Abstract
Recent advances in DNA sequencing technologies, both in the form of high lane-density gels and automated capillary systems, will lead to an increased requirement for sample preparation systems that operate at low cost and high throughput. As part of the development of a fully automated sequencing system, we have developed an automated subsystem capable of producing 10,000 sequence-ready ssDNA templates per day from libraries of M13 plaques at a cost of $0.29 per sample. This Front End has been in high throughput operation since June, 1997 and has produced > 400,000 high-quality DNA templates.
Improvements in electrophoresis technology have been announced recently by various companies (Molecular Dynamics, Perkin-Elmer/ABI, Beckman Coulter). In particular, the 96-sample capillary electrophoresis instruments being released promise increased throughput and decreased cost for DNA sequencing. Despite improved separation time, automation, and sample tracking compared to slab gels, capillary technologies have not gained acceptance in the past because of difficulties in resolving long DNA strands. Recent developments in sequencing reaction purification (Ruiz-Martineź et al. 1998) and their effect on read length have made it more likely that capillary electrophoresis technologies will supplant slab gel-based sequencing instruments in the near future.
Consequently, there is an increasing demand for high-throughput, low-cost methods for the preparation of samples to supply these instruments. Commercially available methods for high-throughput sample purification (Qiagen) are expensive (∼$1 per sample for the purification kit only; we estimate the total cost of sample preparation for most centers to be between $1.50–3.00). The more common centrifugation-based protocols are very labor intensive. The high sample-preparation costs for sequencing centers that rely on these methods will offset savings resulting from advances in electrophoresis systems.
At Stanford, a modular, integrated, and automated system is being developed for shotgun sequencing DNA at a rate of 10,000 sequencing lanes per day, and at a cost of $0.70 per lane. In June of 1997, the first half of this system, here referred to as the “Front End,” was completed. This subsystem produces sequence-ready templates from dishes of M13 plaques at a throughput and cost compatible with the stated goals, and is therefore ideal for use with new, high-throughput electrophoresis technology. The total cost of plating, picking, growth, and template purification with this system is $0.29 per sample, 5–10 times less than the cost of the equivalent processes employed at many sequencing centers. Though other integrated, automated systems (Hawkins et al. 1997) have been constructed for template purification and cycle sequencing, no other proven automated systems exist that are capable of producing templates from dishes of M13 clones at this throughput and cost.
The Front End is operated by a single technician (per 8-hr shift) and produces all the M13 templates required by our production sequencing center. In addition, it is used to produce all the Arabidopsis thaliana templates (Marziali et al. 1997) sequenced by all members of the SPP consortium[Nancy Federspiel (Standford, CA) Sakis Theologis (U.C. Berkeley), and Joe Ecker (University of Pennsylvania, Philadelphia) (http://sequence-www.stanford.edu/ara/SPP.html)]. After a year of operation, >400,000 templates have been produced for Escherichia coli, A. thaliana, Candida albicans, Chlamydia trachomatis, Plasmodium falciparum, and Homo sapiens sequencing projects. This number of templates represents <20% of the available Front End throughput and is only limited by the number of available sequencers.
The Stanford Automated Sequencing System
The Stanford automated sequencing system is an integrated collection of modules, each of which can be operated individually. The modules are interfaced through the use of plastic cassettes containing up to 14 microtiter (96-well) plates. These modules are also designed to link electronically to our sample tracking and quality control database. At the projected throughput of 10,000 samples per day, only eight cassettes need to be manually transferred per instrument per day. This simple operation requires <5 min of the operator’s time.
The modularity of this system differentiates it from other systems designed for high-throughput DNA sequencing. Constructing an integrated system from a collection of stand-alone modules with a common interface results in many benefits over a monolithic, fully automated system. Modularity provides greater flexibility in operation and scheduling of the instruments. It also allows extensive testing of individual modules prior to the completion of adjacent instruments. Similarly, export, maintenance, and piece-wise integration of the modules into a production sequencing center are facilitated.
The automated system is composed of a plaque picker, incubator/shaker, template preparation instrument, thermal cycler, and capillary electrophoresis sequencer. The first three of these modules make up the Front End and have been in operation for over a year. Production-level sequencing of templates produced by the Front End is currently based on Perkin-Elmer Taq-FS Big-Dye Primer and Terminator brews. The terminators are set up manually or with a Robbins Hydra and cycled on MJ Research and PE 9700 cyclers. The Primer reactions are run at one-fourth volume dilution on ABI Catalyst workstations. Electrophoresis is performed on ABI 377 sequencers with 48 lanes per gel. The new thermal cycler and sequencer instruments are being developed.
The Front End process begins with libraries of M13 clones plated with β-galactosidase screen into square petri dishes. After overnight growth, up to 100 dishes are stacked in the input carousel of the plaque picker module (Fig. 1) that picks the selected plaques into 96-well microtiter plates filled previously with media and cells. The picker acquires a plaque dish, images it, and identifies useful plaques; it then retrieves a target plate from a cassette, positions plate and dish under the needle turret, and begins the transfer process. The image processing software discriminates against blue plaques (no insert), irregularly shaped plaques, and plaques that are in close proximity to a white or blue neighbor.
Figure 1.

Automated plaque picker. Plaque dishes are stacked in the carousel (left), picked up by an XY lead screw stage (bottom), imaged over the lighting/imaging station (center), and picked by the main needle turret (center). Plaques are picked into plates retrieved from a carousel of cassettes (right). The instrument is shown here picking plaques from a dish and placing into a microtiter plate held by the server arm. The sterilization coil and wash stations are just visible below and behind the needle turret.
This instrument inoculates ∼20 microtiter plates per hour (average 3 min/plate) and can process up to 84 plates unattended. When the number of plaques per dish is within the desired window of 50–300, >90% of the desired plaques are picked, and >95% of blue plaques are rejected. Less than 2% of picks result in failed templates because of mixed clones, blue plaque, or blank picks.
The plaque picker stores the inoculated plates in cassettes that are transferred to our high-capacity shaker (not shown) for incubation. Up to four cassettes are housed on the shaker in sealed tubes that are flushed with oxygen gas during the growth period. Growth is performed in 350-μl plates filled with 300 μl of growth media per well; this geometry allows good aeration of the surface of the growth media and prevents carbon dioxide build-up as can happen with deep-well blocks. The resulting M13 phage titer (3 × 1012 to 1 × 1013 PFU/ml over 12–16 hr) is four- to five fold higher than that obtained with standard microtiter plate growth methods (data not shown). This enhanced growth allows a sufficient quantity of phage to be produced in the same shallow well plates used by the plaque picker; no pipetting or inoculation into deep-well blocks is required.
After overnight growth, the plates are centrifuged to pellet cells, returned to their cassettes, and placed on the template-preparation module. This instrument performs a modified form of a glass-filter-based single-stranded DNA(ssDNA) purification protocol (Kristensen et al. 1987), which involves precipitating the phage using 20% PEG-8000 in 2.5 m NaCl and collecting it by positive pressure onto a glass-fiber filter plate (Polyfiltronics UN350PSC/GFB/M+D2). The phage are lysed, and the released DNA is bound to the glass fibers using 3 m NaClO4 in 70% ethanol. Salts and contaminants are washed through the filter with 70% ethanol, and the purified product is eluted from the filter using 60 μl of TE buffer.
The template instrument is shown in Figure 2; it consists of a rotating disc of six replaceable filter plates that can be moved under liquid dispensing, pipetting, and pressure filtration stations. Robotic plate servers access carousels of six standard cassettes to provide the instrument with sample and collection plates. The instrument can process six 96-well plates in 1.3 hr, producing 60–70 μl of ssDNA at a concentration of 50–70 ng/μl for a typical input phage density of 3 × 1012 PFU/ml.
Figure 2.
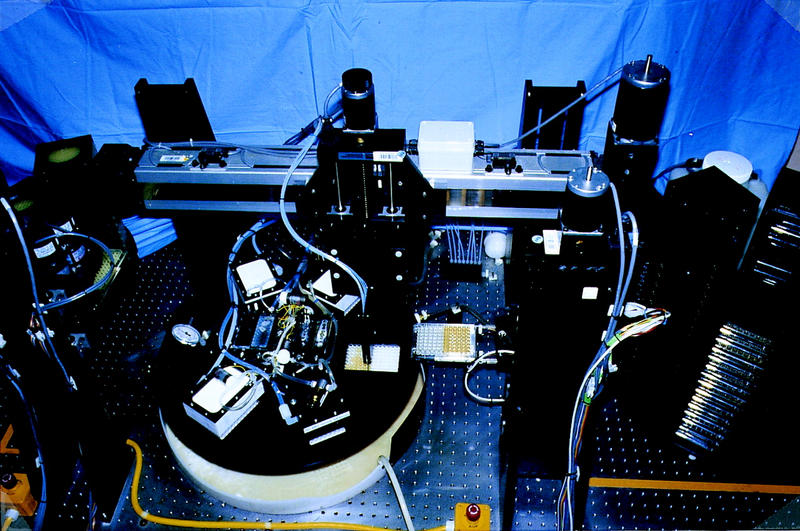
Template preparation instrument. Microtiter plates containing M13 phage supernatant and cell pellets are picked up from cassettes by the server at the right. The supernatant is transferred into white glass filter plates mounted on a rotating disc (center). The disc rotates under the various dispense (white) and pressure (at left of disc) stations to perform the wash protocol. Purified templates are collected by the plate server at the left of the picture and stored in a second set of cassettes.
RESULTS
Template Quality
High-quality templates must contain a sufficient amount of DNA from a unique library clone, and must be of sufficient purity to produce accurate sequence information. This latter aspect of template quality is measured ultimately by the quality of sequencing traces. On the other hand, the quality of the sequencing traces is heavily dependent on success of the electrophoresis and the thermal cycling, both of which are performed with standard commercial instruments. Consequently, production sequence data is not a very informative indicator of template quality. To extract template quality from such data, a separate experiment must be carried out in which a set of sequenced templates is analyzed and the templates resulting in low-quality traces are resequenced to determine whether the failure should be attributed to the sequencing or to the templates themselves. This experiment was carried out on a random set of 721 C. albicans templates, consistently using PE9700 and MJ Research thermal cyclers followed by ABI 377 sequencers.
In an effort to standardize evaluation of sequence quality, the number of bases with Phred (Ewing et al. 1998) quality scores >20 (99% confidence) has been chosen as a measure of sequencing-trace quality. The often quoted “useful read length” for assembly purposes can be considerably greater than the number of good bases (NGB) calculated with this criterium, making this a very stringent, but easily standardized, quality measure.
Figure 3 is a histogram of 721 sequencing traces from the C. albicans sequencing project binned by NGB per trace. To measure template failure rate separately from failures of template sequencing, templates that produced < 100 NGB in production sequencing were resequenced using standard conditions (Perkin Elmer TaqFS BigDye Primer brews, full volume, are cycled in Perkin Elmer 9700 thermocyclers and run on ABI 377 sequencers as per manufacturer’s protocol) (Fig. 3; hatched bars). Templates that yielded <100 NGB after the second sequencing attempt are assumed to be results of Front End failure. Choosing a quality cutoff of 100 NGB the Front-End failure rate is thus calculated to be 2.6%. Agarose gel analysis indicates that ∼25% of the failures are caused by contamination or multiple-clone picks, whereas the rest are caused by missed inoculations, failed template purification, or failed growth. Preliminary data collected in a separate test from templates sequenced using a Molecular Dynamics MegaBACE capillary electrophoresis (CE) sequencing system and associated protocols indicates good compatibility between the Front End templates and CE systems. (Samples are prepared according to the Amersham Dyenamic Direct Kit instructions with added ethanol precipitation and 70% ethanol wash. Products are run on MegaBACE capillaries filled with 2.5% LPA, at 6–9 kV with 20 to 30 sec injection times. MegaBACE is otherwise operated as per manufacturer’s protocol.) Single reads in excess of 900 NGB, and average reads (>96 samples) of 501 NGB, have been obtained with Front End templates in a CE system.(In this case, NGB is based on 98.5% confidence as assigned by the Molecular Dynamics base-caller.)
Figure 3.
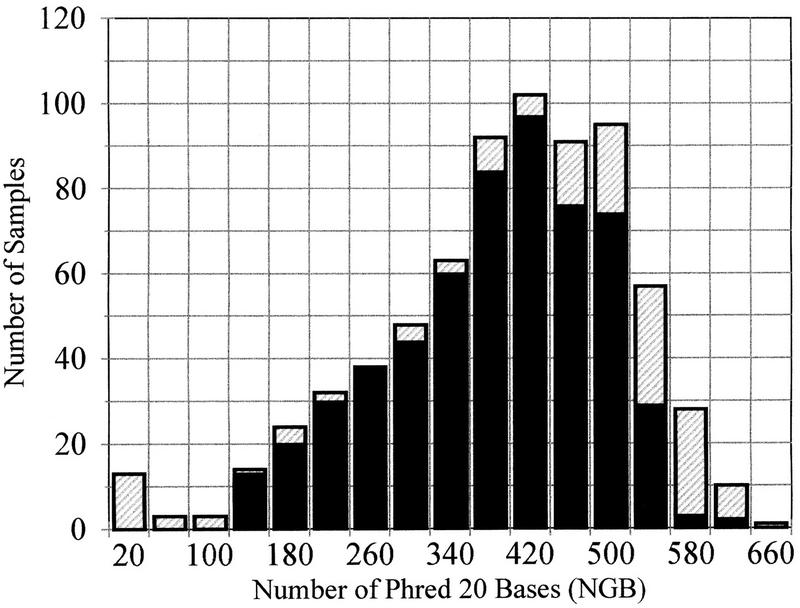
Histogram of template quality measured by number of Phred bases with quality scores of 20 and higher. The shaded regions correspond to templates resequenced to remove effect of sequencing end failures.
Operating Cost
Table 1 contains a summary of the operating costs for the Front End, including reagents, disposables, labor, instrument costs, and maintenance. Costs include all steps in the process from, and including, plating libraries into Petri dishes to collection of purified templates. Not all possible costs have been taken into account; administrative costs and overhead vary widely between research groups and should be factored in separately. In addition to the benefits of low operating costs, it should be noted that the entire system requires only three technicians to operate at a throughput of 100 plates per day, and requires less than 500 ft2 of space. These additional benefits translate to administrative and facilities cost reductions.
Table 1.
Operating Cost Summary
| Reagents and disposables | 0.154 |
| agar, X-gal, IPTG, growth media, oxygen, sodium perchlorate, ethanol, PEG-8000, sodium chloride, TE, square petri dish, polystyrene microtiter plate, polypropylene microtiter plate, glass fiber filter plate | |
| Labor costs | 0.08 |
| three research associates per 100 plates/day. Based on $60K/year salary, including benefits | |
| Instrument cost | 0.04 |
| based on estimated retail cost of $400,000 and full throughput operation over 5 years | |
| Maintenance cost | 0.02 |
| estimated cost of $40,000/year based on full throughput operation | |
| Total operating cost | $0.29/sample |
Operating cost is expressed in dollars per sample.
Reliability
Mechanical reliability of these instruments has been excellent. Mechanical failures occur rarely and are often the result of incorrect usage or power spikes and outages. These failures are usually rectified by the user who simply recalibrates or rehomes the instruments. Since June 1997 there has been a single major mechanical failure on the plaque picker caused by a defective motion stage. There have been no mechanical failures of the shaker, and there have been three valve failures on the template instrument. These failures are not catastrophic and are now being prevented with scheduled maintenance. Maintenance items for these instruments include: picker needle replacement, syringe and valve replacements, and lubrication. As the instruments are now operating on average at 20% of their maximum throughput, scheduled maintenance is required approximately every 6 months.
DISCUSSION
The Front End has decreased the cost and effort of sample preparation at our center greatly and increased its success rate and reproducibility. The instruments described in this paper have now been commercialized (Genemachines Inc.) and are available to any sequencing center. The modular design allows centers to incorporate these instruments into an existing M13 production sequencing operation with minimal disruption. The high-quality templates produced with this system are suitable for both slab–gel-based and capillary-based sequencing.
Though costs for sample preparation at very large sequencing centers are comparable to the operating costs of this system, it should be noted that these centers presumably benefit from economies of scale in purchasing of reagents and disposables. The same savings would be realized for the Front End if it were operated in a higher throughput sequencing center. Furthermore, the lower labor requirement of the automated system will lead to decreased management, administrative, and facilities costs. Though we have made no attempt to calculate such costs in this paper, it is clear that they contribute substantially to the final cost per base.
Sample preparation methods based largely on manual labor are also likely to suffer from failures caused by human error. Poor quality and reproducibility, management difficulties, and problems related to highly repetitive tasks (such as tendinitis) will worsen as labor-based production groups attempt to increase their throughput. Adoption of automated systems such as the Front End will allow scale-up of sequencing throughput within small groups, leading to more cost-effective production of DNA sequence.
METHODS
A detailed description of the protocols used in the Front End follows. Images, video clips, descriptions, and mechanical drawings of the instruments are available at http://sequence-www.stanford.edu/group/techdev/index.html. More information is also available from Genemachines (http://www.genemachines.com).
Plaque Picker
M13 phage libraries are plated onto square petri dishes (Applied Scientific AS-72077) previously filled with 25 ml of sterile LB agar using standard methods (Sambrook et al. 1989). Based on estimates of the library titer, we plate sufficient phage to produce 200–400 plaques per dish. Immediately after overnight incubation, the dishes are arranged in stacks on the automated plaque picker that sequentially retrieves each plate from the stack, images it to determine plaque locations, inserts a sterile tungsten needle in each plaque, and immerses the needle into a standard 96-well microtiter plate previously filled with growth media (300μl/well). The growth media consists of 2× TB (50 grams of Bacto-yeast tryptone, 25 grams of Bacto-tryptone in 900 mL dH2O) + 100 mL of TB salts and 10% (vol/vol) of a fresh DH12S overnight grown in TB + salts to OD550 = 1–1.5. After inoculation, the needles are washed by means of a high-velocity water jet, and sterilized by insertion into a heater coil. Typically, 105–107 phage are transferred to the inoculated plate with each pick.
Shaker/Incubator with Oxygen Enrichment
Shaking and incubation of microtiter plates are performed in cassettes, as used on the plaque picker. After the picker is finished picking, up to four cassettes of plates are removed from the picker and placed directly into the four growth chambers that are then sealed. Shaking is performed without lids on the plates, at 520 rpm with an orbit diameter of .316 in. The entire shaker is located in an incubator held at 37°C internal temperature. Medical grade 100% oxygen gas is injected into the shaker through a manifold with jets over each plate in a 3-sec burst that repeats every 30 sec (∼25 ft2 of gas are used in a 16-hr growth). Waste gas exits at the bottom of the growth chamber. The dry oxygen gas is preheated to 37°C in a heat exchanger and bubbled through a water bath to humidify it. This is done to prevent thermal cycling and evaporation of the samples during oxygen pulses. Growth time is typically 16 hr, resulting in cultures containing from 1012–1013 PFU/ml of phage.
M13 Template Purification Instrument
The protocol to be executed by the instrument is a standard glass-fiber purification protocol based on binding of DNA to glass in a chaotropic salt buffer. This version of the protocol uses a PEG/NaCl buffer for phage precipitation rather than the acidic buffers generally used for this purpose. It has been found that PEG does not clog the filter membrane as thought previously and leads to better discrimination against binding of degraded DNA to the filter matrix during phage precipitation. The protocol is executed as follows:
Add 80 μl of 20% PEG-8000 + 2.5 m NaCl to 260 μl of phage supernatant, mix, and incubate in filter to precipitate phage.
Pass solution through filter to collect phage.
Wash filter with 300 μl of 3 m NaClO4 in 70% ethanol to set up high-salt binding conditions.
Add 300 μl of 3 m NaClO4 in 70% ethanol to filter and incubate for 2 min to lyse phage and bind DNA to glass fibers.
Wash filter six times with 300 μl of 70% ethanol to remove contaminants and salts.
Dry the filter membrane with compressed air to remove traces of ethanol.
Add 60 μl of 1× TE buffer to each well of the filter plate and incubate to elute DNA.
Apply pressure to filter and collect purified templates.
To avoid the added difficulty of separating phage from the cells present in the growth culture, the machine requires that the input plates be manually transferred to a centrifuge to pellet the cells prior to use on the instrument. Pelleting is performed at 3000g for 15 min.
Acknowledgments
We gratefully acknowledge contributions by Rick Norgren and Gad Shelef for the original plaque picker and server arm design; by Michael Proctor for template quality data; by Matthew O’Keefe, Maneesh Jain, Les Roberts, the Stanford DNA Sequencing Group, Farooq Siddiqui, Scott Hunicke-Smith and Genemachines, Polyfiltronics; and by National Human Genome Research Institute for financial support (grant no. P01-HG-00205).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL andre@physics.ubc.ca; FAX (604) 822-5324.
REFERENCES
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assesment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Hawkins TL, McKernan KJ, Jacotot LB, MacKenzie JB, Richardson PM, Lander ES. Magnetic attraction to high-throughput genomics. Science. 1992;276:1887–1889. doi: 10.1126/science.276.5320.1887. [DOI] [PubMed] [Google Scholar]
- Kristensen T, Voss H, Ansorge W. A simple and rapid preparation of M13 sequencing templates for manual and automated dideoxy sequencing. Nucleic Acids Res. 1987;15:5507–15516. doi: 10.1093/nar/15.14.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marziali A, Federspiel N, Davis R. Automation for the Arabidopsis Genome Sequencing Project. Trends Plant Sci. 1997;2:71–74. [Google Scholar]
- Ruiz-Martinez MC, Salas-Solano O, Carrilho E, Kotler L, Karger BL. A sample purification method for rugged and high-performance DNA sequencing by capillary electrophoresis using replaceable polymer solutions. Anal Chem. 1998;70:1516. doi: 10.1021/ac971143f. [DOI] [PubMed] [Google Scholar]
- Sambrook I, Fritsch EF, Maniatis T, editors. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]


