Abstract
Posttranslational modifications (PTMs) are chemical alterations that are critical to protein conformation and activation states. Despite their functional importance and reported involvement in many diseases, evolutionary analyses have produced enigmatic results because only weak or no selective pressures have been attributed to many types of PTMs. In a large-scale analysis of 16,836 PTM positions from 4,484 human proteins, we find that positions harboring PTMs show evidence of higher purifying selection in 70% of the phosphorylated and N-linked glycosylated proteins. The purifying selection is up to 42% more severe at PTM residues as compared with the corresponding unmodified amino acids. These results establish extensive selective pressures in the long-term history of positions that experience PTMs in the human proteins. Our findings will enhance our understanding of the historical function of PTMs over time and help in predicting PTM positions by using evolutionary comparisons.
Keywords: genomics, proteomics, posttranslational modifications, evolution
Introduction
Posttranslational modifications (PTMs) fine-tune biochemical functions in a large percentage of human proteins (Mann and Jensen 2003; Seo and Lee 2004). In particular, phosphorylation and glycosylation of amino acid positions in thousands of human proteins are now known, many of which have been implicated in a number of complex human diseases (e.g., Aly et al. 1992; Lu et al. 1999; Marquardt and Denecke 2003; Lemeer and Heck 2009). In phosphorylation, a negatively charged phosphate group is added by a kinase targeting serine (S), threonine (T), or tyrosine (Y) residues. To date, one-third of known proteins have been shown to contain at least one phosphorylated residue (Mann et al. 2002). In glycosylation, an enzyme covalently attaches a sugar to an amino acid. The asparagine (N)-linked glycosylation is the most well studied and is known to direct protein folding, product secretion, binding affinity, substrate specificity, and enzymatic activity. It is estimated that over 75% of glycosylated proteins contain at least one N-linked glycan (Apweiler et al. 1999; Nakajima et al. 2010). Because of the importance of PTMs in proper protein function and the known implications of their disruption in complex diseases, we set out to quantify the degree of evolutionary selective pressure exerted on PTM positions at the amino acid level and compare and contrast these selective pressures on four major amino acid residues involved in phosphorylations (S*, T*, and Y*) and N-linked glycosylations (N*).
We analyzed 16,836 PTM positions (S*, T*, Y*, and N*) from 4,484 human proteins. A vast majority of proteins are reported to have one or a few PTM residues, but a substantial number of proteins contain >10 PTMs (fig. 1A). About 5% of proteins contain both phosphorylated as well as glycosylated amino acids. For each modified (PTM) and unmodified residue, we estimated the absolute rates of evolution by mapping sequence differences among 44 diverse species onto their evolutionary tree and dividing the amount of change by the total time elapsed over all tree branches (fig. 1B; see Methods). As expected, proteins containing PTMs evolve at vastly different rates (fig. 1C). The fastest 25% evolving proteins evolve at a rate approximately 7 times greater than the slowest 25% evolving proteins. Overall, phosphorylated proteins are more conserved than N-linked glycosylated proteins (20% difference; P << 0.01; t-test with unequal variances). Similarly, different amino acid residues evolve with different rates (fig. 2), with tyrosines showing the most skewed distribution and many more slowly evolving sites than in other amino acid residues.
FIG. 1.
Density and evolutionary patterns of PTMs in human proteins. (A) The log distribution of the number of recorded phosphorylation and N-linked glycosylation events in human proteins. Of the 4,484 proteins that were analyzed, 33% have only one reported modified residue while as many as 77 phosphorylation and glycosylation events were observed on a single protein. (B) Evolutionary timetree of 44 species used for estimating evolutionary rates (Kumar et al. 2009). (C) The frequency distributions of protein evolutionary rates measured in terms of the average rate of amino acids S, T, Y, and N in each protein, only N (N-linked glycosylation), and S, T, and Y (phosphorylations). The average rates for phosphorylation proteins (STY), N-linked glycosylation proteins, and all proteins are 0.82, 1.23, and 1.04 substitutions/site/Byr, respectively.
FIG. 2.
Frequency distributions of evolutionary rates of all Serines (S), Threonines (T), Tyrosines (Y), and Asparagines (N) in proteins with one or more ofPTMs. The average rates are 0.85, 0.95, 0.35, and 1.07 substitutions/site/Byr for S, T, Y, and N, respectively.
In order to examine whether the positions involved in phosphorylation are under additional purifying selection at PTM positions (i.e., more conserved evolutionarily), we compared evolutionary rates of positions harboring phosphorylated residues (phosphosites: S*, T*, and Y*) with those of unmodified residues (S, T, and Y, respectively). For each protein, we calculated the ratio of rates at PTM versus all sites occupied by the given amino acid, α, by dividing the average evolutionary rate at PTM positions (rS*) with the average evolutionary rate at all positions with that residue (rS). For example, αS = rS*/rS for serine phosphorylations in a protein. PTM rate ratios αN, αT, and αY are estimated in the same way for asparagine, threonine, and tyrosine residues, respectively. Biologically, α < 1 indicates additional purifying selection, inferred from greater conservation, on a PTM position as compared with their unmodified counterparts in the same protein. We estimate α for each protein separately because of large differences in average evolutionary rates, and thus constraints, in different proteins (fig. 1C). This was also done for each amino acid type separately because different amino acids evolve with different rates (fig. 2).
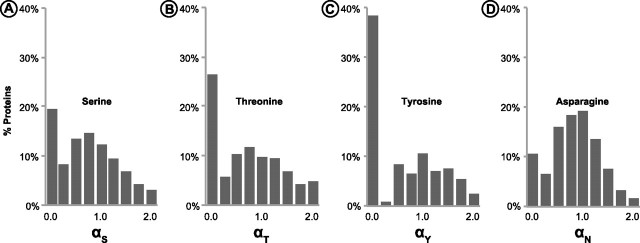
Proteome-wide distributions of α for the four PTM amino acids show many similarities and differences (fig. 3A–D). In each case, there is significant evidence of higher purifying selection at positions harboring PTMs; α is less than 1.0 for 68.1%, 63.6%, 63.5%, and 70.5% of the proteins for S*, T*, Y*, and N*, respectively. Based on these numbers, we tested a simple null hypothesis of no-effect by evaluating whether the fraction of proteins with α < 1 is significantly different from 50%. This null hypothesis is rejected in each case (P << 0.01; Z-test). Overall, 69.6% of all proteins showed α less than 1.0, when all PTMs in every protein were considered together. Because the observed distributions of α are not normal, we use median as a summary statistic. The median values are 0.65, 0.68, 0.62, and 0.73 for S*, T*, Y*, and N*, respectively. This means that a majority of PTM harboring positions have experienced a greater degree of purifying selection, with 27–38% more amino acid mutations eliminated in PTM sites as compared with their non-PTM counterparts in the same proteins. These results are robust to artifacts and distortion caused by potentially unreliable estimates of α produced for proteins that have only a few amino acids of each type. This is because the median (as well as mean) values from all data were very similar to those obtained from the top 25% of data-rich proteins that contained the largest number of unmodified S, T, Y, and N residues.
FIG. 3.
Frequency distributions of relative evolutionary rates (α) of different types of PTM residues (S, T, Y, and N) as compared with all residues of the same type in each protein. α < 1 indicates additional purifying selection on PTM residues. Median (mean) values are 0.65 (0.83), 0.68 (0.87), 0.62 (1.03), and 0.73 (0.78) for S, T, Y, and N, respectively.
The magnitude and direction of selective pressures on PTM positions in our study stands in stark contrast with those reported recently (Landry et al. 2009; Chen et al. 2010; Gnad et al. 2007). For example, Gnad et al. (2007) and Tan et al. (2009) have reported that phosphorylated residues do not show appreciably higher evolutionary conservation than the other residues. We have not only shown that there is significant purifying selection on phosphorylated residues but also that the additional purifying selection is extensive (32–38%). Although some recent studies have supported the existence of purifying selection, there are differences between our and their observations. First, the magnitude of purifying selection we have uncovered is many times larger than that reported by Landry et al. (2009). Second, Chen et al. (2010) did not find purifying selection in phosphorylated tyrosines (see below for a reason). Furthermore, none of these studies explored evolutionary pressures in glycosylated positions, which we have shown to undergo extensive additional purifying selection (27%) at residues with PTMs.
A common feature of many previous studies has been that proteome-wide patterns were generated by pooling evolutionary rates across sites. In many cases, the proteome-wide mean of evolutionary conservation at PTM and unmodified positions were contrasted to assess evolutionary pressures at phosphorylated residues. However, each protein experiences different degrees of evolutionary pressures, which is reflected in the great diversity of evolutionary rates with which they evolve (fig. 1C). Therefore, it is important to compare rates of evolution at PTM and unmodified residues for each protein separately. Otherwise, proteins with many PTM positions and those with dramatically different evolutionary rates will contribute in unexpected ways to the overall summary estimates. This is indeed true in the present data. The estimates of α for different proteins and amino acids clearly lack distributions with a strong central tendency (fig. 3). Furthermore, the distributions are not always unimodal and some have a rather long tail with appreciable frequencies. For example, a large proportion of proteins have αY > 1 for PTMs involving tyrosines, although a rather large number of them show αY = 0 (fig. 3C). When the rates are pooled proteome wide for modified and unmodified residues, one gets a mean estimate of 0.89 by dividing the average rate at Y* residues by the rate at all unmodified Y residues for our data. This would suggest that only 11% additional mutations are eliminated by natural selection at Y* residues, which is much less than that inferred by using the median of protein-by-protein α values (38%). Such dilution of the evolutionary signal contributed to previous conclusions of the lack of purifying selection on Y* residues because selective pressures were not measured for each protein separately. Similarly, per-residue averages computed without regard to the protein-specific patterns produce 21%, 13%, and 20% lower estimates of additional purifying selection at N*, S*, and T* residues, respectively. Therefore, the use of protein as a unit of selection is important in fully incorporating protein-specific evolutionary rate differences as well as the nonsymmetric distributions of α.
Overall, our results suggest that human PTMs are likely to be shared with other species because otherwise PTM and unmodified positions would not show a large difference in long-term evolutionary rates. Independent support for this possibility exists in observations that phosphorylated positions in mouse and human proteins are shared more often than expected by chance (Boekhorst et al. 2008). Greater purifying selection in PTM sites is also consistent with an enrichment of inherited disease mutations at PTM positions (Li et al. 2010), a pattern that is seen for Mendelian disease mutations (Subramanian and Kumar 2006). Concordantly, the neutral mutations will be depleted in positions harboring PTMs. The discovery of higher purifying selection in PTM positions in our study is consistent with the known importance of these positions in protein function and human diseases. And they also establish that trends seen in single-celled organisms, such as yeast (Ba and Moses 2010) and bacteria (Macek et al. 2008), hold true for complex organisms as well. The existence of signatures of additional evolutionary conservation at PTM positions would facilitate greater extrapolation of knowledge from model organisms to understanding human diseases.
Methods
The modified amino acid data set was downloaded from the public database dbPTM (http://dbptm.mbc.nctu.edu.tw/), which is the largest repository of human PTMs (Lee et al. 2006). We only analyzed human proteins containing phosphorylation and N-linked glycosylation sites because data on other types of PTMs are rather small. Each PTM was mapped onto a 44-species alignment available in the University of California–Santa Cruz (UCSC) Genome Browser (Rhead et al. 2010) by mapping RefSeq identifiers and matching reference amino acids (Pruitt et al. 2007). Our final data sets included 16,836 modified residues in 4,484 human proteins, which contained 219,041 unmodified S, T, Y, and N residues. Of 16,836 modified residues, 7,809 were involved in phosphorylation (6,089 S*; 1,322 T*; and 398 Y*) and 9,027 were involved in N-linked glycosylated residues. For each residue, we estimated absolute evolutionary rates using the 44-species amino acid sequence alignment from UCSC following the procedure in Kumar et al. (2009) in which absolute evolutionary rate at each site is calculated separately by mapping the amino acid sequence differences on the well-established tree of species (fig. 1B). The evolutionary rates are in the unit of the number of amino acid substitutions per site per billion years (Byrs).
Acknowledgments
We thank Alan Filipski for helpful discussions and comments pertaining to this study and Revak Raj Tyagi for assistance in data preparation and analysis execution. This research was supported by the National Institutes of Health to S.K. (HG002096 and LM010834).
References
- Aly A, Higuchi M, Kasper C, Kazazian HJ, Antonarakis S, Hoyer L. Hemophilia A due to mutations that create new N-glycosylation sites. Proc Natl Acad Sci U S A. 1992;89:4933–4937. doi: 10.1073/pnas.89.11.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Ba A, Moses A. Evolution of characterized phosphorylation sites in budding yeast. Mol Biol Evol. 2010;27:2027–2037. doi: 10.1093/molbev/msq090. [DOI] [PubMed] [Google Scholar]
- Boekhorst J, van Breukelen B, Heck A, Snel B. Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol. 2008;9:R144. doi: 10.1186/gb-2008-9-10-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chen F, Li W. Phosphorylated and non-phosphorylated serine and threonine residues evolve at different rates in mammals. Mol Biol Evol. 2010;27:2548–2554. doi: 10.1093/molbev/msq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Suleski M, Markov G, Lawrence S, Marco A, Filipski A. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009;19:1562–1569. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C, Levy E, Michnick S. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25:193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Lee T, Huang H, Hung J, Huang H, Yang Y, Wang T. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeer S, Heck A. The phosphoproteomics data explosion. Curr Opin Chem Biol. 2009;13:414–420. doi: 10.1016/j.cbpa.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Li S, Iakoucheva L, Mooney S, Radivojac P. Loss of post-translational modification sites in disease. Pac Symp Biocomput. 2010:337–347. doi: 10.1142/9789814295291_0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Wulf G, Zhou X, Davies P, Lu K. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- Macek B, Gnad F, Soufi B, Kumar C, Olsen J, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- Mann M, Jensen O. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- Mann M, Ong S, Grønborg M, Steen H, Jensen O, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Denecke J. Congenital disorders of glycosylation: review of their molecular bases, clinical presentations and specific therapies. Eur J Pediatr. 2003;162:359–379. doi: 10.1007/s00431-002-1136-0. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Koga T, Sakai H, Yamanaka H, Fujiwara R, Yokoi T. N-Glycosylation plays a role in protein folding of human UGT1A9. Biochem Pharmacol. 2010;79:1165–1172. doi: 10.1016/j.bcp.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Tatusova T, Maglott D. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. (23 co-authors). 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Lee K. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol. 2004;37:35–44. doi: 10.5483/bmbrep.2004.37.1.035. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner M, Jørgensen C, Bader G, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]