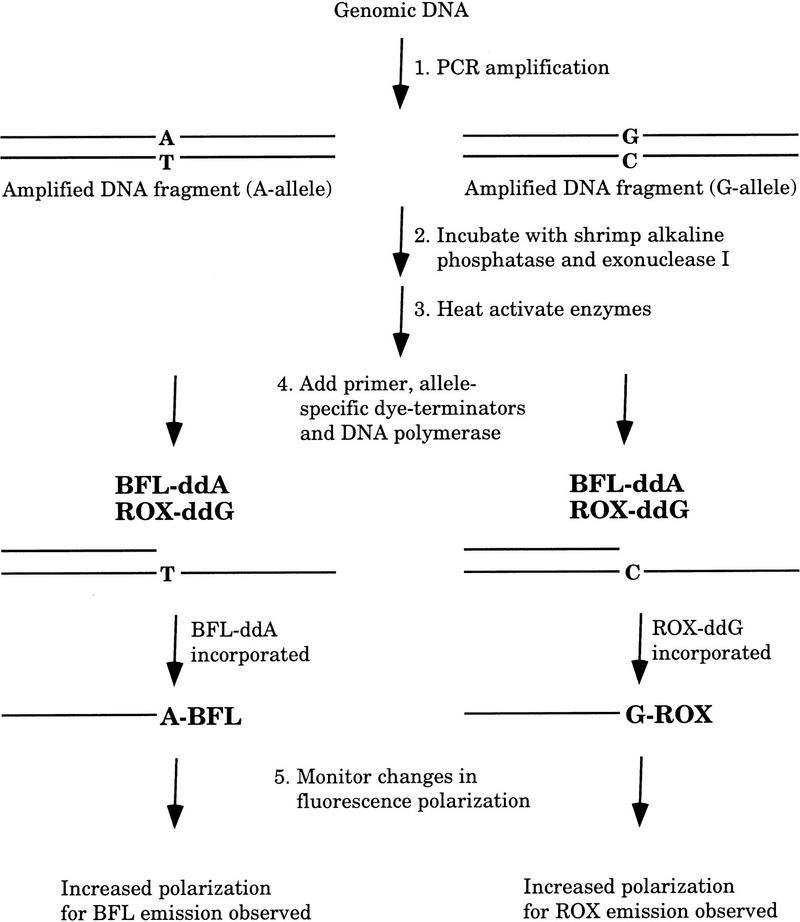
In our method, named template-directed dye-terminator incorporation assay with FP detection (FP–TDI assay), the sequencing primer is an unmodified primer with its 3′ end immediately upstream from the polymorphic or mutation site. When incubated in the presence of ddNTPs labeled with different fluorophores, the allele-specific dye-labeled ddNTP is incorporated onto the TDI primer in the presence of DNA polymerase and target DNA. The genotype of the target DNA molecule can be determined simply by exciting the fluorescent dye in the reaction and determining whether a change in FP is observed (Fig. 2).
Figure 2.
FP–TDI assay.
In this report, we demonstrate that FP is a simple, sensitive, and specific detection method in a homogeneous primer extension genotyping assay. Both single-stranded synthetic DNA oligomers and double-stranded DNA fragments amplified by PCR (Saiki et al. 1988) can be used as templates in this assay. In all cases, the FP–TDI assay proves to be highly sensitive and specific.
RESULTS
Although dye-labeled dideoxy-terminators have been used extensively in sequencing reactions (Parker et al. 1996) and the sensitivity and specificity of template-directed primer extension genotyping methods are well established (Pastinen et al. 1997; Syvanen 1998), the use of FP as a detection method in a primer extension reaction has not been reported before this work was done. Three sets of experiments were performed to show that FP is a simple, highly sensitive and specific detection method in a homogeneous primer extension reaction for single base-pair changes. In the first set of experiments, four synthetic oligonucleotide templates containing the four possible nucleotides at one particular site in the middle of otherwise identical sequence were used to establish the sensitivity and specificity of FP detection of dye-terminator incorporation. In the second set of experiments, several dyes were tested for their utility in this assay. In the third set of experiments, PCR products were used as templates in a dual-color FP–TDI assay to show that accurate genotyping data could be obtained for both alleles of a marker or mutation in a homogeneous assay.
DNA Typing by the FP–TDI Assay with Synthetic Templates
Four synthetic 48-mers with identical sequence except for position 23 were prepared (CF508-48, the variant bases are shown as boldface letters in Table 1). For each synthetic template, one of the four possible bases was found at position 23. The synthetic 48-mer served as template in four separate reactions where it was incubated with the 25-mer TDI primer (CF508-25) and one of the four 5-carboxy-fluorescein (FAM)-labeled terminators in the presence of AmpliTaq DNA polymerase FS. At the end of the TDI reaction, the reaction mixture was diluted and the fluorescence polarization was measured. Table 2 shows the results of these experiments. In all cases, only the terminator complementary to the polymorphic base was incorporated and showed significant FP change, with net gains of FP of at least 50 mP, which is nine more times standard deviation of the controls.
Table 1.
Synthetic Templates and Primers Used in the FP–TDI Studies
| Oligonucleotides | Sequence (5′ to 3′) | |||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthetic templates | ||||||||||||||||||||||||||||||||||||||||||||||||
| CF508–48A | A | T | A | T | T | C | A | T | C | A | T | A | G | G | A | A | A | C | A | C | C | A | A | A | G | A | T | G | A | T | A | T | T | T | T | C | T | T | T | A | A | T | G | G | T | G | C | C |
| CF508–48C | A | T | A | T | T | C | A | T | C | A | T | A | G | G | A | A | A | C | A | C | C | A | C | A | G | A | T | G | A | T | A | T | T | T | T | C | T | T | T | A | A | T | G | G | T | G | C | C |
| CF508–48G | A | T | A | T | T | C | A | T | C | A | T | A | G | G | A | A | A | C | A | C | C | A | G | A | G | A | T | G | A | T | A | T | T | T | T | C | T | T | T | A | A | T | G | G | T | G | C | C |
| CF508–48T | A | T | A | T | T | C | A | T | C | A | T | A | G | G | A | A | A | C | A | C | C | A | T | A | G | A | T | G | A | T | A | T | T | T | T | C | T | T | T | A | A | T | G | G | T | G | C | C |
| PCR primers | ||||||||||||||||||||||||||||||||||||||||||||||||
| C282Y–p1 | T | G | G | C | A | A | G | G | G | T | A | A | A | C | A | G | A | T | C | C | ||||||||||||||||||||||||||||
| C282Y–p2 | C | T | C | A | G | G | C | A | C | T | C | C | T | C | T | C | A | A | C | C | ||||||||||||||||||||||||||||
| D18S8–p1 | T | T | G | C | A | C | C | A | T | G | C | T | G | A | A | G | A | T | T | G | T | |||||||||||||||||||||||||||
| D18S8–p2 | A | C | C | C | T | C | C | C | C | C | T | G | A | T | G | A | C | T | T | A | ||||||||||||||||||||||||||||
| FP–TDI primers | ||||||||||||||||||||||||||||||||||||||||||||||||
| C282Y–31 | G | T | A | C | C | C | C | C | T | G | G | G | G | A | A | G | A | G | C | A | G | A | G | A | T | A | T | A | C | G | T | |||||||||||||||||
| CF508–25 | G | G | C | A | C | C | A | T | T | A | A | A | G | A | A | A | A | T | A | T | C | A | T | C | T | |||||||||||||||||||||||
| D18S8–31 | G | G | A | G | G | C | T | G | A | G | G | C | A | G | G | A | G | A | A | T | T | G | C | T | T | G | A | A | C | C | C | |||||||||||||||||
Synthetic template sequences (bold) are bases to be tested.
Table 2.
FP–TDI Assay with Synthetic Templates Using FAM-Labeled Dye Terminators
| Templates | FAM–ddA (mP)a | FAM–ddC (mP)a | FAM–ddG (mP)a | FAM–ddU (mP)a |
|---|---|---|---|---|
| CF508–48A | 52 | 36 | 54 | 89 |
| 55 | 37 | 41 | 92 | |
| 52 | 39 | 48 | 101 | |
| 50 | 39 | 40 | 93 | |
| CF508–48C | 57 | 37 | 121 | 39 |
| 50 | 37 | 126 | 30 | |
| 55 | 39 | 115 | 40 | |
| 52 | 34 | 117 | 40 | |
| CF508–48G | 52 | 92 | 42 | 42 |
| 63 | 85 | 35 | 32 | |
| 50 | 91 | 40 | 47 | |
| 49 | 103 | 37 | 35 | |
| CF508–48T | 186 | 32 | 48 | 34 |
| 180 | 38 | 63 | 41 | |
| 183 | 36 | 43 | 33 | |
| 179 | 33 | 55 | 45 | |
| Avg. ctrl. | 53 | 36 | 46 | 38 |
| s.d. Ctrl. | 4.0 | 2.5 | 8.3 | 5.3 |
| Avg. net chg.b | 129 | 57 | 74 | 55 |
Positive reactions are in boldface type.
FP measurements for FAM were made with excitation at 485 nm and monitored at 530 nm.
Net change over average of control.
FP–TDI Assay With Different Terminators Labeled with Different Dyes
In an effort to identify different dyes suitable for multicolor detection in the same reaction, a number of different dyes were studied for their FP properties in the FP–TDI assay. With all the combinations of dye-terminators tested, the optimal set of terminators, chosen for minimal standard deviations in the control samples and large net changes in the positive samples, were found to be BODIPY-fluorscein-ddA (BFL–ddA), N,N,N‘,N‘-tetramethyl-6-carboxyrhodamine (TMR–ddC) b-carboxy-x-rhodamine–ddG (ROX–ddg), and BODIPY–Texas Red–ddU (BTR–ddU) (see Table 3). In all of these cases, the net increase in FP exceeded 10 times standard deviation of the mean of the control samples. In addition, BFL–ddC, BFL–ddT, ROX–ddA, BTR–ddC, TMR–ddU, and all FAM terminators also worked well.
Table 3.
FP–TDI Assay with Synthetic Templates Using Different Dye Terminators
| Templates | BFL–ddA (mP)a | TMR–ddC (mP)a | ROX–ddG (mP)a | BTR–ddU (mP)a |
|---|---|---|---|---|
| CF508–48A | 38 | 43 | 77 | 174 |
| 37 | 53 | 73 | 175 | |
| 31 | 36 | 78 | 174 | |
| 35 | 49 | 82 | 170 | |
| CF508–48C | 20 | 50 | 214 | 32 |
| 19 | 37 | 209 | 27 | |
| 20 | 56 | 215 | 25 | |
| 14 | 38 | 207 | 26 | |
| CF508–48G | 23 | 247 | 84 | 23 |
| 24 | 266 | 80 | 30 | |
| 22 | 253 | 75 | 23 | |
| 15 | 262 | 74 | 21 | |
| CF508–48T | 113 | 52 | 81 | 32 |
| 106 | 41 | 68 | 39 | |
| 108 | 59 | 81 | 30 | |
| 103 | 32 | 76 | 28 | |
| Avg. ctrl. | 25 | 46 | 86 | 28 |
| s.d. Ctrl. | 8.2 | 8.8 | 4.6 | 5.0 |
| Avg. net chg.b | 83 | 211 | 134 | 145 |
Positive reactions are in boldface type.
FP measurements for BFL were made with excitation at 480 nm and emission at 520 nm; for TMR the excitation was 535 nm and emission was 590 nm; for BTR the excitation was 591 nm and emission was 635 nm; for ROX the excitation was 580 nm and emission was 620 nm.
Net change over average of control.
Dual Color FP–TDI Assay for Amplified Genomic DNA
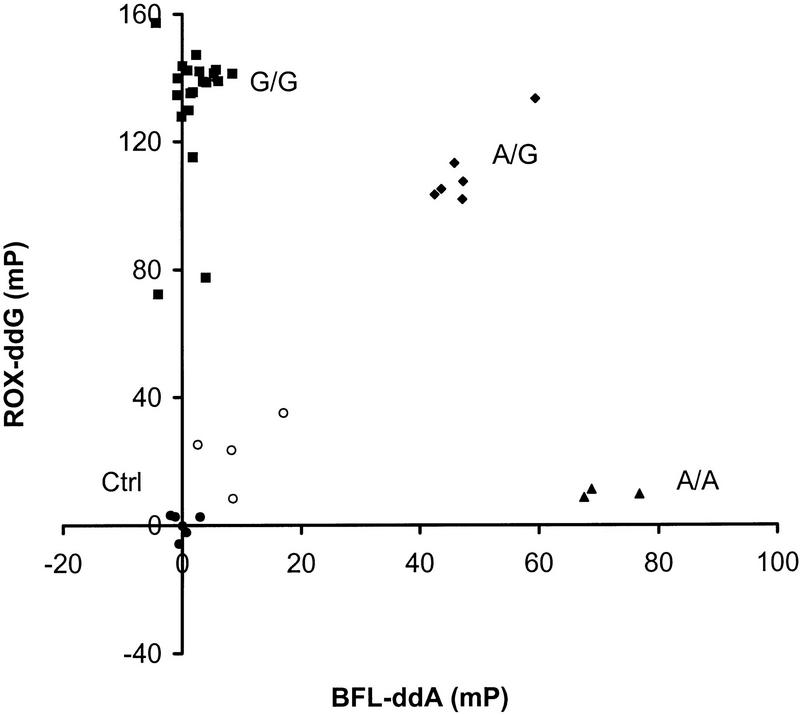
Marker D18S8 (Parry et al. 1991) and the C282Y mutation in the human hereditary hemochromatosis (HFE) gene implicated in hemochromatosis (Feder et al. 1996) were used in FP–TDI assays designed to test for both alleles in the same reaction. For marker D18S8, genomic DNA samples from 34 individuals were amplified and the excess reagents degraded enzymatically as previously reported (Chen et al. 1997) and the samples then incubated with the 31-mer TDI primer (D18S8-31)in the presence of BFL–ddA and ROX–ddG (Chen and Kwok 1997). The FP values of the reaction mixtures were read at the BFL and ROX emission wavelengths, respectively, and the results are plotted and shown in Figure 3. The FP values cluster into four groups. In the upper left corner of the plot, the samples have high FP for ROX–ddG but low FP for BFL–ddA, signifying that they are of homozygous G genotype (█). The heterozygous A/G samples (♦) exhibit high FP values in both BFL–ddA and ROX–ddG and occupy the right upper corner of the plot. The homozygous A/A samples (▴) are found in the lower right corner, with low ROX–ddG but high BFL–ddA FP values. The negative controls (●) and samples with failed PCR reactions (○) occupy the area near the origin with low FP values for both dyes. The positive samples in both the BFL–ddA and the ROX reactions gave FP values that were >40 mP and 100 mP above average of controls, respectively. These values were >20 times standard deviation of the controls and the genotypes of the samples were easily assigned. These results were in complete concordance with those obtained by other genotyping methods (Nickerson et al. 1990; Chen et al. 1997). Of 34 test samples, 4 gave inconclusive results because of PCR failure, which would prevent analysis by any method, including those based on gel electrophoresis.
Figure 3.
Changes in fluorescence polarization for DNA samples genotyped with the FP–TDI assay. The results are plotted in mP units above the average polarization of the negative controls. A change of 40 mP for a dye-terminator is scored as positive. DNA samples from 34 individuals and 6 water blanks were used. (█) Samples positive for the G allele but negative for the A allele (homozygous G); (▴) samples positive for the A allele but negative for the G allele (homozygous A); (♦) samples positive for both alleles (heterozygotes); (●) negative controls; (○) samples with failed PCR amplification. Numerical values for the data are available as online supplementary material at http://www.genome.org.
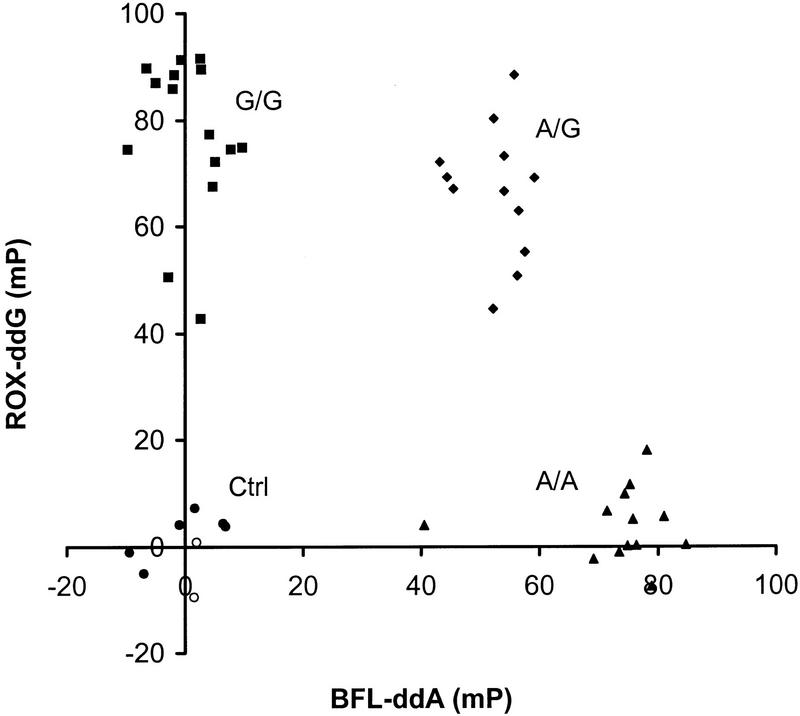
For the C282Y mutation in the HFE gene, genomic DNA samples from 42 patients and healthy controls were tested with the 31-mer TDI primer (C282Y-31) in the presence of BFL–ddA and ROX–ddG. The results are shown in Figure 4. Once again, the FP values cluster into four clearly delineated groups and the genotypes can be assigned easily. In this experiment, two samples gave inconclusive results because of PCR failure and are found near the negative controls at the origin. As before, the genotypes determined by the FP–TDI assay agreed completely with those obtained by other genotyping methods.
Figure 4.
FP–TDI genotyping data for the C282Y mutation in human hereditary hemochromatosis gene. DNA samples from 42 patients and 6 water blanks were tested. (▴) Samples positive for the A allele but negative for the G allele (homozygous A); (♦) samples positive for both alleles (heterozygotes); (●) negative controls; (○) samples with failed PCR amplification.
DISCUSSION
The allele-specific primer extension reaction has been used widely in DNA testing and there are several detection methods based on this core allelic discrimination reaction. We report here a new detection method for the assay based on fluorescence polarization. The usefulness of FP has long been established in clinical assays (Checovich et al. 1995; Heyduk et al. 1996). Recently, FP was used in the amplification-refractory mutation system (ARMS) genotyping assay, where allele-specific PCR products were detected by hybridization of amplicon-specific DNA sequences (Walker and Linn 1996; Gibson et al. 1997). Although hybridization is sensitive and robust in detecting specific DNA sequences, it needs extensive experience in probe design and optimization in genotyping applications. With the primer extension reaction, the reaction conditions are practically universal and almost no optimization is required.
Our data show that when a fluorescent dye-labeled ddNTP is incorporated onto a primer (25–31 bases), the FP value increases dramatically. The net change in FP can be improved further if the free rotation of the dye around one axis of its linkage to the oligonucleotide can be restricted. Even without optimized TDI primers, the net change of FP value in all positive reactions is >40 mP, which is at least seven times standard deviation of the negative controls (>99% significance).
These studies demonstrate that the FP–TDI assay is a robust, homogeneous genetic test that requires no modified primers for its execution, and retains the sensitivity and specificity of the primer extension reaction. Because the unmodified FP–TDI primer cost is only 20% of that of a dye-labeled primer, this new detection method is more cost effective than genotyping assays based on dye-labeled probes. However, this assay still requires relatively costly (at current prices) dye-labeled ddNTPs and FP detection instrumentation. Another advantage of the FP–TDI assay is the potential for testing all four possible alleles in a single reaction. For SNP typing, all one needs is a master mix with four fluorescence dyes each linked to a different base. The standardization of reagent supply will simplify genotyping protocols and reduce human errors.
As the demand for DNA testing (i.e., assaying for the presence or absence of known DNA polymorphisms or mutations) is expected to increase dramatically in the areas of DNA diagnostics, forensics, and population studies, developing a cost-effective and highly scalable application is of great interest to the scientific community. The homogeneous FP–TDI assay is highly suitable for large-scale genetic studies because it is not limited by a particular reaction format. Moreover, it offers the flexibility of using the best markers as they become available for a particular application without redesigning dye-labeled probes or refabricating high-density DNA chips. Furthermore, the FP–TDI assay is simple to set up (by adding the standard reagent mixture to the DNA template), the results are obtained in electronic form minutes after the allele-discriminating reaction is performed, and the genotype can be assigned automatically by use of a simple computer program. Because the principle of FP applies to any fluorescent dye, including those absorbing in the infrared region, studies are now under way to identify a set of four optimal fluorescent dyes to produce a standard set of reaction conditions suitable for a multiplex FP–TDI assay. As DNA diagnostic tests will no doubt be performed more and more by clinical rather than research laboratories, methods (such as the FP–TDI assay) using standard protocols that require minimal laboratory skills or manual handling will be crucial to the clinical practice of medicine in the future.
METHODS
Enzymes
AmpliTaq and AmpliTaq-FS DNA polymerase were obtained from Perkin-Elmer Applied Biosystems Division (Foster City, CA). Shrimp alkaline phosphatase and Escherichia coli exonuclease I were purchased from Amersham (Arlington Heights, IL).
Oligonucleotides
Oligonucleotides used are listed in Table 1. PCR and TDI primers and synthetic template oligonucleotides were obtained from Life Technologies (Grand Island, NY).
Dye-Labeled Dideoxyribonucleoside Triphosphates
Dideoxyribonucleoside triphosphates labeled with FAM, ROX, TMR, BFL, and BTR were generous gifts from NEN Life Science Products, Inc. (Boston, MA). Unlabeled ddNTPs were purchased from Pharmacia Biotech (Piscataway, NJ).
PCR Amplification
Human genomic DNA (20 ng) from 34 unrelated individuals and 6 negative controls (water-blanks) were amplified for the marker D18S8 in 20-μl reaction mixtures containing 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 1.5 mm MgCl2, 0.2 mm dNTP, 1 μm of each primer, and AmpliTaq DNA polymerase (1 unit). The reaction mixture was held at 94°C for 2 min followed by 10 cycles of 94°C for 10 sec, ramping to 60°C >90 sec, held at 60°C for 30 sec, followed by 30 cycles of 94°C for 10 sec, and 53°C for 30 sec. For hemochromatosis mutation C282Y, 42 samples and 6 negative controls were amplified in the same buffer with these cycling conditions: 94°C for 2 min followed by 10 cycles of 94°C for 10 sec, ramping to 68°C >90 sec, held at 68°C for 30 sec, followed by 30 cycles of 94°C for 10 sec, and 62°C for 30 sec. At the end of the reaction, the reaction mixtures were held at 4°C until further use.
Primer and dNTP Degradation
At the end of the PCR assay, 10 μl of an enzymatic cocktail containing shrimp alkaline phosphatase (2 units), E. coli exonuclease I (1 unit) in shrimp alkaline phosphatase buffer [20 mm Tris-HCl (pH 8.0), 10 mm MgCl2] was added to the PCR product. The mixture was incubated at 37°C for 30 min before the enzymes were heat inactivated at 95°C for 15 min. The DNA mixture was kept at 4°C and used in the FP–TDI assay without further quantification or characterization.
Genotyping by the FP–TDI Assay
To the enzymatically treated PCR product was added 10 μl of TDI reaction mixture containing the TDI buffer [50 mm Tris-HCl (pH 9.0), 50 mm KCl, 5 mm MgCl2, 8% glycerol, 0.1% Triton X-100], 1.25 μm TDI primer, 25 nm of each allele-specific dye-labeled ddNTP, 100 nm unlabeled other two ddNTPs, and AmpliTaq DNA polymerase FS (1 unit). The reaction mixtures were incubated at 93° 1 min, followed by 35 cycles of 93° 10 sec and 55° 30 sec. At the end of the reaction, the samples were held at 4°C.
Fluorescence Polarization Measurement
After the primer extension reaction, 100 μl of TDI buffer and 50 μl of methanol were added to each tube before they were transferred to a microtiter plate for FP measurement on a Fluorolite FPM2 instrument (Jolley Consulting and Research, Grayslake, IL) or Analyst fluorescence reader (LJL Biosystems, Sunnyvale, CA). Fluorescence polarization value was calculated using the formula:
 |
where Ivv is the emission intensity measured when the excitation and emission polarizers are parallel and Ivh is the emission intensity measured when the emission and excitation polarizers are oriented perpendicular to each other. The degree of polarization is expressed by the unit mP, or a 0.001 ratio between (Ivv − Ivh) and (Ivv + Ivh).
Genotype Assignment
The average FP value and standard deviation of the negative control samples were determined for each set of experiment. The FP value of the test sample reactions was then compared to the average FP value of the control samples. If the net change is >40 mP (more than seven times the standard deviation of the controls), the test sample is scored as positive for the allele.
Acknowledgments
We thank Dr. Phil Buzby of NEN Life Science Products, Inc. for generous gifts of various dye-labeled ddNTPs used in this study, Dr. Sandy Spurgeon of Perkin-Elmer Applied Biosystems Division for a generous gift of AmpliTaq DNA polymerase FS, Dr. Andi Gnirke for hemochromatosis DNA samples, and Qun Li for technical assistance. This work was supported in part by grants from the National Institutes of Health to P.Y.K. (RO1-HG1720 and RO1-EY12557) and to X.C. (F32-HG156).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sam@psts.wustl.edu; FAX (314) 362-8159.
REFERENCES
- Alford RL, Caskey CT. DNA analysis in forensics, disease and animal/plant identification. Curr Opin Biotechnol. 1994;5:29–33. doi: 10.1016/s0958-1669(05)80066-7. [DOI] [PubMed] [Google Scholar]
- Checovich WJ, Bolger RE, Burke T. Fluorescence polarization—A new tool for cell and molecular biology [published erratum appears in Nature 1995; 375:520] Nature. 1995;375:254–256. doi: 10.1038/375254a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Kwok PY. Template-directed dye-terminator incorporation (TDI) assay: A homogeneous DNA diagnostic method based on fluorescence resonance energy transfer. Nucleic Acids Res. 1997;25:347–353. doi: 10.1093/nar/25.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zehnbauer B, Gnirke A, Kwok PY. Fluorescence energy transfer detection as a homogeneous DNA diagnostic method. Proc Natl Acad Sci. 1997;94:10756–10761. doi: 10.1073/pnas.94.20.10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Clayton JF. DNA polymorphism and the study of disease associations. Hum Genet. 1988;78:299–312. doi: 10.1007/BF00291724. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Smith BA, Cooke HJ. An estimate of unique DNA sequence heterozygosity in the human genome. Hum Genet. 1985;69:201–205. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr, Ellis MC, Fullan A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nature Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- Gibson NJ, Gillard HL, Whitcombe D, Ferrie RM, Newton CR, Little S. A homogeneous method for genotyping with fluorescence polarization. Clin Chem. 1997;43:1336–1341. [PubMed] [Google Scholar]
- Heyduk T, Ma Y, Tang H, Ebright RH. Fluorescence anisotropy: Rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 1996;274:492–503. doi: 10.1016/s0076-6879(96)74039-9. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Marmaro J, Todd JA. Towards fully automated genome-wide polymorphism screening. Nature Genet. 1995;9:341–342. doi: 10.1038/ng0495-341. [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Kaiser R, Lappin S, Stewart J, Hood L, Landegren U. Automated DNA diagnostics using an ELISA-based oligonucleotide ligation assay. Proc Natl Acad Sci. 1990;87:8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov TT, Rendle RB, Goelet P, Rogers YH, Kotewicz ML, Anderson S, Trainor GL, Knapp MR. Genetic bit analysis: A solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res. 1994;22:4167–4175. doi: 10.1093/nar/22.20.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LT, Zakeri H, Deng Q, Spurgeon S, Kwok PY, Nickerson DA. AmpliTaq DNA polymerase, FS dye-terminator sequencing: Analysis of peak height patterns. Biotechniques. 1996;21:694–699. doi: 10.2144/96214rr02. [DOI] [PubMed] [Google Scholar]
- Parry PJ, Markie D, Fearon ER, Nigro JM, Vogelstein B, Bodmor WF. PCR-based detection of two MspI polymorphic sites at D18S8. Nucleic Acids Res. 1991;19:6983. doi: 10.1093/nar/19.24.6983-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T, Kurg A, Metspalu A, Peltonen L, Syvanen AC. Minisequencing: A specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 1997;7:606–614. doi: 10.1101/gr.7.6.606. [DOI] [PubMed] [Google Scholar]
- Perrin F. Polarization de la lumiere de fluorescence. Vie moyenne de molecules dans l’etat excite. J Phys Radium. 1926;7:390–401. [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Saiki R K, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Syvanen AC. Detection of point mutations in human genes by the solid-phase minisequencing method. Clin Chim Acta. 1994;226:225–236. doi: 10.1016/0009-8981(94)90217-8. [DOI] [PubMed] [Google Scholar]
- ————— Solid-phase minisequencing as a tool to detect DNA polymorphism. Methods Mol Biol. 1998;98:291–298. doi: 10.1385/0-89603-443-7:291. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- Walker GT, Linn CP. Detection of Mycobacterium tuberculosis DNA with thermophilic strand displacement amplification and fluorescence polarization. Clin Chem. 1996;42:1604–1608. [PubMed] [Google Scholar]
- Walker GT, Linn CP, Nadeau JG. DNA detection by strand displacement amplification and fluorescence polarization with signal enhancement using a DNA binding protein. Nucleic Acids Res. 1996;24:348–353. doi: 10.1093/nar/24.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Yershov G, Barsky V, Belgovskiy A, Kirillov E, Kreindlin E, Ivanov I, Parinov S, Guschin D, Drobishev A, Dubiley S, Mirzabekov A. DNA analysis and diagnostics on oligonucleotide microchips. Proc Natl Acad Sci. 1996;93:4913–4918. doi: 10.1073/pnas.93.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]