Abstract
A number of biophysical and population-genetic processes influence amino acid substitution rates. It is commonly recognized that proteins must fold into a native structure with preference over an unfolded state, and must bind to functional interacting partners favourably to function properly. What is less clear is how important folding and binding specificity are to amino acid substitution rates. A hypothesis of the importance of binding specificity in constraining sequence and functional evolution is presented. Examples include an evolutionary simulation of a population of SH2 sequences evolved by threading through the structure and binding to a native ligand, as well as SH3 domain signalling in yeast and selection for specificity in enzymatic reactions. An example in vampire bats where negative pleiotropy appears to have been adaptive is presented. Finally, considerations of compartmentalization and macromolecular crowding on negative pleiotropy are discussed.
Keywords: molecular evolution, binding specificity, substitution rate, sequence–structure–function relationships
1. Introduction
Selective pressures on protein-encoding genes include selection on factors that lead to proper function of the protein. This involves selection of a protein to fold into a structure that will enable its function, binding and, in some cases, catalysis. It therefore also involves selection to enable inter-molecular interaction with partners to maintain fitness. Recent discussion in the molecular evolution literature has focused on robustness to translation errors as an important constraint on amino acid substitution rates [1]. What may also be an important part of selective pressure on sequences is selective pressure on what not to bind to. This involves both selective pressures on enzymes to catalyse a reaction only on substrates where catalysis is not deleterious and on signalling proteins to interact only with other proteins where interaction is not deleterious. One line of supporting evidence for translational robustness as a hypothesis is the correlation between expression level (concentration) of the gene/protein and evolutionary rate, but this correlation is also expected when specificity of interaction is a selective constraint [2]. As the concentration of a protein increases, so does the potential for non-specific interactions.
At a biophysical level, high-affinity binding is easy to achieve. It can be accomplished through hydrophobic interactions, where the non-specific exclusion of solvent drives binding. However, protein–protein interfaces are not simple hydrophobic patches, but also include charged residues and residues capable of other specific interactions, and are regions of proteins that evolve much more slowly than if they just maintained unspecified hydrophobic residues [3].
Thus, it may be that NOT statements are important determinants of evolutionary rates, and correspondingly of selective constraints on proteins, driven by which proteins they should not bind. The consequences of this in molecular evolution and comparative genomics remain to be explored.
2. Pleiotropy and not statements
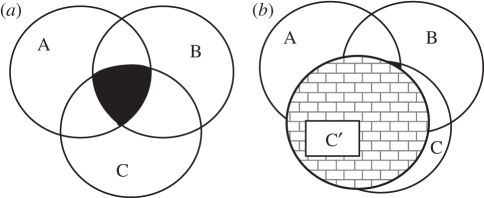
Pleiotropy has long been recognized as an important evolutionary constraint on genes and proteins. At a biophysical level, pleiotropy can reflect the necessity for a protein to bind to multiple interacting partners. This is easy to accomplish if there is no NOT statement, as a hydrophobic patch will work. This first type of pleiotropy might be termed as positive pleiotropy and this becomes harder in the presence of NOT statements, which is subsequently termed as negative pleiotropy. Negative pleiotropy also places a constraint upon sequences. If one considers sequence space in the context of Venn diagrams (figure 1), then positive pleiotropy would reflect the intersection of sequences that favourably interact with all partners that confer fitness. As suggested for protein–protein interactions, hydrophobic-rich sequences will form a large intersecting space. However, the introduction of negative pleiotropy will severely restrict this space and force it to regions of the spaces of each individual binding sequence that are less likely to intersect, making positive pleiotropy more constraining. In figure 1b, positive pleiotropy of involving A + B with C becomes almost impossible because of the negative pleiotropy of interacting with C′.
Figure 1.
(a) A Venn diagram reflects the sequence space accessible for binding three proteins (A, B and C). The black area in the centre reflects the area of overlap, where positive pleiotropic constraint restricts the area available to sequences that can bind all three interacting partners. (b) If C is duplicated to generate C′, a paralogous protein that now acts in a different pathway and is deleterious to activate, then the overlap between sequences that bind A + B and those that bind C but NOT C′ is severely reduced, as denoted by the small black area.
3. The expected interplay between negative pleiotropy, protein fold and system-level constraints
Both positive and negative thresholds for interactions are determined (Boltzmann distribution) by constraints on physical parameters. Two factors play into such constraints. One constraint is the protein fold, which dictates the orientation and size of the binding interface, further constrained by binding interface interactions with the shell residues of the hydrophobic core. The composition of the binding interface under this constraint dictates the potential for interaction with different partners with different affinities based upon accessible amino acid composition. This is reflected in the Venn diagrams in figure 1. Folds differ in the number of sequences that will fold into a structure, and this is related to the size and the thermostability of the protein [4,5], where more stable proteins can explore larger parts of sequence space while maintaining a properly folded structure. Similarly, folds with larger surface areas can more readily evolve new binding interactions and are also more likely to be under selective constraint to restrict these. Larger neutral walks through sequence space have been linked to the evolvability of proteins [6], and these are likely to be restricted by the actions of negative pleiotropy.
The second link is that between the actual physical constants (binding constants for signalling networks, binding and enzyme constants for metabolic pathways) and the overarching system-level constraints. Each link in a pathway will have a range of physical constants that give optimal flow through the pathway, where mutations that lead to deviation from this range will generally be deleterious. The size of the range of values will be very different depending upon the position in the pathway and the physical constants of other members [7].
Negative pleiotropy will reflect the opposite. It reflects a selective pressure to not interact in a pathway. Therefore, at the system level, this will be reflected in a threshold of physical constants that are deleterious to cross. Activating the wrong signalling cascade or metabolizing the wrong substrate can have clear deleterious effects on a cell and organism [7].
The evolution of pathway structure in Escherichia coli has been found to have been dominated by a process where duplicate enzymes change their specificity, while retaining a catalytic profile [8]. This is consistent with the process by which bacteria evolve the capabilities of metabolizing anthropogenic compounds [9,10]. While the pathways are not initially particularly efficient, it is clear that this occurs with a relative evolutionary ease when there has not been active selection against binding a particular compound, as moonlighting reactions in enzymes appear to be common [11,12]. Given this, there is an evolutionary potential for evolving new pathways at high rates, but this contrasts with the relative conservation of pathways over very long evolutionary periods, especially in multicellular eukaryotes [13].
4. The expected interplay between negative pleiotropy, mutation rate, population size and ease of neofunctionalization
It is thought that neofunctionalization is hard to achieve, especially for orthologous proteins involving a build-up of (positive) pleiotropic constraint. This is evidenced by the small fraction of orthologous gene tree lineages showing positive selection [14,15]. Even for duplicates, the neofunctionalization process is dependent upon the waiting time for acceptable beneficial changes, and most duplicates are non-functionalized [16–19]. It is expected that negative pleiotropy is at least partially responsible for the difficulty in neofunctionalizing, given the restrictions to sequence space placed by its constraints.
Negative pleiotropy as an active selective pressure is distinct from neutral loss of a binding interaction, as in the subfunctionalization model of duplicate gene retention [20]. In this case, even before the binding interaction is lost, the duplicate is no longer under selective pressure to bind to an interacting partner so long as the other copy still does. When the interaction between two partners is neutral, there is no restriction on the available sequence space to prevent the re-emergence of the interaction, unlike in the negative pleiotropy case.
When negative pleiotropy is considered, the population genetic underpinnings of neofunctionalization become important. Finding pockets of sequence space that yield neofunctionalized proteins may be dependent upon sampling of variants that contain multiple co-segregating mutations or that find their way through bottlenecks in sequence space. It is expected that organisms with larger mutation rates and higher population sizes would be better able to evolve rapidly in this context. Metazoans with generally low mutation rates and small population sizes would seem to have the most constrained networks with the strongest selective pressures on NOT statements. It might be expected, then, that these regulatory cascades emerged earlier in metazoan evolution and have then been relatively conserved as population sizes decreased in the evolution of chordates. In fact, many regulatory and metabolic pathways are indeed highly conserved and slow-evolving within the chordates. This is evidenced by the conserved domain structures through metazoans of many signalling proteins (for example [21]; or, more generally, SH2 and SH3 domain trees in Pfam [22]).
SH2 and SH3 domains will be used as examples in this paper. These are important proteins mediating signalling through specific protein–protein interactions in eukaryotic systems. SH2 domains bind to phosphorylated tyrosines, dependent upon the amino acids surrounding the tyrosine to generate specificity [23]. SH3 domains, which are also found in prokaryotes, also play an important role in signalling specificity, binding to proline-rich sequences in a PPII helical structure with specificity driven by interactions with non-proline residues [24].
5. Negative pleiotropy in simulated evolution
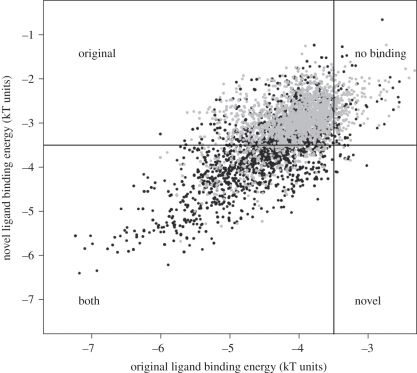
A hypothesis has been generated that describes an important role for negative pleiotropy as an evolutionary constraint on sequence evolution. A sequence simulation framework has previously been developed that enables evolution of sequences in a population with a designated mutation rate constrained to fold into a given structure and bind to a given ligand. In an evaluation of SH2 sequences that were selected to bind to an original ligand (figure 2), the sequences were evolved under this constraint and the mutations in the next generation of random sampling were evaluated for their ability to also bind a second ligand. It is observed that relatively few (but some) sequences would have been specific for this second ligand, but that evolution of binding to both ligands is easy in the absence of negative pleiotropy. In this system, it is too easy for neofunctionalization to occur. It is the case that this ease decreases depending upon the difference in binding energy between the original and new ligands, but, biologically, changes in specificity frequently involve changes between chemically related binding partners (see [23] for a phylogenetic analysis of SH2 domain-binding specificities from the human proteome).
Figure 2.
In two simulations (see [25] for details of the simulation, which involved use of Bastolla et al.'s informational matrix [26]), an SH2 domain was evolved under negative selective pressure to fold and bind to an original ligand. In one simulation, there was also a selective pressure not to bind to a second ligand. After 30% overall sequence divergence, binding affinities for a third novel ligand never seen by the protein were evaluated. It is clear that neutral evolution of binding to the third ligand via natural moonlighting is severely reduced. Black dots represent positive pleiotropy only and grey dots represent positive + negative pleiotrophy.
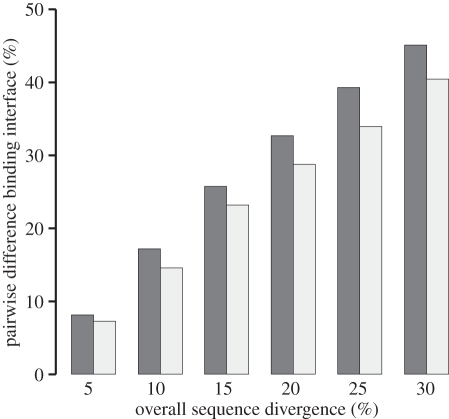
To evaluate the effect of negative pleiotropy on sequence diversity, SH2 domains were simulated as above with and without negative pleiotropies on binding, and the ultimate sequence diversity that was sampled is compared (figure 3). It is clear that negative pleiotropy for binding inhibits progression through sequence space and ultimately substitution rates. An example of the structural underpinning of this effect is shown in figure 4. This constraint may in fact enforce more compensatory co-variation than would occur in more neutral scenarios. One general rule is that charge seems to play a role in granting specificity, where matched charge–charge interactions complement affinities generated through hydrophobic interactions. An aspect of this simulation that should be noted is that only one binding partner evolved, whereas coevolution occurs in naturally evolving systems.
Figure 3.
Using the same simulation scheme as figure 2, the average pairwise sequence difference at the binding interface of the SH2 domain is compared with overall sequence diversity in the proteins as the average divergence within the population of molecules at various points of the simulation from the initial sequence. Error bars (too small to be visible) reflect standard error of the mean and are based upon greater than 200 000 sequences. Simulation with negative pleiotropy clearly reduces accessible diversity at the binding interface with evolutionary time and, correspondingly, with overall sequence diversity. The reduction of sequence diversity with negative pleiotropy is expected to be dependent upon many factors, including the binding site size and composition, the mutation rate, the effective population size and the identities of the ligands. Black bars represent positive pleiotropy only and grey bars represent positive + negative pleiotrophy.
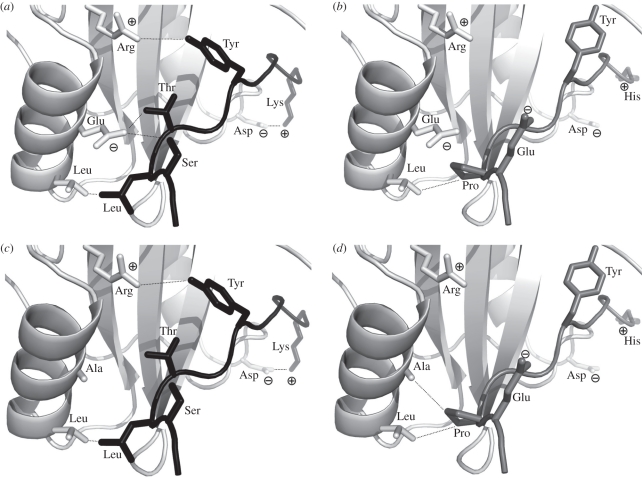
Figure 4.
From the SH2 domain simulation in figure 3, a structural example of a mutation underpinning the negative pleiotropy effect is shown with the SH2 domain in white, the ligand in black and decoy ligands in grey. (a) The native interaction is shown, where residues critical for interaction are shown as sticks, and interactions are marked with dotted lines. (b) Potential binding of the native sequence to the decoy ligand is shown, with residues differing from the native ligand shown as sticks. The hydrophobic patch on the bottom left is partially preserved, but a repulsive charge–charge interaction between two Glu residues prevents a favourable binding interaction. (c) An E17A mutation slightly alters the interaction interface, but would still bind favourably under the simulation model, suggesting an accepted mutation if only positive pleiotropy were considered. (d) When the interaction of the E17A mutant with the decoy ligand is considered, the hydrophobic interaction patch is strengthened and the repulsive Glu–Glu interaction is removed, generating a favourable binding interaction. Under selective pressure for negative pleiotropy, this less specific protein sequence would be rejected.
The protein design community has also noticed the importance of negative design (negative pleiotropy) in optimizing sequences. It is suggested that negative pleiotropy restricts sequence space, but owing to the metastable nature of energetic distributions of protein folding it does so without a major deviation from optimal energetics [27,28]. In fact, sequences with lower overall affinities are less likely to enable non-specific interactions. Correspondingly, the importance of negative design (in protein design, corresponding with negative pleiotropy in evolution) increases with the contact density, as these are the interactions that are most likely to lead to high-affinity non-specific interactions [29].
6. Enzymes and negative pleiotropy
Enzymes have tight control over substrate specificity and there are many classic cases of negative pleiotropy among enzymes. Alcohol dehydrogenases in primates include five classes with different physiological roles, and corresponding differences in substrate specificities, expression profiles and kinetic constants [30]. Differences in specificity involve the length of the chain on the alcohol, with preferences for short-chain versus long-chain alcohols. Ethanol is a short-chain alcohol, while retinol is a long-chain alcohol. Retinol is oxidized ultimately to retinoic acid, a transcription factor, when bound by the retinoic acid receptors. These transcriptional activators regulate a number of developmentally important pathways. Specific enzymes have evolved to independently oxidize ethanol and retinol as cross-talk between the pathways is expected to be deleterious, although the physiological extent of cross-talk in various tissues is unclear ([31,32]; Matthew Carrigan & Steven A. Benner 2011, unpublished data and personal communication).
More generally, optimization of binding affinities or kinetic constants can be seen as a quantitative version of a NOT statement (although not formally negative pleiotropy), whereby an enzyme that is very active or binds a necessary partner very tightly will be deleterious and restrict sequence diversity in a similar manner to selective pressures not to bind something. This would be the case when there is a NOT statement against binding and the concentration of a binding partner is very low. However, owing to the metastable nature of evolutionary selection, this effect may be smaller [27].
While preventing cross-talk is important, negative pleiotropy can drive adaptation as well. In vampire bats, the most basal species, Diphylla ecaudata, has only a single plasminogen activator (inhibiting blood clotting) that can bind mammalian-specific plasminogen activator inhibitor I (PAI-1), enabling it to feed only on bird blood. The common vampire bat, Desmodus rotundus, has evolved plasminogen activator paralogues that have lost PAI-1 inhibition [33], and this negative pleiotropy for not binding to PAI-1 enables using mammalian blood as a food source. While adaptation is achieved through domain loss in this particular case, it reflects the mechanism by which addition of negative pleiotropy can drive evolution. Processes like this (through whatever mechanism is most easily accessible evolutionarily) may be common in organismal coevolutionary scenarios.
7. Sh3 domains and negative pleiotropy
While there are many examples of negative pleiotropy acting on enzymes, Zarrinpar et al. [34] present an equally compelling case for signalling via protein–protein interactions, using SH3 domains. The Saccharomyces cerevisiae proteome contains 27 different SH3 domains, each involved in specific signalling interactions. In this case, the SH3 domain from Sho1 is highly specific for its interaction with Pbs2. Replacing the Sho1 SH3 domains with less specific SH3 domains that introduced cross-talk generated a fitness cost, which was not owing to a defect in the native pathway. The SH3 domains that could reconstitute pathway activity were also shown to bind Pbs2 in vitro. Thus, there is biological evidence of a major role for negative pleiotropy in signalling as well as metabolism.
8. Compartmentalization, the intracellular milieu and solution thermodynamics
Eukaryotic organisms have other mechanisms of dealing with negative pleiotropy as well. Compartmentalization is a solution that prevents proteins that would interact from seeing each other and having the ability to interact. When proteins are expressed in different compartments (either in different cell types or in different intracellular compartments within a cell type), they will not experience selection for negative pleiotropy. Proteins expressed at different times will also not be subject to selection for negative pleiotropy.
Another consideration is that binding interactions in vivo are not the same as binding interactions in vitro [35]. The intracellular milieu is extremely dense with proteins that are restricted in their diffusion. This results in excluded volume effects, where effective local concentrations of molecules about each other will be higher, potentially leading to stronger non-specific interactions. In addition to altering the affinities of molecules for each other, however, regional differences in effective local concentrations of molecules about each other can also serve as a type of compartmentalization, even in bacterial cells. Ultimately, molecules behave very differently in intracellular environments, but the effects of this on selection for negative pleiotropy are not all unidirectional.
With these caveats, it is still clear that negative pleiotropy or NOT statements play a role in governing selective pressures on sequences. It is evident in patterns of substitution and in selective pressures on protein functions. As molecular evolution increasingly integrates principles from systems biology and biophysical chemistry into models, negative pleiotropy will be an important consideration, and an important hypothesis to consider.
Acknowledgements
We thank Matthew Carrigan, Ashley Teufel, Jan Kubelka and Jessica Siltberg-Liberles for helpful discussions. D.A.L. receives funding from NSF DBI-0743374 and by NIH-INBRE award P20 RR016474, which also funds J.A.G. and M.D.M.T.
References
- 1.Drummond D. A., Wilke C. O. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 10.1016/j.cell.2008.05.042 (doi:10.1016/j.cell.2008.05.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf Y. I., Gopich I. V., Lipman D. J., Koonin E. V. 2010. Relative contributions of intrinsic structural-functional constraints and translation rate to the evolution of protein-coding genes. Genome Biol. Evol. 2, 190–199 10.1093/gbe/evq010 (doi:10.1093/gbe/evq010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pechmann S., Levy E. D., Tartaglia G. G., Vendruscolo M. 2009. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc. Natl Acad. Sci. USA 106, 10 159–10 164 10.1073/pnas.0812414106 (doi:10.1073/pnas.0812414106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dokholyan N. V., Shakhnovich E. I. 2001. Understanding hierarchical protein evolution from first principles. J. Mol. Biol. 312, 289–307 10.1006/jmbi.2001.4949 (doi:10.1006/jmbi.2001.4949) [DOI] [PubMed] [Google Scholar]
- 5.Bloom J. D., Wilke C. O., Arnold F. H., Adami C. 2004. Stability and the evolvability of function in a model protein. Biophys. J. 86, 2758–2764 10.1016/S0006-3495(04)74329-5 (doi:10.1016/S0006-3495(04)74329-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soskine M., Tawfik D. S. 2010. Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 11, 572–582 10.1038/nrg2808 (doi:10.1038/nrg2808) [DOI] [PubMed] [Google Scholar]
- 7.Soyer O. S., Kuwahara H., Csikász-Nagy A. 2009. Regulating the total level of a signaling protein can vary its dynamics in a range from switch like ultrasensitivity to adaptive responses. FEBS J. 276, 3290–3298 10.1111/j.1742-4658.2009.07054.x (doi:10.1111/j.1742-4658.2009.07054.x) [DOI] [PubMed] [Google Scholar]
- 8.Light S., Kraulis P. 2004. Network analysis of metabolic enzyme evolution in Escherichia coli. BMC Bioinform. 5, 15. 10.1186/1471-2105-5-15 (doi:10.1186/1471-2105-5-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copley S. D. 2000. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 25, 261–265 10.1016/S0968-0004(00)01562-0 (doi:10.1016/S0968-0004(00)01562-0) [DOI] [PubMed] [Google Scholar]
- 10.Copley S. D. 2009. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 5, 559–566 10.1038/nchembio.197 (doi:10.1038/nchembio.197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copley S. D. 2003. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 7, 265–272 10.1016/S1367-5931(03)00032-2 (doi:10.1016/S1367-5931(03)00032-2) [DOI] [PubMed] [Google Scholar]
- 12.Bloom J. D., Arnold F. H. 2009. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl Acad. Sci. USA 106(Suppl. 1), 9995–10 000 10.1073/pnas.0901522106 (doi:10.1073/pnas.0901522106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim W. A., Pawson T. 2010. Phosphotyrosine signaling: evolving a new cellular communication system. Cell 142, 661–667 10.1016/j.cell.2010.08.023 (doi:10.1016/j.cell.2010.08.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liberles D. A., Schreiber D. R., Govindarajan S., Chamberlin S. G., Benner S. A. 2001. The adaptive evolution database (TAED). Genome Biol. 2, RESEARCH0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth C., Betts M. J., Steffansson P., Saelensminde G., Liberles D. A. 2005. The adaptive evolution database (TAED): a phylogeny based tool for comparative genomics. Nucleic Acids Res. 33, D495–D497 10.1093/nar/gki090 (doi:10.1093/nar/gki090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch M., Conery J. S. 2003. The origins of genome complexity. Science 302, 1401–1404 10.1126/science.1089370 (doi:10.1126/science.1089370) [DOI] [PubMed] [Google Scholar]
- 17.Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. 2005. Modeling gene and genome duplications in eukaryotes. Proc. Natl Acad. Sci. USA 102, 5454–5459 10.1073/pnas.0501102102 (doi:10.1073/pnas.0501102102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes T., Liberles D. A. 2007. The pattern of evolution of smaller-scale gene duplicates in mammalian genomes is more consistent with neo- than subfunctionalisation. J. Mol. Evol. 65, 574–588 10.1007/s00239-007-9041-9 (doi:10.1007/s00239-007-9041-9) [DOI] [PubMed] [Google Scholar]
- 19.Rastogi S., Liberles D. A. 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 5, 28. 10.1186/1471-2148-5-28 (doi:10.1186/1471-2148-5-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M., O'Hely M., Walsh B., Force A. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159, 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cetkovic H., Grebenjuk V. A., Müller W. E., Gamulin V. 2004. Src proteins/src genes: from sponges to mammals. Gene 342, 251–261 10.1016/j.gene.2004.07.044 (doi:10.1016/j.gene.2004.07.044) [DOI] [PubMed] [Google Scholar]
- 22.Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 10.1093/nar/gkp985 (doi:10.1093/nar/gkp985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H., et al. 2008. Defining the specificity space of the human SRC homology 2 domain. Mol. Cell Proteomics 7, 768–784 [DOI] [PubMed] [Google Scholar]
- 24.Li S. S. 2005. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem. J. 390, 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rastogi S., Reuter N., Liberles D. A. 2006. Evaluation of models for the evolution of protein sequences and functions under structural constraint. Biophys. Chem. 124, 134–144 10.1016/j.bpc.2006.06.008 (doi:10.1016/j.bpc.2006.06.008) [DOI] [PubMed] [Google Scholar]
- 26.Bastolla U., Farwer J., Knapp E. W., Vendruscolo M. 2001. How to guarantee optimal stability for most representative structures in the Protein Data Bank. Proteins 44, 79–96 10.1002/prot.1075 (doi:10.1002/prot.1075) [DOI] [PubMed] [Google Scholar]
- 27.Taverna D. M., Goldstein R. A. 2002. Why are proteins marginally stable? Proteins 46, 105–109 10.1002/prot.10016 (doi:10.1002/prot.10016) [DOI] [PubMed] [Google Scholar]
- 28.Fromer M., Shifman J. M. 2009. Tradeoff between stability and multispecificity in the design of promiscuous proteins. PLoS Comput. Biol. 5, e1000627. 10.1371/journal.pcbi.1000627 (doi:10.1371/journal.pcbi.1000627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noivirt-Brik O., Horovitz A., Unger R. 2009. Trade-off between positive and negative design of protein stability: from lattice models to real proteins. PLoS Comput. Biol. 5, e1000592. 10.1371/journal.pcbi.1000592 (doi:10.1371/journal.pcbi.1000592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persson B., Hedlund J., Jörnvall H. 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol. Life Sci. 65, 3879–3894 10.1007/s00018-008-8587-z (doi:10.1007/s00018-008-8587-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lidén M., Eriksson U. 2006. Understanding retinol metabolism: structure and function of retinol dehydrogenases. J. Biol. Chem. 281, 13 001–13 004 10.1074/jbc.R500027200 (doi:10.1074/jbc.R500027200) [DOI] [PubMed] [Google Scholar]
- 32.Parés X., Farrés J., Kedishvili N., Duester G. 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell Mol. Life Sci. 65, 3936–3949 10.1007/s00018-008-8591-3 (doi:10.1007/s00018-008-8591-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tellgren-Roth A., Dittmar K., Massey S. E., Kemi C., Tellgren-Roth C., Savolainen P., Lyons L. A., Liberles D. A. 2009. Keeping the blood flowing: plasminogen activator genes and feeding behavior in vampire bats. Naturwissenschaften 96, 39–47 10.1007/s00114-008-0446-0 (doi:10.1007/s00114-008-0446-0) [DOI] [PubMed] [Google Scholar]
- 34.Zarrinpar A., Park S. H., Lim W. A. 2003. Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 10.1038/nature02178 (doi:10.1038/nature02178) [DOI] [PubMed] [Google Scholar]
- 35.Betts M. J., Russell R. B. 2007. The hard cell: from proteomics to a whole cell model. FEBS Lett. 581, 2870–2876 10.1016/j.febslet.2007.05.062 (doi:10.1016/j.febslet.2007.05.062) [DOI] [PubMed] [Google Scholar]