Abstract
In complex organisms, neutral evolution of genomic architecture, associated compensatory interactions in protein networks and emergent developmental processes can delineate the directions of evolutionary change, including the opportunity for natural selection. These effects are reflected in the evolution of developmental programmes that link genomic architecture with a corresponding functioning phenotype. Two recent findings call for closer examination of the rules by which these links are constructed. First is the realization that high dimensionality of genotypes and emergent properties of autonomous developmental processes (such as capacity for self-organization) result in the vast areas of fitness neutrality at both the phenotypic and genetic levels. Second is the ubiquity of context- and taxa-specific regulation of deeply conserved gene networks, such that exceptional phenotypic diversification coexists with remarkably conserved generative processes. Establishing the causal reciprocal links between ongoing neutral expansion of genomic architecture, emergent features of organisms' functionality, and often precisely adaptive phenotypic diversification therefore becomes an important goal of evolutionary biology and is the latest reincarnation of the search for a framework that links development, functioning and evolution of phenotypes. Here I examine, in the light of recent empirical advances, two evolutionary concepts that are central to this framework—natural selection and inheritance—the general rules by which they become associated with emergent developmental and homeostatic processes and the role that they play in descent with modification.
Keywords: natural selection, inheritance, genome architecture, development, emergence, modularity
1. Outstanding problems in historical context
Over the past decade, progress in evolutionary biology has been shaped by the realization that a significant portion of genomic architecture and developmental processes arises by processes other than natural selection [1–3], that connectivity and conservation of protein networks and emergent developmental and homeostatic processes determine directionality of contemporary adaptive evolution [4–7], that a majority of current adaptations and novelties are rearrangements of processes that have evolved in a different context, some billions of years ago (e.g. [8]), that tremendous diversification of metazoans uses a limited set of deeply conserved gene networks and associated core set of molecular and cellular processes [9–11], and that prevalence of symbiosis in eukaryotes and horizontal gene transfer in prokaryotes questions the tree of life view of adaptive evolution, except near terminal branches [12,13].
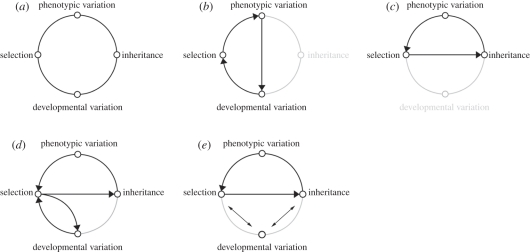
Taken together, these discoveries reveal that, at many levels of organization, the mechanisms that generate diversity of organismal forms are commonly distinct from those that account for their subsequent modification and maintenance. In particular, remarkable coexistence of conserved generative processes with exceptional diversification in morphological, physiological and behavioural systems, calls for re-evaluation of our understanding of basic evolutionary principles that link them [14–18]. It also emphasizes the importance of understanding the relationship between the generation of developmental variation and its modification and evolutionary retention, the link that was once the very motivation behind the growth of evolutionary biology, and is still its most central question [19–22]. I suggest that in particular need of updating is the role of two main concepts in evolutionary processes—natural selection and inheritance—especially in relation to development and functioning of organisms (figure 1).
Figure 1.
Historical changes in the concepts of selection and inheritance in an evolutionary framework. (a) A complete framework. (b) Darwinian concept establishes the primacy and uniqueness of natural selection in production and directing the evolution of traits, but leaves out inheritance. (c) Modern Synthesis sets aside developmental and physiological variations, reinstates inheritance in the framework, and directly links selection to it. (d) Framework that charges natural selection with both direct shaping of developmental variation and subsequent sorting of this variation. (e) Summary of modern views that either combine development with selection (as in ‘selection favouring evolution of traits that indicate a particular function’) or combine development with inheritance (as in ‘inherited developmental toolkits’). The difficulties in incorporating development (a highly contingent process) into an evolutionary framework are due to a lack of rules of development on par with rules of inheritance and natural selection (both treated as patterns).
(a). Historical modifications of evolutionary framework
Over the history of evolutionary theory, the main conceptual difficulty in envisioning the links between development, selection, phenotypic variation and inheritance has centred on reconciliation of the stability of phenotypes across generations with obvious environmental contingency of within-generation development. It has long been recognized that such a reconciliation would require an evolutionary framework that includes explicit causal links between development, natural selection, phenotypic variation and inheritance (figure 1a)—a framework, in which inheritance (which, by definition, limits variation) coexists with variability and plasticity, and where natural selection arises as a result of changes in developmental resources and ecological contexts between the generations. However, coming up with a framework that includes all of these components has been a challenge. The two most successful attempts—the theory of evolution by natural selection [23] and the Modern Evolutionary Synthesis [24] accomplished the continuity of proposed evolutionary frameworks only after eliminating some of these components (figure 1).
Darwin [23] famously set aside the details of inheritance, borrowing the concept of direct inheritance from Lamarck [25], and thus connecting directly the phenotypic variation favoured by natural selection to its development in subsequent generations. Such direct linkage elevated natural selection to the role of driver of both local adaptation and evolutionary diversification (figure 1b) and thereby clearly linked organismal functioning with its evolution, eliminating the duality of heritable and non-heritable components of organism's development and functionality. This, however, side-stepped generative processes in evolution by essentially merging the concept of natural selection with the process of development, a view under which selection shapes developmental variation that it subsequently sorts.
Such a creative role of natural selection in evolution required that it acted on heritable traits, setting an initial stage for links between natural selection and inheritance, and eventually culminating in the population genetics definition of evolution (either by random drift or natural selection) as the change in genotypic frequencies and distributions (e.g. [26]). Another legacy is the view of natural selection as the main provider of rules and directionality in evolutionary processes, such that, under his view, development is best understood as a collection of past adaptations while heredity is best understood as cumulative long-term natural selection.
The framers of the Modern Synthesis similarly fully recognized the difficulty of explicitly linking inheritance (a stable transgenerational transmission in the population genetics' framework) and development (a highly environmentally contingent process). The solution was ‘to put aside all developmental-physiological questions […] and concentrate strictly on the problems of transmission’ [27, p. 832]. Explicit linkage of natural selection and inheritance (figure 1c)—whereby selection accumulates successful variations and channels future genetic and phenotypic variability (either through accumulated variations or by making some combinations of successful traits more likely than others: [28,29])—gave evolutionary biology a powerful foundation for understanding the maintenance and diversification of existing adaptations. But, simultaneously, it explicitly constrained generation of novel variation to modification of already existing structures. It also had an unfortunate side effect of diminishing the evolutionary role of the phenotype, with all of its homeostatic, behavioural and functional features and abilities [15,30–34], thereby setting the stage for debates on the nature and the importance of phenotypic plasticity, and the role of physiology and behaviour in evolution. Most importantly, however, it similarly side-stepped generative processes, instead merging the phenomenon of inheritance with the process of development. The modern day legacy of such a merger is the view of development as a collection of ‘inherited developmental toolkits’, the perspective that opens the stage for debates on the origin of these toolkits (neutral versus adaptive), on the primacy of selective versus developmental processes in evolution, and on the nature and existence of the rules of development on par with the rules of natural selection. Yet, because much of organismal functionality and adaptability is attributable to emergent features of development and homeostasis, the rules by which an association between these processes and inheritance is formed becomes the central, but not addressed, question in this framework.
Other evolutionary frameworks that linked functionality, development and inheritance, (that is, attempted to envision the evolution of organismal systems that enable both continuing novel input and homeostatic stability of already evolved structures [35]), suggested that the key requirements of adaptive evolution by natural selection—the heritable basis of selected traits and associated strong effects of natural selection on phenotypes (both requiring some degree of developmental and genetic modularity)—might, in fact, be highly derived conditions [36–38]. So the Darwinian model of evolution might be most suited for late evolutionary stages, such as adaptive radiation, while the origination and initial diversification of traits is commonly a result of autonomous and emergent processes that are only later stabilized by natural selection or fixed via genetic drift [2,39].
Indeed many patterns of development are generated by autonomous biochemical and physical mechanisms with their often complex conditional behaviours and nonlinear dynamics [3,17,40–43], and fixation of such novel elements by random drift appears to be a dominant and an ongoing process in the evolution of organismal complexity, at least in small populations of eukaryotes [44–46]. Findings of ubiquitous convergent evolution with limited genetic homology, the historical contingency of genomic and developmental pathways, frequent recruitment of conserved core processes for a diverse set of phenotypic functions and integration of newly arising genes into existing functional complexes [11,47–49] all seem to suggest that while natural selection for stability and canalization of phenotypes contributed strongly to the maintenance of a correspondence between genotypes and phenotypes, its role in the origination of this correspondence is limited.
(b). Reconciling neutral genomic processes, emergent self-regulatory mechanisms and adaptive evolution
In complex organisms, neutral evolution of genomic architecture, associated compensatory interactions in protein networks and emergent developmental processes can ultimately delineate the directions of adaptive evolution and determine the opportunity for natural selection [4,7,50,51]. Such effects are reflected in the evolution of developmental programmes that link genomic architecture with a corresponding functioning phenotype. Establishing the causal reciprocal links between genomic architecture and phenotypic diversification is an important goal of evolutionary biology, and is the latest reincarnation of the search for a framework that links development, functioning and evolution of phenotypes (figure 1). This search, however, had been greatly reshaped by several recent insights. First is the finding that a high dimensionality of genotypes results in vast areas of nearly neutral variation and equal fitness [5], emphasizing the importance of continuity and compensatory interactions in protein networks (e.g. [52–55]). Second is the realization that step-wise evolution of developmental programmes that link genotype and phenotype, with each step providing incremental fitness benefits or drifting into fixation, is far less likely than the context- and taxa-specific regulation of pre-existing gene networks [8,56,57]. Third is a finding that a significant portion of developmental systems are guided by autonomous properties of their components and their capacity for self-organization and environmental response and not by genetically determined programmes [2,3,58].
(c). Evolving concepts of natural selection and inheritance
Because ‘biologists tend to ask the questions they can answer’ (R. Lewontin in [59]), the concepts of natural selection and heredity underwent their own evolution over the history of biology, changing from abstract concepts, to convenient empirical measures, to definitions, each viewpoint ultimately directing the growth of the field (figure 1). One of the major issues in this evolution is the static versus dynamic views of natural selection and inheritance, i.e. in treatment of these concepts as patterns versus processes and as causes versus outcomes.
Under the static view, selection is a post-production checkpoint that sorts among developmental outcomes and whose evolutionary significance depends on the extent to which these developmental outcomes are heritable and modular. Such a view not only necessitates a direct link between selection and inheritance (figure 1), but, importantly, also assumes that certain stages of development are excluded from selection and some elements of functionality are excluded from inheritance. Such often inconsistent designation was criticized by some of the earliest contributors to the field, who called for explicit separation of ‘the fact of survival’ (captured by natural selection) from ‘the means of survival’ (captured by developmental and functional homeostasis), noting that existence is not the same as natural selection and that survival and reproduction are not the same as differential survival and differential reproduction [19–22,60,61]. Importantly, however, viewing natural selection and inheritance as discrete static terms necessitates the construction of process-like links between them (figure 1); some of these links transcend generations, some stay within generations, some operate at the level of the individual and some at the population or species level.
Alternations between the pattern versus process view of natural selection and inheritance, which is most clearly evident across different biological disciplines (e.g. developmental evolution versus behavioural ecology, palaeontology versus functional ecology), have left us with a number of unanswered questions. How do emergent features of development and functioning become associated with heritable aspects of the phenotype? How are selectively advantageous features generated in development, i.e. what is the link between developmental construction and adaptiveness of traits? What is the role of the organism in generating variation available for selection? Are unique phenotype–genotype associations a product or prerequisite of evolution by natural selection? How to account for similarity of proximate mechanisms that modify a trait within generation and trait change over evolutionary time? How to reconcile deep conservation of genetic networks in morphologically disparate taxa with often remarkably precise context-dependency in their regulation? What is the evolutionary relationship between core conserved elements of gene networks and network regulators? Are epigenetic and genetic effects historical stages of the same process or fundamentally distinct phenomena? And what are the causes of evolution and what is the relationship between those and natural selection?
The main difficulty in answering these questions is understanding the general rules by which the phenomena of natural selection and inheritance are incorporated into developmental and generative processes and the role that they, together with other evolutionary forces, play in descent with modification.
2. General themes and key problems
(a). Natural selection depends on reliability of life cycles
Natural selection—variation among organisms in a population in fit to their environment—occurs, proximately, when there is a mismatch between the environment of development and the environment of functioning or, more generally, a mismatch between the ranges of current and historical environments experienced by a lineage of organisms. Depending on an organism's complexity, such mismatches can be due to mistakes in the processes of development (including mutation and recombination), replication or in environmental variation between generations, such that in complex multi-cellular organisms, changes in life-stage-specific reliability of resources during an organism's existence lead to natural selection. In a population of complex organisms, therefore the issue is not the avoidance of natural selection, but the ability to incorporate a greater range of environments into development to lessen selection's probability and capitalize to a greater extent on environmentally dependent contribution to the next generation [62,63]. A particular outcome is the evolution of conditional expression, when a trait is expressed only when advantageous, thereby accomplishing conditional environment–phenotype matching [15]. Correspondingly, evolution in a highly reliable environment is often associated with the loss of developmental plasticity and the loss of ability to explore different environments. Two classical illustrations of the power of environmental contingency in development are the frequency and extent of reversion to wild-type once artificial selection ceases and developmental diversification that accompanies colonizations of competitor- and predator-free environments [64–66]. This view, therefore, limits the role of natural selection to stabilization and integration of developmental variability to a prevalent ecological context, while leaving the origination and production of actual adaptive solutions to the specific properties of development, including emergent and self-organizing features (§2c below).
Because the most reliable (e.g. those important over wider range of environments over evolutionary time) organism–environment associations are, by definition, most recurrent in an organism's developmental history, they are most likely to become heritable and expressed at earlier embryological stages and shared between related lineages ([23], for recent empirical work see [67]), and this can account for the evolution of the link between natural selection and inheritance. Such a view of natural selection clarifies several issues. First, it shows that natural selection can be viewed not as an additional external force in charge of filtering, sorting and eliminating developmental outcomes (see also [68–70]), but as a process that continuously and directly arises from the generation of organismal forms within a population. Second, it links explicitly three major players in evolution: emergence of novel variation, functionality arising from generative and exploratory processes and adaptation resulting from and retained by current natural selection. Third, it explains how ubiquitous emergent, compensatory and homeostatic elements of developmental processes can facilitate the appearance of novel adaptations during episodes of significant environmental change (i.e. strong natural selection).
(b). Inclusion of a wider range of environments in organismal production and associated longer generations lead to weaker natural selection and accumulation of developmental complexity
From §2a, we can expect a general relationship between the duration and complexity of development, environmental variability and the probability of natural selection, e.g. longer development and associated historical experience with a greater environmental range should lead to lesser probability of natural selection, particularly in fully grown phenotypes. In turn, organisms with longer development and greater complexity commonly have smaller populations, and longer generations that further lessen the intensity and effectiveness of natural selection by minimizing differences in fitness among individuals in relation to stochastic forces of random genetic drift [44,71]. The effect of such weak natural selection is twofold: first, it facilitates accumulation of organismal complexity; second, it has a progressively weaker ability to streamline such complexity and redundancy for contemporary functions; both of these effects further limit the strength of natural selection. Indeed, continuous accumulation and modification of developmental pathways (rather than adaptive changes in regulatory genetic architecture and reuse of older functional modules) seem to be the dominant path of evolution of organismal complexity in small populations of multi-cellular organisms [9,72,73]. Faster evolution under relaxed natural selection in smaller mammalian populations [74], including humans (e.g. [75,76]), might be an example of such a process.
(c). Weaker selection and greater developmental complexity channel expression of novel developmental variation and facilitate its genetic accommodation
An important consequence of accumulated organismal complexity (§2b) is its channelling effect on the expression of newly arising modifications. That accumulated organismal complexity (neutral or adaptive in origin) facilitates, channels and accommodates novel genomic and developmental modifications that make an organism fit in its present environment, has been repeatedly suggested for more than 100 years [15,27,61,77–80]. Four of the most insightful predictions of these theories stated that (i) novel adaptations often include modifications of existing structures that shape both genetic and developmental variations, such that neither is randomly expressed, (ii) accumulated genetic variance of complex structures can include developmental components of individual accommodations, providing initial standing genetic variance for evolutionary retention of novel modifications, (iii) any novel character necessarily includes new heritable variation in the expression of its phenotype, leading to acquisition of novel developmental functions by the genome over the course of evolution, and (iv) developmental mechanisms that ultimately confer stabilization and robustness of phenotypes are the same mechanisms that generate novel modifications. That is why, these theories suggest, historical experience of an organismal lineage shapes the pathways available for its evolution [19], such that ‘the history of current adaptation cannot be uncoupled from the history of the organism’ [21, p. 136]. Dobzhansky [28, pp. 315–316] further extended this view by stating that the ‘mutational repertoire of a gene is a function of its structure and hence billions of years of evolution’, such that expression of mutational variance ‘is channeled and conditioned by gene's and organisms' histories of natural selection’. These insights are corroborated by empirical documentations of historical contingency of genomic pathways, mutational effects and developmental networks [48,73,81–83].
Either directional mutation input or physical properties of development can facilitate accumulation of complexity in developmental networks under weak natural selection [9,45,84,85]. Redundancy of such complex networks harbours unexpressed variation, and expression and channelling of this variation under environmental or internal perturbations can be a further source of novelties (e.g. [86], reviewed in [87]). This source is particularly important for evolutionary change because many individuals show similar modifications, making these patterns of concordant variation particularly susceptible for subsequent genetic capture and stabilization [85,88–90].
Such a mode of accumulation and expression of developmental and genomic complexities are expected to produce spurts of emergent variation and exploratory evolution associated with physico-chemical features and exploratory behaviours followed by adaptive diversification, specialization and stabilization of lineages by natural selection in the range of environments with greatest fit (e.g. greatest utilization of resources and lowest potential for change) [13,21,37,91,92], leading to increasing complexity that ultimately results in accumulation of reproductive incompatibilities, speciation and evolutionary diversification [2,93,94]. Thus, at the core of this process is a cycle of weaker genetic control and greater role of environmental induction that accompanies exploratory evolution by emergent processes alternating with genetic assimilation and greater genetic stabilization that accompanies adaptive diversification and specialization under natural selection.
(d). Unique phenotype–genotype associations for traits imply a recurrent environment during development. Such association can be a result of natural selection, but can also emerge without direct selection
The stable genotype–phenotype association is an implicit prerequisite for differential survival and reproduction to affect gene frequency. Furthermore, traits with closest genotype–phenotype association are thought to facilitate evolution and diversification because their modularity limits interference with existing functions and reduces the number of regulatory changes needed for viable phenotypic variation [47,58,95,96]. A crucial question is whether such association is a derived condition—a de novo product of natural selection, a result of autonomous developmental processes secondarily stabilized by natural selection for reliability of developmental outcomes—or modifications of genomic architecture through degenerative mutations of regulatory networks, and gene duplication [97–102].
From a developmental perspective, close phenotype–genotype association requires recurrent organism–environment complexes over evolutionary time, a requirement that limits evolutionary stability of such an association. Indeed, population genetic considerations suggest that the evolution of phenotype–genotype modularity is a slow process even in very large populations [101,103,104], unless such modularity is strongly and immediately advantageous and expressed in many individuals, a condition that, in principle, can be facilitated by channelling and integrative effects of developmental complexity (e.g. [88,105,106]). However, even strong functional integration (co-selection) between traits seems to rarely leads to genetic integration because the developmental modularity that would be needed to link the two is hard to maintain (see also [107,108])—it requires evolutionary recurrence of developmental contexts. Furthermore, consistent selection for stable co-expression depletes variance in regulatory elements of developmentally linked traits resulting in their lesser sensitivity to environmental conditions, which, in turn, progressively weakens ability of natural selection to maintain such modularity [109,110].
(e). Natural selection and organism functioning create heritable variation; elements of inheritance can result in selection
Treated as a process, the phenomenon of inheritance is the transference of genomic and developmental resources for reconstruction of the phenotype in subsequent generations [111,112]. Some transferred elements are genomic determinants, some are contexts and templates that deliniate their expression, some are elements of a modified environment. When there is a mismatch between any of such transferred resources and the context of development between generations (§2a), inherited elements themselves generate natural selection (reviewed in [113]). Considering that the most historically consistent organism–environment interactions are likely to be the most heritable [114–117], stabilizing natural selection, in a historical sense, is the process behind patterns of inheritance. Indeed, the original argument for a creative role of natural selection in evolution was built on the view of selection as an integrating and facilitating factor that brings together genetic architecture, current functional organization and its evolutionary persistence across historical ecological contexts [21], setting general ‘boundary condition’ [11] for emergent processes. This view of inheritance raises two questions. First, what is the connection between inheritance (of genes, epigenetic states or environment) and the actual generation of a phenotype? That is, what delineates inherited determinants to produce a specific phenotype in response to particular environmental or genetic change? Second, what is the evolutionary relationship between epigenetic effects and genetic inheritance?
The answer to both of these questions depends on the mechanisms by which novel emergent and exploratory processes spread through population and become genetically integrated over historical time—a process in which epigenetic inheritance plays a central role. Empirical studies suggest that epigenetic integration often precedes genetic integration of novel traits [15,118], such that novel traits originating in the interaction of organism with its environment can remain epigenetically integrated for prolonged time before acquiring greater genetic integration through natural selection and becoming incorporated into developmental repertoire of a lineage (e.g. during domestication [66], or range expansion [119]). These results are supported by computer simulations showing a tendency for emergent processes to gradually form hierarchical controls (e.g. [120]). As developmental complexity of local adaptations increases, this tendency results in greater genetic determination of upstream developmental stages and stronger epigenetic and environmental influences on later developmental stages.
This view is corroborated by observations that some currently genetically heritable norms of reaction can be traced to emergent patterns (such as in animal migrations, exploratory behaviour and diet-derived pigmentation); the steps between the two reflecting successive accumulation of organism–environment associations that build contingency in developmental pathways and that can be expressed rapidly when the present and past environmental conditions match [15,115,116,121].
Such a view of inheritance emphasizes that historical stability of phenotypes within lineages proximately means reliable ontogenetic reconstructions across environments and generations, such that incorporation of a wider temporal and spatial range of heterogeneous environments into the species ontogeny (§2a) is central for stasis, diversification and extinction in lineages [122,123]. Therefore, proximately, evolution of local adaptation requires reliable integration of effects of developmental and molecular levels into ecological contexts—a process accomplished by inheritance. An important question is whether stages of such integration represent stages of an evolutionary continuum of inheritance systems that retain, accumulate and pass on the most recurrent organism–environment associations [21,80,114,124], or whether heritable components arise from modification of regulatory elements, regardless of recurrence of ecological contexts, leading to retention and inheritance of a particular phenotypic construction independently of changes in their molecular and developmental composition [11,125].
3. Evolution as ‘a historical self-directed movement of organism–environment associations’ and as ‘the history of ontogenetic accommodations of earlier generations’
Findings that evolutionary change proceeds by converting conserved core processes at the genomic and molecular level into genetic and evolutionary change by both neutral processes and natural selection, that increasing complexity of organismal forms is associated with vast areas of neutrality on fitness landscapes at both the phenotypic and genetic level, and that the sequence of development arises as a combination of ongoing neutral divergence stabilized by both emergent processes and natural selection, echo the visionary predictions of founders of evolutionary theory highlighted in this section's title ([21], p. 372 and [19], p. 447, respectively). But these findings also bring forth several outstanding phenomena that need to be re-evaluated in the light of recent discoveries.
First, it is becoming increasingly clear that in multi-cellular complex organisms, the evolutionary consequences of transmission of developmental contexts (that modulate time- and stage-specific gene effects) are often equal to those of transmission of genes themselves [126,127], raising the question of the evolutionary link between epigenetic and genetic effects and the wisdom of treating them as static evolutionary phenomena [113,118,128]. Whether we consider epigenetic inheritance that consolidates environmental perturbations a potential stage in the evolution of inheritance systems or as a collection of disjointed short-term ‘non-genetic’ effects determines how we view the origin and evolution of developmental innovation. Second, in similar need of re-evaluation is the original view on the links between functionality (produced primarily by plastic and emergent features of phenotype) and inheritance that preoccupied evolutionary thinkers since the birth of the theory of evolution (reviewed in [27]). This re-evaluation is particularly important in the light of recent realizations that homeostatic accommodation of plasticity is a likely path for evolution of novel adaptations in complex multi-cellular organisms [15,17]. The central question that remains, however, is how does inheritance become associated with emergent and locally contingent processes of development and functioning—a link that Darwin considered to be central for his evolutionary theory [23]. Fundamentally, these questions emphasize that an explicit consideration and periodic updating of two classical concepts of evolutionary theory—natural selection and inheritance—in the light of recent empirical discoveries could provide an important insight into the central, but still the most elusive relationship in biology—the one between genotype and phenotype.
Acknowledgements
I am grateful to two anonymous reviewers and many colleagues for insightful comments on previous versions of the manuscript, as well as to the US National Science Foundation (DEB-0075388, DEB-0077804, and IBN-0218313), Fulbright Fellowship and the David and Lucille Packard Foundation Fellowship for partially funding this work.
References
- 1.Lynch M. 2007. The origins of genome architecture. Sunderland, MA: Sinauer Associates [Google Scholar]
- 2.Reid R. G. B. 2007. Biological emergences: evolution by natural experiment. Vienna Series in Theoretical Biology Cambridge, MA: MIT Press [Google Scholar]
- 3.Forgacs G., Newman S. A. 2005. Biological physics of the developing embryo. Cambridge, UK: Cambridge University Press [Google Scholar]
- 4.Maynard Smith J. 1970. Natural selection and the concept of a protein space. Nature 225, 563–564 10.1038/225563a0 (doi:10.1038/225563a0) [DOI] [PubMed] [Google Scholar]
- 5.Gavrilets S. 2004. Fitness landscapes and the origin of species Princeton, NJ: Princeton University Press [Google Scholar]
- 6.Wagner A. 2005. Robustness and evolvability in living systems. Princeton, NJ: Princeton University Press [Google Scholar]
- 7.Pagel M., Pomiankowski A. (eds) 2008. Evolutionary genomics and proteinomics. Sunderland, MA: Sinauer Associates [Google Scholar]
- 8.Shubin N., Tabin C. J., Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457, 818–823 10.1038/nature07891 (doi:10.1038/nature07891) [DOI] [PubMed] [Google Scholar]
- 9.Wilkins A. S. 2001. The evolution of developmental pathways. Sunderland, MA: Sinauer Associates [Google Scholar]
- 10.Davidson E. H. 2006. The regulatory genome: gene regulatory networks in development and evolution. San Diego, CA: Academic Press [Google Scholar]
- 11.Müller G. B. 2007. Evo-devo: extending the evolutionary synthesis. Nat. Rev. Genet. 8, 943–949 10.1038/nrg2219 (doi:10.1038/nrg2219) [DOI] [PubMed] [Google Scholar]
- 12.Maddison W. P. 1997. Gene trees in species trees. Syst. Biol. 46, 523–536 10.1093/sysbio/46.3.523 (doi:10.1093/sysbio/46.3.523) [DOI] [Google Scholar]
- 13.Koonin E. V., Wolf Y. I. 2009. The fundamental units, processes and patterns in evolution, and the Tree of Life conundrum. Biol. Direct 4, 33. 10.1186/1745-6150-4-33 (doi:10.1186/1745-6150-4-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller G. B., Newman S. 2003. Origination of organismal form: beyond the gene in developmental and evolutionary biology. Cambridge, MA: The MIT Press [Google Scholar]
- 15.West-Eberhard M. J. 2003. Developmental plasticity and evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 16.Schlichting C. D., Murren C. J. 2004. Evolvability and the raw materials for adaptation. In Plant adaptation: molecular genetics and ecology (eds Cronk Q. C. B., Whitton J., Ree R. H., Taylor I. E. P.), pp. 18–29 Ottawa, Ontario: NRC Research Press [Google Scholar]
- 17.Kirschner M., Gerhart J. C. 2005. The plausibility of life: resolving Darwin's dilemma. New Haven, CT: Yale University Press [Google Scholar]
- 18.Pigliucci M., Müller G. B.(eds) 2010. Evolution: the extended synthesis. Cambridge, MA: The MIT Press [Google Scholar]
- 19.Baldwin J. M. 1902. Development and evolution. New York, NY: Macmillan [Google Scholar]
- 20.Dobzhansky T. 1937. Genetics and the origin of species. New York, NY: Columbia University Press [Google Scholar]
- 21.Schmalhausen I. I. 1938. Organism as a whole in individual development and history. Leningrad: Academy of Sciences, USSR [Google Scholar]
- 22.Cope E. D. 1887. The origin of the fittest: essays on evolution. New York, NY: Appleton [Google Scholar]
- 23.Darwin C. 1859. The origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr E., Provine W. B. (eds) 1980. The Evolutionary synthesis: perspectives on the unification of biology. Cambridge, MA: Harvard University Press [Google Scholar]
- 25.Lamarck J. B. 1809. Philosophie zoologique. Paris: Duminil-Lesueur [Google Scholar]
- 26.Crow J. F., Kimura M. 1970. An introduction to population genetics theory. New York, NY: Harper and Row [Google Scholar]
- 27.Mayr E. 1982. The growth of biological thought. Cambridge, MA: Belknap Press, Harvard University Press [Google Scholar]
- 28.Dobzhansky T. 1974. Chance and creativity in evolution. In Studies in the philosophy of biology (eds Ayala F., Dobzhansky T.), pp. 307–338 London: McMillan [Google Scholar]
- 29.Jacob F. 1977. Evolution and tinkering. Science 196, 1161–1166 10.1126/science.860134 (doi:10.1126/science.860134) [DOI] [PubMed] [Google Scholar]
- 30.Lewontin R. C. 1983. Gene, organism and environment. In Evolution: from molecules to men (ed. Bendall D. S.), pp. 273–285 Cambridge, UK: Cambridge University Press [Google Scholar]
- 31.Schlichting C. D., Pigliucci M. 1998. Phenotypic evolution: a reaction norm perspective. Sunderland, MA: Sinauer Associates [Google Scholar]
- 32.Hall B. K., Pearson R. D., Müller G. B. (eds) 2004. Environment, development and evolution: toward a synthesis. Vienna Series in Theoretical Biology. Cambridge, MA: MIT Press [Google Scholar]
- 33.Duckworth R. A. 2009. The role of behavior in evolution: a search for mechanism. Evol. Ecol. 23, 513–531 10.1007/s10682-008-9252-6 (doi:10.1007/s10682-008-9252-6) [DOI] [Google Scholar]
- 34.Piersma T., van Gils J. A. 2010. The flexible phenotype: a body-centered integration of ecology, physiology, and behaviour. Oxford, UK: Oxford University Press [Google Scholar]
- 35.Callebaut W., Müller G. B., Newman S. A. 2007. The organismic systems approach. Streamlining the naturalistic agenda. In Integrating evolution and development. From theory to practice (eds Sansom R., Brandon R. N.), pp. 25–92 Cambridge, MA: MIT Press [Google Scholar]
- 36.Müller G. B., Newman S. A. 2005. The innovation triad: an EvoDevo agenda. J. Exp. Zool. (Mol Dev Evol) 304B, 487–503 10.1002/jez.b.21081 (doi:10.1002/jez.b.21081) [DOI] [PubMed] [Google Scholar]
- 37.Newman S. A. 2005. The pre-Mendelian, pre-Darwinian world: shifting relations between genetic and epigenetic mechanisms in early multicellular evolution. J. Biosci. 30, 75–85 10.1007/BF02705152 (doi:10.1007/BF02705152) [DOI] [PubMed] [Google Scholar]
- 38.Newman S. A., Bhat R. 2008. Dynamical patterning modules: physico-genetic determinants of morphological development and evolution. Phys. Biol. 5, 1–14 10.1088/1478-3975/5/1/015008 (doi:10.1088/1478-3975/5/1/015008) [DOI] [PubMed] [Google Scholar]
- 39.Koonin E. V., Wolf Y. I. 2009. Is evolution Darwinian or/and Lamarkian? Biol. Direct 4, 42. 10.1186/1745-6150-4-42 (doi:10.1186/1745-6150-4-42) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kauffman S. A. 1993. The origins of order: self organization and selection in evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 41.Newman S. A. 2006. The developmental-genetic toolkit and the molecular homology-analogy paradox. Biol. Theory 1, 12–16 10.1162/biot.2006.1.1.12 (doi:10.1162/biot.2006.1.1.12) [DOI] [Google Scholar]
- 42.Montell D. J. 2008. Morphogenetic cell movements: diversity from modular mechanical properties. Science 322, 1502–1509 10.1126/science.1164073 (doi:10.1126/science.1164073) [DOI] [PubMed] [Google Scholar]
- 43.Kondo S., Miura T. 2010. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 10.1126/science.1179047 (doi:10.1126/science.1179047) [DOI] [PubMed] [Google Scholar]
- 44.Lande R. 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30, 314–334 10.2307/2407703 (doi:10.2307/2407703) [DOI] [PubMed] [Google Scholar]
- 45.Lynch M., Conery J. S. 2003. The origins of genome complexity. Science 302, 1401–1404 10.1126/science.1089370 (doi:10.1126/science.1089370) [DOI] [PubMed] [Google Scholar]
- 46.Whitlock M. C. 2003. Fixation probability and time in subdivided populations. Genetics 164, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carroll S. B. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3, e245. 10.1371/journal.pbio.0030245 (doi:10.1371/journal.pbio.0030245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bridgham J. T., Ortlund E. A., Thornton J. W. 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461, 515–519 10.1038/nature08249 (doi:10.1038/nature08249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S., Zhang Y. E., Long M. 2010. New genes in Drosophila quickly become essential. Science 330, 1682–1685 10.1126/science.1196380 (doi:10.1126/science.1196380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinreich D. M., Watson R. A., Chao L. 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59, 1165–1174 [PubMed] [Google Scholar]
- 51.Newman S. A. 2010. Dynamic patterning modules. In Evolution: the extended synthesis (eds Pigliucci M., Muller G. B.), pp. 281–306 Cambridge, MA: The MIT Press [Google Scholar]
- 52.van Nimwegen E., Crutchfield J. P. 2000. Metastable evolutionary dynamics: crossing fitness barriers or escaping via neutral paths. Bull. Math. Biol. 62, 799–848 10.1006/bulm.2000.0180 (doi:10.1006/bulm.2000.0180) [DOI] [PubMed] [Google Scholar]
- 53.Poelwijk F. J., Kiviet D. J., Weinreich D. M., Tans S. J. 2007. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445, 383–386 10.1038/nature05451 (doi:10.1038/nature05451) [DOI] [PubMed] [Google Scholar]
- 54.Meer M. V., Kondrashov A. S., Artzy-Randrup Y., Kondrashov F. A. 2010. Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness. Nature 464, 279–282 10.1038/nature08691 (doi:10.1038/nature08691) [DOI] [PubMed] [Google Scholar]
- 55.Povolotskaya I. S., Kondrashov F. A. 2010. Sequence space and the ongoing expansion of the protein universe. Nature 465, 922–926 10.1038/nature09105 (doi:10.1038/nature09105) [DOI] [PubMed] [Google Scholar]
- 56.Carroll S. B., Grenier J. K., Weatherbee S. D. 2001. From DNA to diversity: molecular genetics and the evolution of animal design. Oxford, UK: Blackwell Science [Google Scholar]
- 57.Monteiro A., Podlana O. 2009. Wings, horns, and butterfly eyespots: how do complex traits evolve? PLoS Biol. 7, 209–216 10.1371/journal.pbio.1000037 (doi:10.1371/journal.pbio.1000037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerhart J., Kirschner M. 2007. The theory of facilitated variation. Proc. Natl Acad. Sci. USA 104, 8582–8589 10.1073/pnas.0701035104 (doi:10.1073/pnas.0701035104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oyama S. 2000. The ontogeny of information: developmental systems and evolution Durham, NC: Duke University Press [Google Scholar]
- 60.Severtsov A. H. 1934. Principal directions of the evolutionary process. Moscow: Leningrad: Biomedgiz [Google Scholar]
- 61.Gould S. J. 2002. The structure of evolutionary theory. Cambridge, MA: Belknap Press, Harvard University Press [Google Scholar]
- 62.Odling-Smee F. J., Laland K. N., Feldman M. W. 2003. Niche construction: the neglected process in evolution. Monographs in Population Biology Princeton, NJ: Princeton University Press [Google Scholar]
- 63.Young R. L., Badyaev A. V. 2007. Evolution of ontogeny: linking epigenetic remodeling and genetic adaptation in skeletal structures. Integr. Comp. Biol. 47, 234–244 10.1093/icb/icm025 (doi:10.1093/icb/icm025) [DOI] [PubMed] [Google Scholar]
- 64.Van Valen L. 1965. Morphological variation and width of the ecological niche. Am. Nat. 94, 377–390 [Google Scholar]
- 65.Losos J. B., Queiroz K. D. 1997. Evolutionary consequences of ecological release in Caribbean Anolis lizards. Biol. J. Linn. Soc. 61, 459–483 [Google Scholar]
- 66.Trut L. N. 1999. Early canid domestication: the farm-fox experiment. Anim. Sci. 87, 160–169 [Google Scholar]
- 67.Kalinka A. T., Varga K. M., Gerrard D. T., Preibisch S., Corcoran D. L., Jarrells J., Ohler U., Bergman C. M., Tomancak P. 2010. Gene expression divergence recapitulates the developmental hourglass model. Nature 468, 811–814 10.1038/nature09634 (doi:10.1038/nature09634) [DOI] [PubMed] [Google Scholar]
- 68.Eldredge N. 1985. Unfinished synthesis: biological hierarchies and modern evolutionary thought. Oxford, UK: Oxford University Press [Google Scholar]
- 69.Reid R. G. B. 1985. Evolutionary theory: the unfinished synthesis. Beckenham, UK: Crom Helm [Google Scholar]
- 70.Endler J. A. 1986. Natural selection in the wild. Princeton, NJ: Princeton University Press [Google Scholar]
- 71.Caballero A. 1994. Developments in the prediction of effective population size. Heredity 73, 657–679 10.1038/hdy.1994.174 (doi:10.1038/hdy.1994.174) [DOI] [PubMed] [Google Scholar]
- 72.Nowak M. A., Boerlijst M. C., Cooke J., Smith J. M. 1997. Evolution of genetic redundancy. Nature 388, 167–171 10.1038/40618 (doi:10.1038/40618) [DOI] [PubMed] [Google Scholar]
- 73.Assis R., Kondrashov A. S., Koonin E. V., Kondrashov F. A. 2007. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 24, 475–478 10.1016/j.tig.2008.08.003 (doi:10.1016/j.tig.2008.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Valen L. M. 1985. Why and how do mammals evolve unusually rapidly? Evol. Theory 7, 127–132 [Google Scholar]
- 75.Khaitovich P., Wieiss G., Lachmann M., Hellmann I., Enard W., Muetzel B., Wirkner U., Ansorge W., Paabo S. 2004. A neutral model of transcriptome evolution. PLoS Biol. 2, 682–689 10.1371/journal.pbio.0020132 (doi:10.1371/journal.pbio.0020132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mikkelsen T. S., Hillier L. W., Eichler E. E., Zody M. C. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 10.1038/nature04072 (doi:10.1038/nature04072) [DOI] [PubMed] [Google Scholar]
- 77.Baldwin J. M. 1896. A new factor in evolution. Am. Nat. 30, 441–451 10.1086/276408 (doi:10.1086/276408) [DOI] [Google Scholar]
- 78.Morgan C. L. 1896. Habit and instinct. London, UK: Arnold [Google Scholar]
- 79.Osborn H. F. 1896. A mode of evolution requiring neither natural selection nor the inheritance of acquired characteristics. Trans. NY Acad. Sci. 15, 141–142 [Google Scholar]
- 80.Schmalhausen I. I. 1969. Problems of darwinism. Leningrad: Nauka [Google Scholar]
- 81.Yampolsky L. Y., Stoltzfus A. 2001. Bias in the introduction of variation as an orienting factor in evolution. Evol. Dev. 3, 73–83 10.1046/j.1525-142x.2001.003002073.x (doi:10.1046/j.1525-142x.2001.003002073.x) [DOI] [PubMed] [Google Scholar]
- 82.Blount Z. D., Borland C. Z., Lenski R. E. 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 7899–7906 10.1073/pnas.0803151105 (doi:10.1073/pnas.0803151105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Badyaev A. V., Oh K. P. 2008. Environmental induction and phenotypic retention of adaptive maternal effects. BMC Evol. Biol. 8, e3. 10.1186/1471-2148-8-3 (doi:10.1186/1471-2148-8-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hermisson J., Wagner G. P. 2004. The population genetic theory of hidden variation and genetic robustness. Genetics 168, 2271–2284 10.1534/genetics.104.029173 (doi:10.1534/genetics.104.029173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rice S. H. 2004. Evolutionary theory: mathematical and conceptual foundations. Sunderland, MA: Sinauer Associates [Google Scholar]
- 86.True H. L., Berllin I., Lindquist S. L. 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431, 184–187 10.1038/nature02885 (doi:10.1038/nature02885) [DOI] [PubMed] [Google Scholar]
- 87.Badyaev A. V. 2005. Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proc. R. Soc. B 272, 877–886 10.1098/rspb.2004.3045 (doi:10.1098/rspb.2004.3045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arthur W. 2001. Developmental drive; an important determinant of the direction of phenotypic evolution. Evol. Dev. 3, 271–278 10.1046/j.1525-142x.2001.003004271.x (doi:10.1046/j.1525-142x.2001.003004271.x) [DOI] [PubMed] [Google Scholar]
- 89.Hall B. K. 2001. Organic selection: proximate environmental effects on the evolution of morphology and behaviour. Biol. Philos. 16, 215–237 10.1023/A:1006773408919 (doi:10.1023/A:1006773408919) [DOI] [Google Scholar]
- 90.West-Eberhard M. J. 2005. Phenotypic accommodation: adaptive innovation due to developmental plasticity. J. Exp. Zool. (Mol Dev Evol) 304B, 610–618 10.1002/jez.b.21071 (doi:10.1002/jez.b.21071) [DOI] [PubMed] [Google Scholar]
- 91.Gould S. J., Eldredge N. 1977. Punctuated equilibrium: the tempo and mode of evolution reconsidered. Paleobiology 3, 115–251 [Google Scholar]
- 92.Fontana W., Schuster P. 1998. Continuity in evolution: on the nature of transitions. Science 280, 1451–1455 10.1126/science.280.5368.1451 (doi:10.1126/science.280.5368.1451) [DOI] [PubMed] [Google Scholar]
- 93.Gavrilets S. 1999. A dynamical theory of speciation on holey adaptive landscapes. Am. Nat. 154, 1–22 10.1086/303217 (doi:10.1086/303217) [DOI] [PubMed] [Google Scholar]
- 94.Gravner J., Pitman D., Gavrilets S. 2007. Percolation on fitness landscapes: effect of correlation, phenotype, and incompatibilities. J. Theor. Biol. 248, 627–645 10.1016/j.jtbi.2007.07.009 (doi:10.1016/j.jtbi.2007.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King M. C., Wilson A. C. 1975. Evolution at two levels in humans and chimpanzees. Science 188, 107–116 10.1126/science.1090005 (doi:10.1126/science.1090005) [DOI] [PubMed] [Google Scholar]
- 96.Olson E. C., Miller R. L. 1958. Morphological integration. Chicago, IL: University of Chicago Press [Google Scholar]
- 97.Force A. G., Cresko W. A., Pickett F. B. 2004. Information accretion, gene duplication, and the mechanisms of genetic module parcellation. In Modularity in development and evolution (eds Schlosser G., Wagner G. P.), pp. 315–338 Chicago, IL: University of Chicago Press [Google Scholar]
- 98.Schlosser G., Wagner G. P. 2004. Modularity in development and evolution. Chicago, IL: University of Chicago Press [Google Scholar]
- 99.Kashtan N., Alon U. 2005. Spontaneous evolution of modularity and network motifs. Proc. Natl Acad. Sci. USA 102, 13 773–13 778 10.1073/pnas.0503610102 (doi:10.1073/pnas.0503610102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wagner G. P., Mezey J. G., Callabretta R. 2005. Natural selection and the origin of modules. In Modularity: understanding the development and evolution of complex systems (eds Callebaut W., Raskin-Gutman D.), Cambridge, MA: MIT Press [Google Scholar]
- 101.Wagner G. P., Pavlicev M., Cheverud J. M. 2007. The road to modularity. Nat. Rev. Genet. 8, 921–931 10.1038/nrg2267 (doi:10.1038/nrg2267) [DOI] [PubMed] [Google Scholar]
- 102.Walsh J. B. 1995. How often do duplicated genes evolve new functions? Genetics 139, 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rice S. H. 2001. The evolution of developmental interactions; epistasis, canalization, and integration. In Epistasis and the evolutionary process (eds Wolf J. B., Brodie E. D., I, Wade M. J.), pp. 82–98 New York, NY: Oxford University Press [Google Scholar]
- 104.Welch J. J., Waxman D. 2003. Modularity and the cost of complexity. Evolution 57, 1723–1734 [DOI] [PubMed] [Google Scholar]
- 105.Young R. L., Badyaev A. V. 2010. Developmental plasticity links local adaptation with evolutionary diversification in foraging morphology. J. Exp. Zool. (Mol. Dev. Evol.) 314B, 434–444 10.1002/jez.b.21349 (doi:10.1002/jez.b.21349) [DOI] [PubMed] [Google Scholar]
- 106.Waddington C. H. 1961. Genetic assimilation. Adv. Genet. 10, 257–290 10.1016/S0065-2660(08)60119-4 (doi:10.1016/S0065-2660(08)60119-4) [DOI] [PubMed] [Google Scholar]
- 107.Wainwright P. C. 2007. Functional versus morphological diversity in macroevolution. Annu. Rev. Ecol. Evol. Syst. 38, 381–401 10.1146/annurev.ecolsys.38.091206.095706 (doi:10.1146/annurev.ecolsys.38.091206.095706) [DOI] [Google Scholar]
- 108.Badyaev A. V. 2010. The beak of the other finch: coevolution of genetic covariance structure and developmental modularity during adaptive evolution. Phil. Trans. R. Soc. B 365, 1111–1126 10.1098/rstb.2009.0285 (doi:10.1098/rstb.2009.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bulmer M. G. 1971. The effect of selection on genetic variability. Am. Nat. 105, 201–211 10.1086/282718 (doi:10.1086/282718) [DOI] [Google Scholar]
- 110.Slatkin M., Frank S. A. 1990. The quantitative genetic consequences of pleiotropy under stabilizing and directional selection. Genetics 125, 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oyama S. 1988. Stasis, development and heredity. In Evolutionary processes and metaphors (eds Ho M.-W., Fox S. W.), pp. 255–274 London, UK: John Wiley & Sons Ltd [Google Scholar]
- 112.Jablonka E. 2001. The systems of inheritance. In Cycles of contigency: developmental systems and evolution (eds Oyama S., Griffiths P. E., Gray R. D.), pp. 99–116 Cambridge, MA: MIT Press [Google Scholar]
- 113.Badyaev A. V., Uller T. 2009. Parental effects in ecology and evolution: mechanisms, processes, and implications. Phil. Trans. R. Soc. B 364, 1169–1177 10.1098/rstb.2008.0302 (doi:10.1098/rstb.2008.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chetverikov S. S. 1926. On certain aspects of the evolutionary process from the standpoint of modern genetics. J. Exp. Biol. Ser. A 2, 1–40 [Google Scholar]
- 115.Wagner A. 2003. Risk management in biological evolution. J. Theor. Biol. 225, 45–57 10.1016/S0022-5193(03)00219-4 (doi:10.1016/S0022-5193(03)00219-4) [DOI] [PubMed] [Google Scholar]
- 116.Badyaev A. V. 2007. Evolvability and robustness in color displays: bridging the gap between theory and data. Evol. Biol. 34, 61–71 10.1007/s11692-007-9004-5 (doi:10.1007/s11692-007-9004-5) [DOI] [Google Scholar]
- 117.Caporale L. H. 1999. Chance favors the prepared genome. Ann. N Y Acad. Sci. 870, 1–21 10.1111/j.1749-6632.1999.tb08860.x (doi:10.1111/j.1749-6632.1999.tb08860.x) [DOI] [PubMed] [Google Scholar]
- 118.Jablonka E., Raz G. 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity. Q. Rev. Biol. 84, 131–176 10.1086/598822 (doi:10.1086/598822) [DOI] [PubMed] [Google Scholar]
- 119.Badyaev A. V. 2009. Evolutionary significance of phenotypic accommodation in novel environments: an empirical test of the Baldwin effect. Phil. Trans. R. Soc. B 364, 1125–1141 10.1098/rstb.2008.0285 (doi:10.1098/rstb.2008.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salazar-Ciudad I., Newman S. A., Sole R. V. 2001. Phenotypic and dynamical transitions in model genetic networks I. Emergence of patterns and genotype-phenotype relationships. Evol. Dev. 3, 84–94 10.1046/j.1525-142x.2001.003002084.x (doi:10.1046/j.1525-142x.2001.003002084.x) [DOI] [PubMed] [Google Scholar]
- 121.Gilbert S. F. 2001. Ecological developmental biology: developmental biology meets the real world. Dev. Biol. 233, 1–12 10.1006/dbio.2001.0210 (doi:10.1006/dbio.2001.0210) [DOI] [PubMed] [Google Scholar]
- 122.Thompson J. N. 2004. The coevolutionary process. Chicago, IL: University of Chicago Press [Google Scholar]
- 123.Eldredge N., et al. 2005. The dynamics of evolutionary stasis. Paleobiology 31, 133–145 10.1666/0094-8373(2005)031[0133:TDOES]2.0.CO;2 (doi:10.1666/0094-8373(2005)031[0133:TDOES]2.0.CO;2) [DOI] [Google Scholar]
- 124.Waddington C. H. 1941. Evolution of developmental systems. Nature 147, 108–110 10.1038/147108a0 (doi:10.1038/147108a0) [DOI] [Google Scholar]
- 125.Wagner G. P. 2007. The developmental genetics of homology. Genetics 8, 473–479 [DOI] [PubMed] [Google Scholar]
- 126.Mousseau T. A., Fox C. W. 1998. (eds) Maternal effects as adaptations. Oxford, UK: Oxford University Press [Google Scholar]
- 127.Allis C. D., Jenuwein T., Reinberg D., Caparros M. L. (eds) 2007. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- 128.Bird A. 2007. Perceptions of epigenetics. Nature 447, 396–398 10.1038/nature05913 (doi:10.1038/nature05913) [DOI] [PubMed] [Google Scholar]