Abstract
Today, electron cryomicroscopy (cryo-EM) can routinely achieve subnanometer resolutions of complex macromolecular assemblies. From a density map, one can extract key structural and functional information using a variety of computational analysis tools. At subnanometer resolution, these tools make it possible to isolate individual subunits, identify secondary structures, and accurately fit atomic models. With several cryo-EM studies achieving resolutions beyond 5 Å, computational modeling and feature recognition tools have been employed to construct backbone and atomic models of the protein components directly from a density map. In this chapter, we describe several common classes of computational tools that can be used to analyze and model subnanometer resolution reconstructions from cryo-EM. A general protocol for analyzing subnanometer resolution density maps is presented along with a full description of steps used in analyzing the 4.3 Å resolution structure of Mm-cpn.
1. INTRODUCTION
Electron microscopy has played an increasingly important role in understanding the structure and function of macromolecular assemblies that contribute to numerous biological processes. It offers an advantage over other structural techniques, like X-ray crystallography, by imaging macromolecular assemblies in near-native conditions (Baumeister and Steven, 2000; Frank, 2002). Even at non-atomic resolutions, three-dimensional (3D) reconstructions (volumetric density maps) from electron microscopy can describe the size, shape, and composition of a macromolecular assembly.
In 1975, the first subnanometer resolution electron microscopy data was derived from regularly arrayed 2D crystals of bacteriorhodopsin by Henderson and Unwin (1975). At 7 Å resolution, the seven transmembrane helices of bacteriorhodopsin were clearly visible. Fifteen years later, Henderson et al. (1990) reported the first atomic model constructed directly from an electron microscopy density map. Another milestone was achieved in 2005, as water molecules were clearly resolved in the 1.9 Å resolution structure of aquaporin-0 (Gonen et al., 2005).
In single-particle electron cryomicroscopy (cryo-EM), a macromolecular assembly does not need to form a regular array. Rather, images of randomly orientated particles are processed to generate a 3D density map (Cong and Ludtke, 2010). Early 3D reconstructions achieved relatively low resolutions due to several factors including small data sets, sample heterogeneity, and technical limitations of the microscopes. In 1997, a significant milestone was achieved in single-particle cryo-EM; two reconstructions of the Hepatitis B virus obtained subnanometer resolutions (9 and 7.4 Å resolution; Böttcher et al., 1997; Conway et al., 1997). From these density maps, it was possible for the first time to clearly identify the core capsid protein and visualize rod-like structures corresponding to α-helices. Using the connectivity of the helices, the overall fold of the 183 amino acid core capsid protein was proposed. Several years later, a reconstruction of the rice dwarf virus (6.8 Å resolution) was the first cryo-EM reconstruction to clearly resolve β-sheets as flat planes of density (Zhou et al., 2001). Like the density maps from the Hepatitis B reconstructions, this resolution allowed for the description of the protein fold, though no models were constructed. Subsequent atomic models confirmed the proposed folds for both Hepatitis B (Wynne et al., 1999) and rice dwarf virus (Nakagawa et al., 2003). Today, technical advances in computing, specimen preparation and data acquisition have made it possible to routinely achieve subnanometer resolutions on a wide variety of specimens with single-particle cryo-EM (Chiu et al., 2005).
A decade after the first subnanometer resolution cryo-EM structures, another milestone was reached; the first near-atomic resolution structures were produced by single-particle cryo-EM. In reconstructions of rotavirus (3.88 Å; Zhang et al., 2008), GroEL (4.2 Å; Ludtke et al., 2008), cytoplasmic polyhedrosis virus (4.0 Å; Yu et al., 2008), and bacteriophage ε15 (4.5 Å; Jiang et al., 2008), the pitch of α-helices and the separation of β-strands were visualized. Though these structures did not have the resolution to utilize standard X-ray crystallographic tools for model construction (typically starting at ~3.5 Å resolution), de novo Cα backbone models were built from the cryo-EM density maps of cytoplasmic polyhedrosis virus, GroEL, and bacteriophage ε15 using a combination of computational and geometric tools (Baker et al., 2007, Ju et al., 2007). The de novo models built directly from these density maps relied almost entirely on visual interpretation of the density and manual structure assignment. Several recent state-of-the-art reconstructions have now resolved sidechain densities and allowed for the construction of complete atomic models directly from the density map (Cong et al., 2010; Yu et al., 2008; Zhang et al., 2010).
2. FEATURES IN A SUBNANOMETER RESOLUTION DENSITY MAP
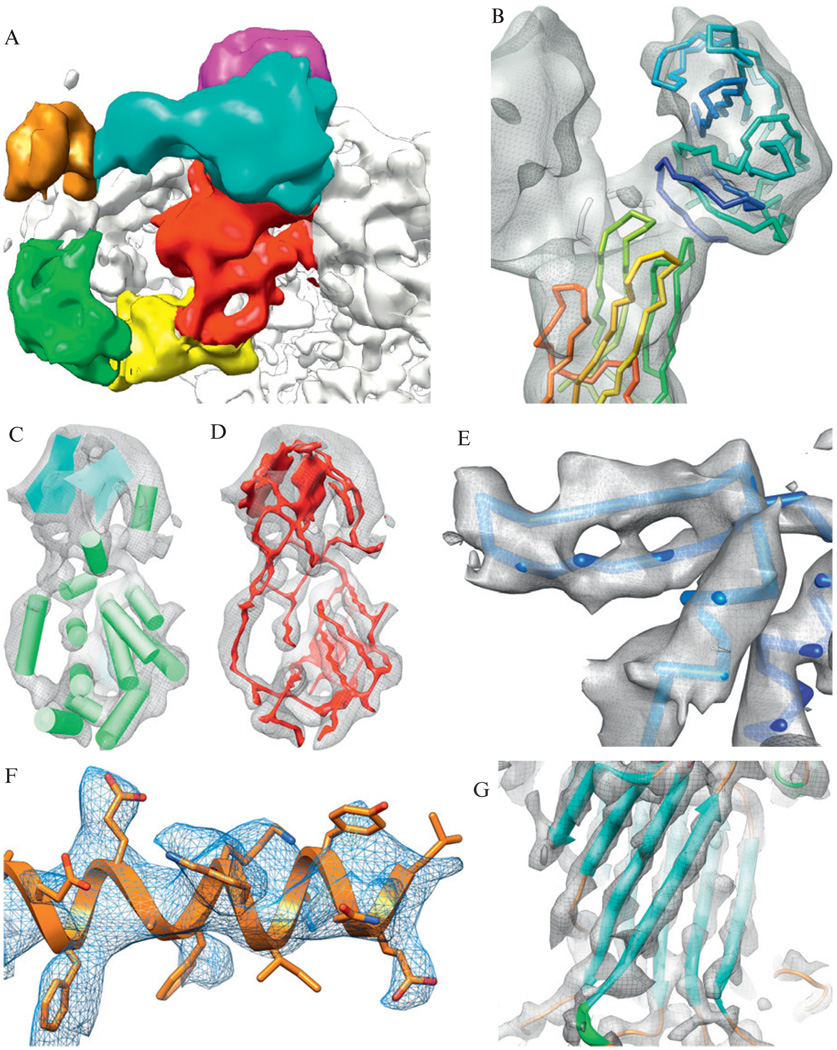
Regardless of the resolution, computational tools are critical in analyzing, interpreting, and annotating structural information in cryo-EM density maps. As such, a number of specialized computational tools have been developed. However, before describing these tools and the general protocol for analyzing a cryo-EM density map, it is important to establish an understanding of the features visible in a density map as a function of resolution (Fig. 1.1). Apparent in many of the density maps deposited in the EMDB (http://emdatabank.org), subnanometer resolution structures have distinct boundaries that allow for subunits and individual domains to be identified and segmented from the entire map (Fig. 1.1A). These well-defined densities help to accurately fit known structural models to a density map (Fig. 1.1B). α-Helices appear as long rod-like densities, and β-sheets appear as thin, continuous planes at ~8 Å resolution (Fig. 1.1C). At slightly higher resolutions, connectivity between the secondary structure elements (SSEs) becomes evident (Fig. 1.1D). α-Helices begin to develop features and the pitch becomes visible at ~5 Å resolution. Beyond 4.5 Å resolution, the thin, flat planes of β-sheets become broken, as individual strands are resolved (Fig. 1.1E). By 4 Å resolution, sidechain densities become recognizable (Fig. 1.1F) and a relatively unambiguous trace of a protein backbone can be seen (Fig. 1.1G).
Figure 1.1.
Features at subnanometer resolutions. A gallery of structural features from cryo-EM reconstructions is shown. (A) Domains in the clamp region of the 9.5 Å resolution reconstruction of RyR1 can be observed (Serysheva et al., 2008). (B) The atomic models of VP5* (lower left) and VP8* (upper right) are fit to the density map corresponding to the VP4 spikes in the 9.5 Å resolution structure of rotavirus (Li et al., 2009). (C) At slightly higher resolutions, secondary structures (α-helices are depicted as cylinders and β-sheets are depicted as planes) can be clearly seen in the capsid proteins of rice dwarf virus at 6.8 Å resolution (Zhou et al., 2001). (D) Around this resolution, possible connections between secondary structure elements can be identified computationally using density skeletonization (red), again as seen in the 6.8 Å resolution rice dwarf virus capsid protein. (E) Increasing resolution reveals the separation of β-strands in GroEL at 4.2 Å resolution (Ludtke et al., 2008). (F) Large, bulky sidechains begin to appear in TriC reconstruction at 4.0 Å resolution (Cong et al., 2010). (G) An unambiguous backbone is apparent in VP6 of rotavirus at ~3.8 Å resolution (Zhang et al., 2008).
3. TOOLS: ANALYZING A SUBNANOMETER RESOLUTION DENSITY MAP
While subnanometer resolutions span a wide range of detectable features, tools for analyzing their structure can be grouped into two classes: feature recognition and data integration. In principle, many of these tools are not restricted to subnanometer resolutions, although higher resolution features do provide significant advantages in validation. Feature recognition tools, however, are generally resolution-specific. The following briefly describes - some “standard” tools and techniques for analyzing subnanometer resolution cryo-EM density maps (Table 1.1).
Table 1.1.
Programs for analyzing subnanometer resolution cryo-EM density maps
Common computational tools used in the analysis of subnanometer resolution cryo-EM density maps are listed. Numerous other algorithms have also been published, though only currently downloadable tools are listed.
3.1. Fitting atomic models
Perhaps the most common method for analyzing a subnanometer resolution density map is fitting a known atomic model into a density map. There are a number of different approaches to fit atomic models to the density map, though an exhaustive rotational and translational search of the model within the map is generally used (reviewed in Rossmann et al., 2005). “Rigid-body” fitting attempts to identify the maximum overlap of the model with the density and minimize the amount of model unaccounted for within the density. Various fitting programs report scores differently, so it is important to consult the program documentation, as well as visually inspect the fit of a model to the density map. Aside from program-specific fitting scores, tools are available for independently reporting the quality of fit for a model within a density map (Serysheva et al., 2005; Volkmann, 2009).
In addition to fitting an atomic model as a “rigid-body” within the density map, atomic models can be morphed, or flexibly fit, into the density map (Schröder et al., 2007; Suhre et al., 2006; Tama et al., 2004; Tan et al., 2008; Topf et al., 2008; Trabuco et al., 2009). In these types of programs an atomic model is allowed to relax or bend at certain points to better fit the density map. This is particularly useful when fitting structures of different conformations or homologous structures.
3.2. Constrained modeling with cryo-EM density
If a structure for one or more of the protein subunits in the assembly is not known, computational modeling approaches can be used to generate homology models or ab initio models for proteins. In these cases, the cryo-EM density map can be used directly to facilitate the construction and evaluation of a protein/domain structural model (Baker et al., 2006a; DiMaio et al., 2009; Topf et al., 2005, 2006; Zhu et al., 2010). In a constrained comparative modeling approach, an initial sequence–structure alignment is allowed to evolve, simultaneously improving the homology model and its fit to the density map (Topf et al., 2005, 2006). This type of approach can also be used to improve local regions of a model within the density map. When a template structure is not known, constrained ab initio modeling may be used to build domains or small proteins (Baker et al., 2006a). In this approach, a gallery of models are built computationally and then later evaluated by their fit to the density map. The resolution of the density map is key in determining the accuracy of the models (Topf et al., 2005). While not restricted to subnanometer resolutions, models built within a subnanometer resolution density map will likely have the correct fold, though atom placement may only be approximate.
3.3. Extracting protein subunits from a density map
Like fitting atomic models to a density map, density segmentation, the process of identifying and isolating a single protein or domain from the cryo-EM density map, is not exclusive to subnanometer resolutions. However, at subnanometer resolutions, cryo-EM density maps move from being “blob-like” to having distinct features, including sharp drop-offs in the density that characterizes the edges and boundaries between subunits (Chiu et al., 2005). The actual process of segmenting a subunit can be done using interactive tools commonly found in visualization programs such as Chimera (Pettersen et al., 2004), Amira (Visage Imaging, GmbH), and Avizo (VSG, France) or computational techniques such as those based on the watershed transform (Ludtke et al., 1999; Pintilie et al., 2010; Volkmann, 2002).
3.4. Secondary structure identification
In cryo-EM, feature recognition tools have been typically used to identify SSEs within a subnanometer resolution density map. At subnanometer resolutions, helices are clearly resolved as long cylinders with relatively high density. Helixhunter (Jiang et al., 2001) was the first tool designed to computationally detect α-helices in a density map using a simple cross-correlation search with a prototypical helix template over three translational and two rotational degrees of freedom. β-Sheets, which resemble thin planes, are more diverse in their structure and not amenable to correlation-based approaches. Rather, morphological analysis of the density was used to first localize β-sheets and strands in subnanometer resolution density maps (Kong and Ma, 2003; Kong et al., 2004). Later developments lead to SSEHunter, a single tool to detect both α-helices and β-sheets (Baker et al., 2007). With SSEHunter, the helix correlation routine is paired with a local geometry analysis and a density skeleton to detect SSEs. With any of these feature recognition tools, it is advisable to operate on segmented density maps rather than the entire map when detecting SSEs.
3.5. De novo modeling
In conjunction with development of SSEHunter, a new density skeletonization routine (Ju et al., 2007) was developed that preserves both the features and the topology in a density map. This density skeleton provided clear connections between observable SSEs. Coupling density skeletonization with the aforementioned feature recognition tools has given rise to a new protocol for de novo structural modeling (Jiang et al., 2008; Ludtke et al., 2008). The de novo method attempts to construct a model directly from a density map without the aid of an existing structural template. Borrowing methods for constructing atomic models from X-ray crystallographic density maps, SSEs in the sequence and the density map are correlated, providing initiation points for model construction (Abeysinghe et al., 2008). This sequence-to-structure correspondence assigns residue positions to SSEs in the map.
Once SSE anchor points are established, models can be constructed using various X-ray crystallographic model building toolkits, such as O (Jones et al., 1991) and Coot (Emsley and Cowtan, 2004), both of which offer tools for placing atoms within a cryo-EM density map. Gorgon (http://gorgon.wustl.edu), a molecular modeling toolkit tailored to subnanometer resolution cryo-EM density maps, provides a comprehensive suite of utilities to analyze subnanometer resolution density maps, including the ability to generate secondary structure assignments, a sequence-to-structure correspondence routine and Cα model construction utilities.
4. PROTOCOL: FROM DENSITY MAP TO ATOMIC MODEL
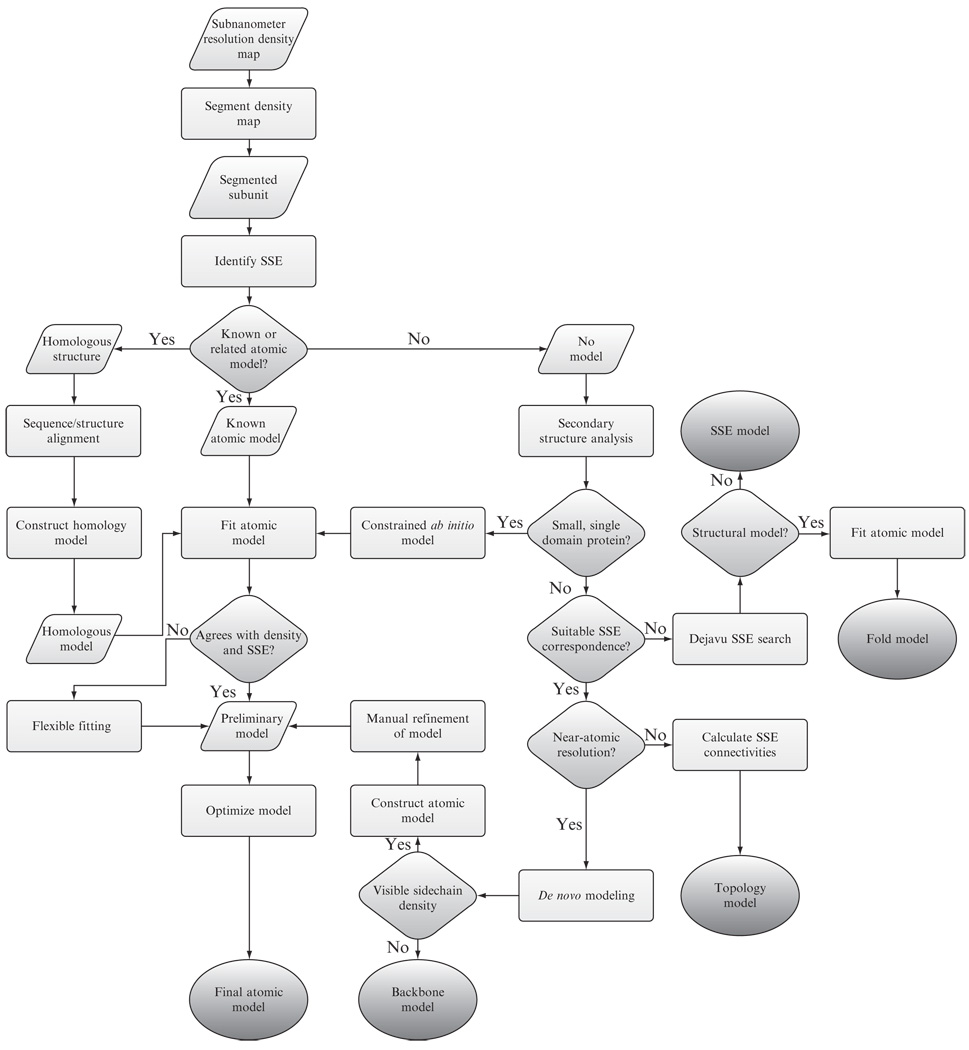
With an extensive software library (Table 1.1), analyzing a subnanometer resolution density map can be an overwhelming task. Simply identifying the proper tools and understanding the expected results can be difficult. Figure 1.2 provides a simplified flowchart describing the assorted paths and decisions one may encounter in analyzing a subnanometer resolution density map.
Figure 1.2.
Analyzing a subnanometer resolution cryo-EM density map. A general scheme for analyzing subnanometer resolution cryo-EM density maps is depicted. Different projects may take advantage of additional information during the analysis process and thus deviate from the overall scheme.
In the following sections, we describe a general protocol (based on Fig. 1.2) for analyzing a subnanometer resolution density map. In a subsequent section, we also detail the analysis of the 4.3-Å resolution structure of Methanococcus maripaludis chaperonin (Mm-cpn), a group II chaperonin. It should be noted that this protocol is not restricted to single-particle cryo-EM and can be used to analyze any type of subnanometer resolution density map.
The steps described in this protocol illustrate a complete pathway for constructing an atomic model from a cryo-EM density map; however, most of the steps can be used independently depending on numerous constraints, such as resolution and available structural information. As such, consulting the various user-guides and documentation for the individual software packages will help the user to optimally select parameters for each particular project.
In this protocol, several image processing, visualization and modeling software packages are utilized, including Coot (Emsley and Cowtan, 2004), EMAN/EMAN2 (Ludtke et al., 2005; Tang et al., 2007), Gorgon (http://gorgon.wustl.edu), Rosetta (Bradley et al., 2005), and UCSF Chimera (Pettersen et al., 2004). Other programs may be substituted, though their exact usage, including inputs and outputs, will vary. Additionally, the amount of time and computational resources required by each of these tools will vary considerably based on the project and experience of the user.
4.1. Segmentation
To begin modeling an individual protein subunit, one subunit/domain must be extracted from the macromolecular assembly. Generally, individual protein densities within subnanometer resolution reconstructions have relatively sharp fall-offs at their boundaries. Identifying the boundaries between subunits can be accomplished using a variety of different approaches. Manual segmentation requires the user to visually identify and demarcate the subunits. By adjusting the isosurface display threshold in visualization programs, such as Chimera and Gorgon, these boundaries can usually be seen clearly. The user can then create a mask as either a set of 2D slices or a single 3D volume, or “erase” spurious density to produce an initial segmentation of a protein subunit. The initial segmentation is usually relatively crude to avoid removing too much density but still adequate for analysis. Once all subunits have been segmented, the user can then check to see that all density has been assigned a segment and no overlapping segments are present. This may require several iterations to improve the initial segmentation. Alternatively, automated approaches like EMAN’s segment3d (Ludtke et al., 1999) and Segger (Pintilie et al., 2010) will cluster every voxel in a density map based on a set of user-provided parameters (e.g., number of subunits, symmetry, etc.). Visual inspection and manual adjustment of the subunits may be required.
4.2. Identifying secondary structure elements
Once the map has been segmented, SSEs are then identified using SSEHunter (Baker et al., 2007). This step requires a cubic density map with an even number of voxels. Additionally, to maximize compatibility with other modeling programs, the origin of the segmented subunit should be reset to zero. To accomplish this, proc3d from EMAN or e2proc3d.py from EMAN2 can be used with the “clip” and “origin” options.
SSEHunter can be executed in three different ways: (1) directly from the command line (ssehunter3.py in EMAN and e2ssehunter.py in EMAN2), (2) as a plug-in to UCSF’s Chimera, or (3) within Gorgon. The first two options require the installation of EMAN or EMAN2, while Gorgon’s version of SSEHunter is built-in. In each of these cases, the user is required to provide the resolution in Å, the sampling of the map (Å/pixel) and a threshold corresponding to the highest isosurface value at which all density of the segmented subunit appears to be connected (obtained visually, ~2–4σ above the mean density).
Map sizes between 483 and 1603 generally require less than 15 min to run on a modern desktop and return a set of pseudoatoms, typically corresponding to ~50% of the total number of amino acids represented in the segmented volume. Encoded in the B-factor column of the SSEHunter PDB file are the per-pseudoatom SSEHunter scores, which range between −3 and 3. These values represent the likelihood of a density region to be either α-helix (0 −3) or β-sheet (−3 −0). In Gorgon, the pseudoatoms are automatically colored from blue (−3, β-sheet) to white (0) to red, (3, α-helix) where the intensity of the color reflects the score and confidence of the prediction.
In addition to these pseudoatoms, a density skeleton is calculated with SSEHunter. As mentioned previously, this skeleton is a simplified geometrical representation of the density map that preserves both features and topology. Gorgon offers an improved threshold-free, grayscale density skeleton that can be substituted place of the SSEHunter skeleton in later steps for identifying the connectivity between SSEs.
4.3. Secondary structure annotation
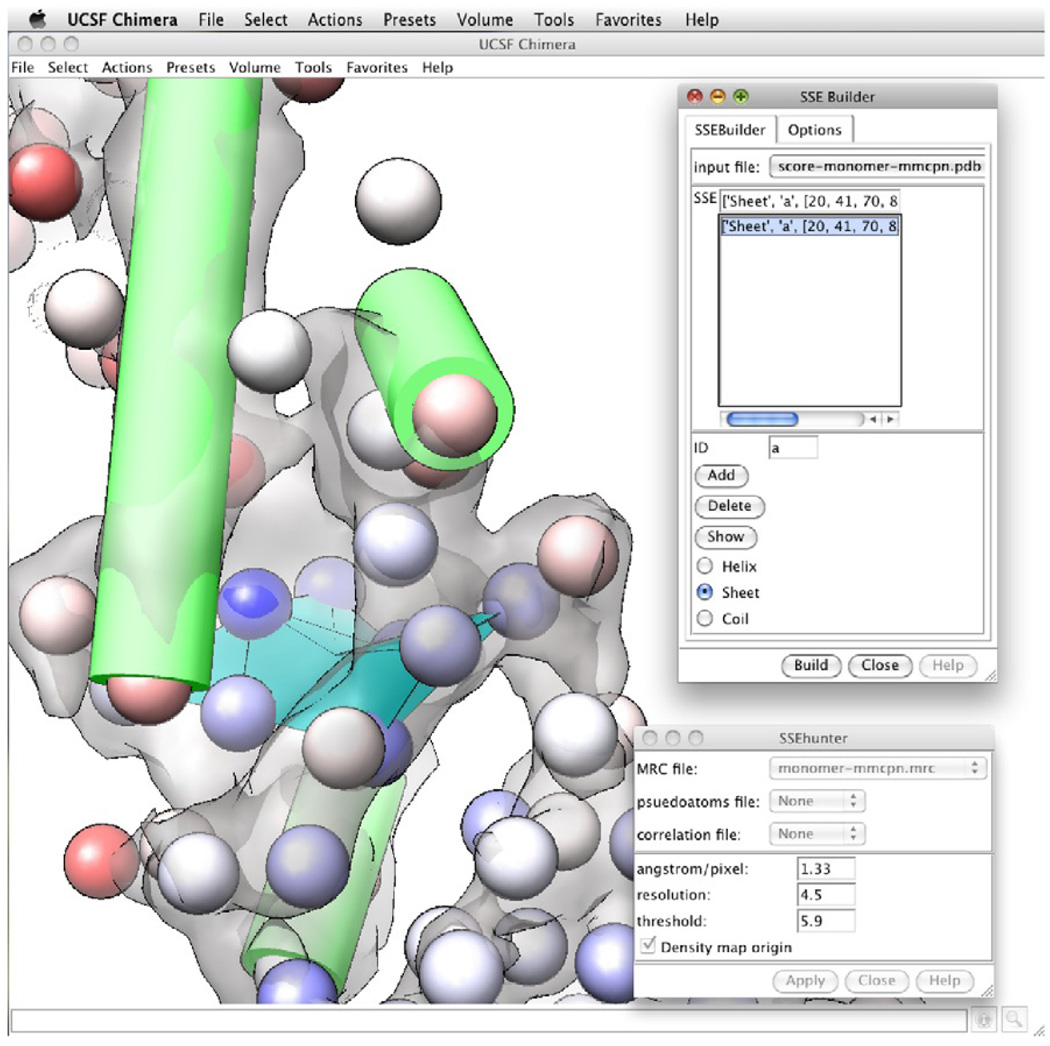
Next, pseudoatoms of similar values are grouped into their respective SSEs using SSEBuilder (Baker et al., 2007; Fig. 1.3). Like SSEHunter, SSEBuilder can be accessed from within Chimera or Gorgon. In SSEBuilder, pseudoatoms with similar scores are manually grouped into individual SSEs and then automatically constructed as VRML objects. The process continues until all visible SSEs are assigned. When selecting SSEs, generally, the minimum size is three pseudoatoms for an α-helix and five pseudoatoms for a β-sheet. False positivesmay occur; therefore, only groups of pseudoatoms that resemble SSEs in the density (α-helices appear as long cylinders, β-sheets resemble thin, curved surfaces) should be annotated.
Figure 1.3.
Secondary structure identification. SSEHunter and SSEBuilder, both EMAN programs, can be run as plug-ins to UCSF’s Chimera. The results for the apical domain of Mm-cpn are shown: red spheres represent helix like regions and the blue spheres represent sheet like regions. Regions of similar scoring pseudoatoms from SSEHunter are grouped and built using SSEBuilder. Helices are depicted as cylinders and sheets are depicted as planes.
4.4. Structural homologues from sequence
Until this point, analysis of the density map has focused on observing features within the density map itself. However, it is possible that an atomic model for one or more of the protein components in the density map is known. This may be a single protein, a domain from the macromolecular assembly or a related structure.
While it is relatively straightforward to search and retrieve a protein or domain with a known structure from the Protein Data Bank (http://pdb.org), identifying and building structural models from related structures is more complicated. Homologous structures can be identified using a number of sequence-based search methods including BLAST (Altschul et al., 1990) and FASTA (Lipman and Pearson, 1985). These types of searches generally return a sequence alignment between the sequence of interest and a sequence with a known structure from which a homologous model can be constructed. A number of tools are available for this procedure, ranging from the fairly automated Swiss-Model web service (Arnold et al., 2006) to more flexible modeling suites like Modeller (Sali et al., 1995) and Rosetta (Bradley et al., 2005).
In addition to these tools, there are several web-based tools that integrate the search and model building steps including Phyre (Kelley and Sternberg, 2009), Fugue (Shi et al., 2001), and 3D Jigsaw (Bates et al., 2001). Simplifying the process even further, meta-prediction servers, like BioInfoBank (Ginalski et al., 2003), provide a convenient way to submit a sequence to multiple prediction servers and view the results. Regardless of the method or service used, fairly reliable atomic models can be produced when a suitable template structure is identified. As each method reports potential models differently, it is important to consult the documentation for each of the tools in determining the validity of the model.
4.5. Identifying structural homologues from SSEs
If structural homologues cannot be identified from the sequence, it is still possible to detect homologues based on the locations and orientations of the SSEs (Baker et al., 2003, 2005). The identified SSEs can be used as inputs into two structural similarity search programs, DejaVu (Kleywegt and Jones, 1997) and COSEC (Mizuguchi and Go, 1995). Results obtained from SSEHunter and SSEBuilder are compatible with either program; an additional format conversion utility is provided with EMAN (dejavu2sse.py). While a sequence-based search will return a sequence alignment, a structure-based search does not. Rather, a structure-based search returns a list of possible folds that match a set of SSEs regardless of sequence.
4.6. Fitting atomic models in cryo-EM density maps
Once a suitable atomic model has been found or constructed, the model can be fit into the density map with any of the aforementioned tools. Fitting can be done either as a rigid body or flexibly fit, where the model is allowed to morph to better fit the density. To avoid mis- or overinterpretation, it is important to note the features in the density map. The shape and overall fold are likely to be recognizable in the segmented density map at subnanometer resolutions. More specifically, the atomic model should correspond well to the observed SSEs. Independent validation tools (fh-stat.py in EMAN; Serysheva et al., 2005) and confidence intervals (Volkmann, 2009) may also be used to assess the statistical likelihood of the model positions in the map.
4.7. Predicting SSEs from sequence
When no known or homologous structure is available, it is still possible to construct structural models for individual proteins or subunits de novo. In this case, a combination of secondary structure prediction and feature recognition can assign sequence elements, such as helices and strands, to SSEs identified within the density map, a process termed SSE correspondence (Abeysinghe et al., 2008; Ludtke et al., 2008).
Having identified the positions of the SSEs within a segmented density map, the next step is to define the SSEs within the sequence of interest. A number of web-based programs can be used to predict secondary structure including SSPro (Pollastri et al., 2002), JPred (Cole et al., 2008), and PsiPred (McGuffin et al., 2000). These programs generally provide a secondary structure assignment and confidence score to each amino acid. Due to errors in prediction, a consensus alignment built from multiple predictions may be better than the results from a single secondary structure prediction server. For convenience, Gorgon contains a tool that will remotely run the sequence prediction, retrieve the predictions, and format the results for the subsequent SSE correspondence routine.
4.8. SSE correspondence
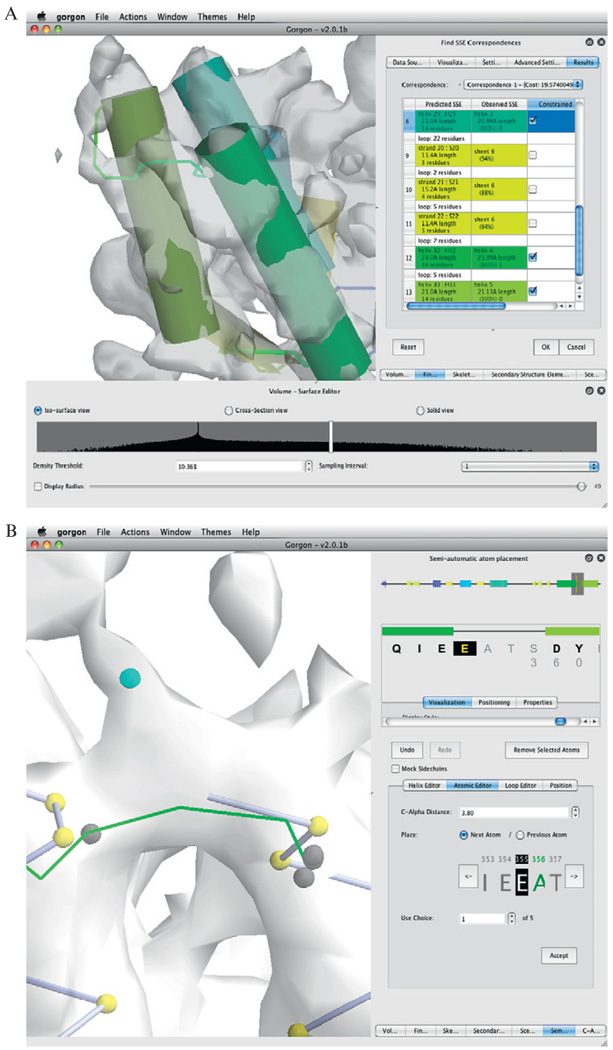
Once SSEs have been identified in both the sequence and density map, a correspondence search can be preformed within Gorgon (Fig. 1.4A). This step provides the initial anchor points to place Cα atoms and construct a protein backbone. Determining the SSE correspondence requires four inputs: helix and sheet locations produced by SSEBuilder, the cryo-EM skeleton from SSEHunter or Gorgon, and the sequence/prediction from the previous step. Once entered, a sequence–structure correspondence is calculated and displayed graphically. In Gorgon, the SSE correspondence results are shown as a list ranked in order from best to worst correspondence. To assess individual correspondences, the lengths of the SSEHunter/SSEBuilder helices are compared to the sequence-predicted helices. Ideally, the lengths of correctly matched SSEs should not differ by more than three amino acids. In a correspondence, one or more SSEs may have been incorrectly assigned. If this occurs, the user may constrain the correct individual SSE correspondences by selecting only correctly paired SSEs and re-running the correspondence search. This process is relatively quick (< 5 s for most cases) and can be done repeatedly until the user is satisfied with the correspondence.
Figure 1.4.
Model construction with Gorgon. The results from the SSE correspondence search on the apical domain of the 4.2-Å resolution structure of GroEL in Gorgon are shown in (A). Helices are shown as cylinders, while sheets are shown as planes. Potential connectivity is depicted as solid lines. A corresponding color scheme for these elements is shown in the SSE correspondence window on the right. (B) Gorgon contains several methods for assigning atoms to the density in the semi-automated atom placement tool. The atomic editor function illustrates the addition of Cα atoms along a density skeleton (not shown). The user can cycle through the possible locations, select the desired position and proceed to the next residue.
4.9. Cα placement
From the previous step, an initial topology for the protein structure is established. Starting from this topology assignment, placement of the Cα backbone atoms begins with helices, followed by sheets and loops. The following steps are described using Gorgon.
4.10. Assigning Cα positions in helices
Based on the chosen SSE correspondence, Cα positions of the helices are registered with the corresponding sequence. This process and the following modeling steps can be done with the “Semi-Automatic Atom Placement” tool found in Gorgon. Possible errors in the SSE correspondence may require the user to adjust helix length and directionality when assigning the helix residues. Helices are initially represented as cylinders; manual adjustment of the helix position may be necessary to best fit the density. Bulky sidechains in the helices provide visual cues and help anchor the position and pitch of the helix within the density (Jiang et al., 2008; Ludtke et al., 2008; Yu et al., 2008). Again, Gorgon contains a variety of mechanisms for optimizing helix position, including an option to show relative sidechain size at Cα atom positions. Once the assignment of helices is complete, strands and loops can be assigned.
4.11. Assigning Cα positions in sheets and loops
For the purposes of modeling a Cα backbone, β-strands are treated as loops. Extending from the assigned residues in the α-helices, the remaining residues can be assigned with two unique options found in Gorgon’s “Semi-Automatic Atom Placement” tool. With the “Atomic Editor” the last assigned residue before an unassigned section of sequence is selected. The possible positions for the next unassigned amino acid are shown in the density map along the skeleton for which the Cα–Cα distance is satisfied (Fig. 1.4B). The user then interactively selects a position for the unassigned amino acid. The model is updated and possible positions for the next amino acid are shown. This process continues until a previously assigned Cα is joined. Alternatively, loops may be assigned using the “Loop Editor.” With the endpoints of the loops selected, the “Loop Editor” allows the user to sketch out the approximate path through the density. Unlike the “Atomic Editor,” no Cα–Cα distance constraints are enforced, though Gorgon provides a convenient way to visualize Cα–Cα distances. Red bond distances are too long, blue bonds are too short, and white bonds are approximately the correct length (3.8 ± 0.5 Å). To aid in the placement of Cα atoms, the density skeleton, calculated by SSEHunter or in Gorgon, can provide possible paths between the SSEs. Using these two techniques, Cα atoms are placed for any unassigned amino acids.
4.12. Fixing an atomic model
In models utilizing a known or homologous structure, adjustment of the Cα positions begins with the SSEs. Entire SSEs are first moved to register with the corresponding features in the density map using modeling programs such as Coot, Gorgon, or Chimera. Once the SSEs of the model have been fit to the density map, the remaining Cα atoms are moved individually to best fit the density. It is possible that some portions of the atomic model structure may be absent. If this is the case, the previous steps can be used to build any missing residues before proceeding.
4.13. Cα optimization
After all Cα atoms have been assigned, the next step is to adjust the atom positions to optimally fit the density while maintaining reasonable Cα–Cα bond distances (~3.8 Å) and angles (~60–120° between three consecutive Cαs), proper secondary structure features and no atom/bond clashes. Optimization of Cα positions begins with helices; the pitch of the model helices should register well with the pitch of the helix observed in the density. In the event that β-strands are resolved, the distance between neighboring strand Cα atoms should be between 4.5 and 5 Å. Once Cα positions in SSEs have been optimized Cα positions in the loops can be adjusted.
4.14. Building a macromolecular model
Following the initial construction of a Cα model, the models are then placed back into the context of the full cryo-EM density map with all other models. As previously mentioned, a number of fitting routines may be used for this step.
At this point, the Cα models are assessed by visually examining how well they fit into the density map. A “good model” will occupy the entire subunit density map and have no clashes with neighboring subunits. Residues in neighboring subunit models should also be readjusted so that the minimum distance between any Cα atoms is ~4.5 Å. The last two steps will likely be iterated multiple times to improve the model until no inter- and intrasubunit clashes are evident and the models account for all the density in the reconstruction. The final refined backbone model is then saved as a PDB file.
4.15. Map rescaling
To enhance the high-resolution information in the density maps, such as sidechains, for subsequent map building, the density map can be rescaled in Fourier space using the Cα backbone model (Fernández et al., 2008; Zhang et al., 2010). The model is first blurred to approximately the same resolution as the original density map. Structure factors can be calculated from this blurred map and applied to the original density map in EMAN (proc3d or e2proc3d.py in EMAN2). A low-pass filter is then applied to the rescaled map at the calculated resolution and normalized so that the positive density values are between 0 and 1.
4.16. Cα to atomic model
An approximate atomic model can be reconstructed from the Cα model using a Cα-to-backbone builder such as the web-based SABBAC (Maupetit et al., 2006). It is likely that the atomic model produced will have some missing or unassigned residues. If residues are missing, they can be manually added and fit to the density using X-ray crystallographic modeling packages like Coot.
It is important to note that building atomic models may not be possible even at near-atomic resolutions. Features in the map dictate the possibility and level of accuracy in model construction. Further model optimization and refinement requires a significant number of sidechain densities to be recognizable.
4.17. Model optimization
At near-atomic resolutions, all sidechains larger than Valine should be evident in the density map though this is generally not the case in practice. Therefore, precedence is typically given to mainchain atom positions within the density. However, sidechain density associated with positively charged and aromatic amino acids are discernable more frequently than other amino acids (Cong et al., 2010; Kimura et al., 1997; Zhang et al., 2010) and thus provide landmarks for optimizing the model in the density map. Conversely, Proline and Glycine are almost always marked by weak or broken density, though this can also be used to register the model to the density map as well.
In model optimization, small stretches of residues in the atomic model are adjusted to better fit the density map while maintaining good stereochemistry and enforcing sidechain and mainchain restraints. This process is generally interactive and begins in well-resolved regions such as α-helices, where the pitch of the helix and large, positively charged sidechains provide sufficient points to anchor the placement of atoms. Real-space refinement options found in computational modeling software, like Coot, allow the user to manually adjust atom positions while maintaining realistic biochemical properties.
Additionally, sidechain positions can be optimized using a rotamer search such that the corresponding atoms occupy any visible sidechain density. To achieve optimal rotamer assignment, mainchain atom position may need to be altered; optimization at this point should be done only over a small (3–5) number of amino acids. The entire model optimization procedure is relatively subjective and will likely be iterated until all the atoms are placed in the density map and registered with any visible features.
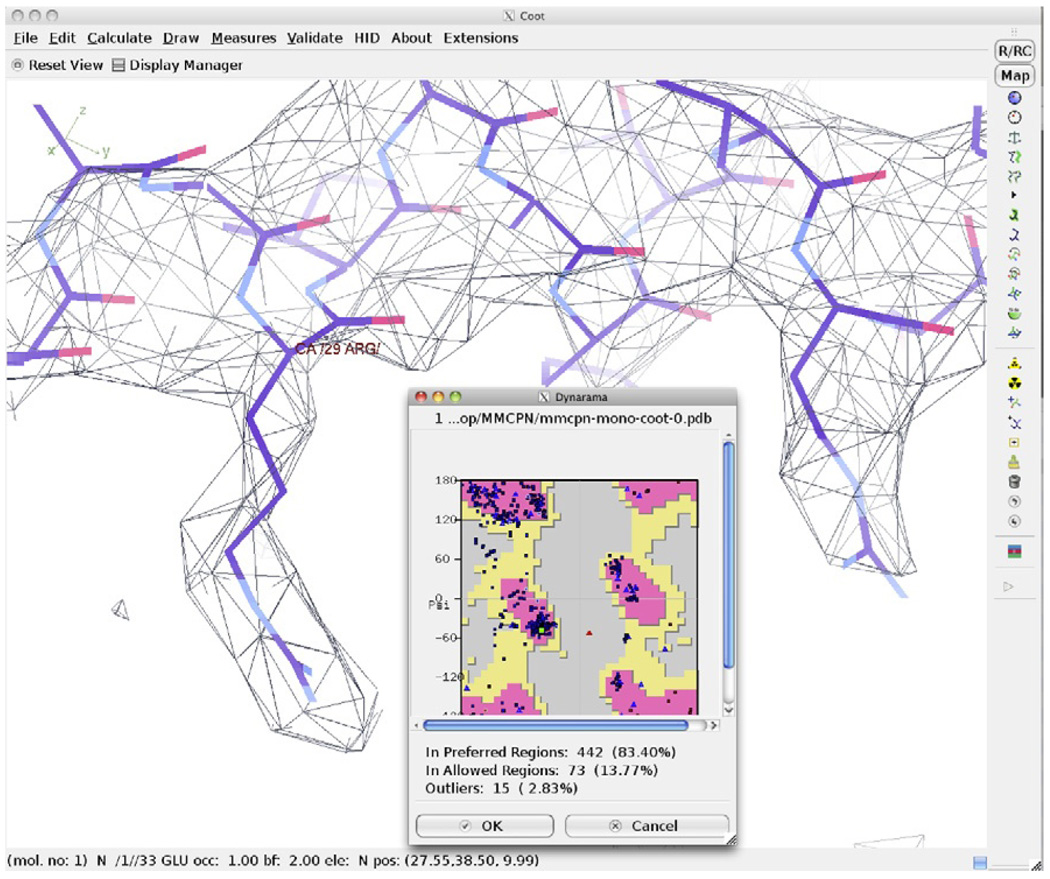
4.18. Monitoring model quality
Results of the previous optimization step can be monitored using a Ramachandran plot (Fig. 1.5). When completed, all amino acids should fall in favorable or acceptable positions on the Ramachandran plot. Several iterations may be required to optimally assign all atoms. Once the final structure has been achieved, the model may be fit back into the entire assembly along with any other components.
Figure 1.5.
Model optimization and validation. Coot can be used to adjust mainchain and sidechain atom positions, optimizing the fit of the atomic model in the density map. A Ramachandran plot is shown overlaid with the map and model of Mm-cpn after optimization.
4.19. Cα to atomic model optimization with Rosetta
The process of model optimization described above is relatively interactive. Alternatively, computational modeling is capable of semiautomatically optimizing an initial model, bypassing much of the user-intensive optimization steps. Rosetta (Bradley et al., 2005) can refine structures constrained by experimental electron density maps by optimizing an all-atom energy function that includes both statistical and chemical potential energy terms. When refining a model, a scoring term that assesses the fit of a model to the density map is simultaneously optimized with Rosetta’s standard energy function (DiMaio et al., 2009).
From a Cα trace, Rosetta’s rebuilding-and-refinement protocol is used to refine the structure. The Rosetta protocol ca_to_allatom infers an all-atom model and performs structure refinement. The protocol generates models by sampling different conformations of individual SSEs. The Cα positions in the starting model guide placement of the initial structure with a user-controllable parameter specifying how far Cαs are allowed to deviate from the starting model.
In the next stage, each atom is explicitly modeled and then evaluated using the complete all-atom energy function. Loops are rebuilt, sidechains are placed on the structure and the entire structure is relaxed with Rosetta’s high-resolution energy function. Throughout the entire process, harmonic constraints keep Cα positions from deviating too far from their initial positions. This protocol generally requires significant sampling, on the order of thousands to tens of thousands of models. In addition, even the best models produced may still have loops and other features outside of the density contours. Thus, it is often necessary to follow this protocol with iterative rebuilding using Rosetta’s loopmodel. A final all-atom optimization with a high-resolution energy function is performed to sample less-common sidechain rotamers and sidechain torsions.
4.20. Model optimization with Rosetta
Not limited to near-atomic resolutions, the aforementioned Rosetta protocols can also be adapted for use with lower resolution structures assuming α-helices can be identified within the density and a clear correspondence can be established with the predicted secondary structure. Similarly, Rosetta’s relax and rebuilding-and-refinement protocols may also be applied to homology models to improve model accuracy and fit into a subnanometer resolution density map.
5. CASE STUDY: MM-CPN
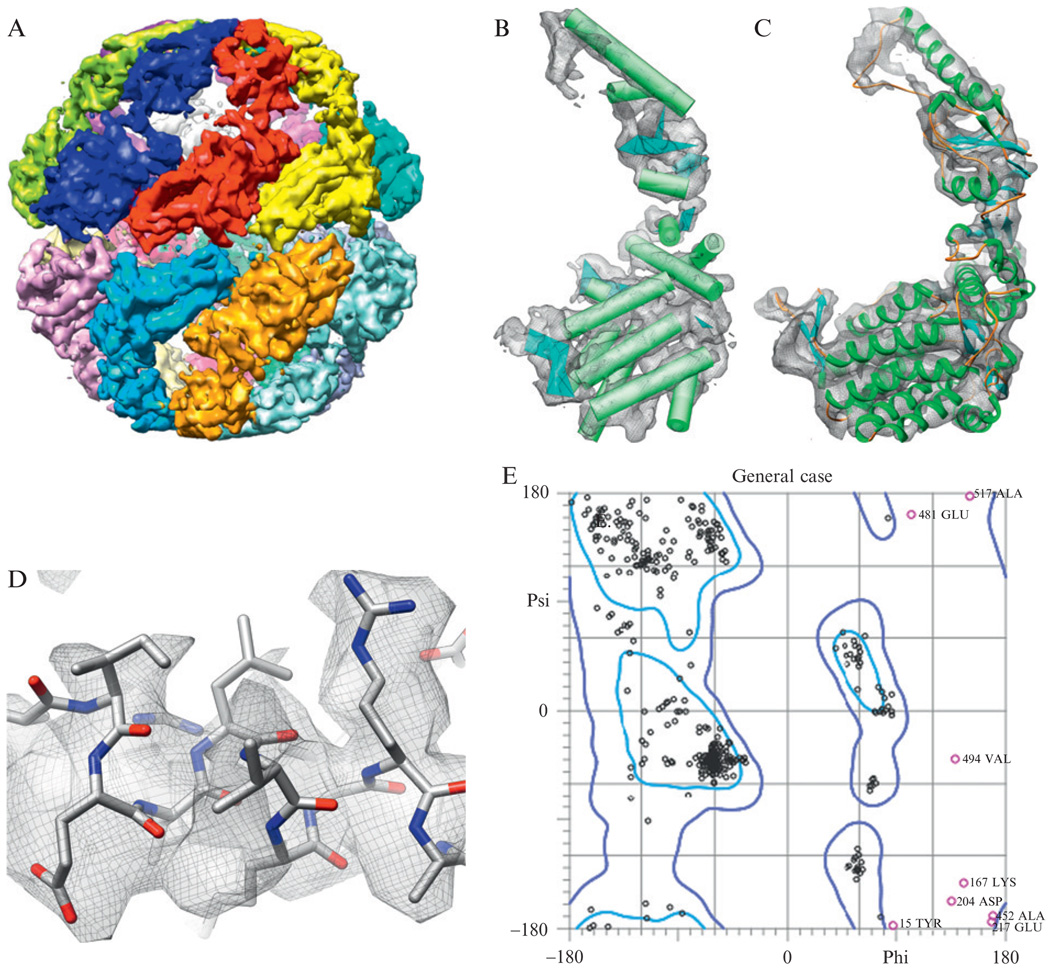
To illustrate the above process, we have chosen to detail the process of constructing an atomic model directly from a cryo-EM density map, as done for the 4.3 Å resolution structure of Mm-cpn (Fig. 1.6A; Zhang et al., 2010). Where appropriate, specific reference is made to the programs, parameters and results obtained for Mm-cpn using this protocol.
Figure 1.6.
Structure of Mm-cpn. (A) The 4.3-Å resolution structure of Mm-cpn is shown (Zhang et al., 2010). (B) Using SSEHunter, the secondary structure elements in the Mm-cpn subunit were identified: α-helices are shown as cylinders and β-strands are shown as planes. (C) Using the de novo modeling approach, an atomic model (residues 1–532) was constructed for one subunit of Mm-cpn. (D) Large, bulky sidechains in the model could be seen in the density. (E) The Ramachandran plot of an Mm-cpn monomer shows greater than 98% of all residues with allowable phi–psi angles.
It should be noted that the level of detail found in the Mm-cpn density map is not required for analyzing and annotating subnanometer resolution protein structure. Rather, Mm-cpn simply provides a convenient and accessible vehicle to describe the variety of tools available for analyzing subnanometer resolution protein structure.
Sixteen subunits were isolated using segment3d from EMAN (segment3d mmcpn.mrc segmented-mmcpn.mrc nseg = 16 split apix = 1.33 sym = d8), which uses a K-means approach to identify subunits (Fig. 1.6A).
For Mm-cpn, a single subunit was padded to a 1283 density map and the origin was reset to zero with EMAN using the following command: proc3d mmcpn1.mrc mmcpn-monomer128.mrc clip = 128,128,128 origin = 0,0,0. Note: mmcpn1.mrc is one the 16 segmented monomers from the prior step.
SSEHunter was used to identify SSEs using the following command in EMAN: ssehunter3.py mmcpn-monomer128.mrc 1.33 4.5 0.4. A single Mm-cpn subunit (543 amino acids per subunit) returned 201 pseudoatoms.
The results were loaded into Chimera along with the density map. Pseudoatoms were represented as spheres and bonds were hidden. The pseudoatoms were colored using Chimera’s “Render by Attribute” option such that the most negative value in the B-factor column (−3) was set to blue, the most positive value (3) was set to red, and zero was set to white.
For a single Mm-cpn subunit, five β-sheets and 17 α-helices were identified and built using SSEBuilder (Fig. 1.6B).
The structure of a related chaperone, the thermosome KS-1 (PDB ID: 1Q3Q; Shomura et al., 2004), was used to construct a homology model for a single Mm-cpn subunit as done in a previous study (Booth et al., 2008).
The homology model for the Mm-cpn subunit was fit to the segmented density map using Foldhunter (Jiang et al., 2001) from EMAN with the following command: foldhunter.py mmcpn-monomer128.mrc mmcpn-homology-model.pdb res = 4.3 apix = 1.33. The resulting transformed PDB model was then loaded into Chimera to verify the fit to the density map and the agreement with the SSEs identified in the previous steps.
The fitted homology model was adjusted in Coot and Gorgon such that the model agreed with the α-helices identified by SSEHunter. The helices required only slight rotations and translations.
Once the helix positions were optimized strands and loops were adjusted to optimally fit the density while maintaining ~3.8 Å Cα–Cα distances. The density skeleton was used to identify potential paths through the density.
In the Mm-cpn homology model, ~30 amino acids were unresolved at the termini. In Coot, Cα atoms were added consecutively to the model starting at the ends of the model until the N-terminus was reached or until the density was not visible (C-terminus). The final model for the Mm-cpn monomer contained residues 1–532 (Fig. 1.6C).
After the addition of missing residues, optimization of the complete Cα model was performed to maximize the occupancy of the model in the subunit density map, while maintaining appropriate distance constraints.
Sixteen copies of the Mm-cpn Cα model were loaded into Chimera, manually moved into a subunit and fit to the density using the “Fit in map” option (Pettersen et al., 2004).
After eight iterations of refinement and fitting (steps 11 and 12) of the entire macromolecular assembly, all clashes were eliminated and model occupancy was optimized. A single PDB file was saved containing all 16 copies of the optimized Mm-cpn Cα model as separate chains.
- From the full Mm-cpn Cα model, the structure factors for the model were calculated and applied to the original density map using EMAN (detailed description of the parameters can be found in the EMAN documentation).
- pdb2mrc mmcpn.pdb mmcpn-simulated.mrc res = 4.3 apix = 1.33 box = 192
- proc3d mmcpn-simulated.mrc junk.mrc calcsf = mmcpn-sf.txt apix = 1.33
- proc3d mmcpn.mrc mmcpn-rescaled.mrc setsf = mmcpn-sf.txt apix = 1.33
- proc3d mmcpn-rescaled.mrc mmcpn-lp.mrc lp = 4.3 apix = 1.33
- proc3d mmcpn-lp.mrc mmcpn-final-map.mrc mult = 0.3
Using the SABBAC web server, the Cα model for one subunit was transformed into an atomic model. The resulting atomic model was loaded into Coot along with the rescaled density map.
Portions of the Mm-cpn atomic model created by SABBAC contained breaks in the polypeptide chain. The “Model/Fit/Refine” tools in Coot were used to move/add residues such that a complete polypeptide chain was constructed.
Once a complete all-atom model for an Mm-cpn subunit was produced, the “Model/Fit/Refine” tools in Coot were used to move the mainchain atoms in the density map. Small stretches of residues, from three amino acids to entire helices, were fit to the density such that (1) the mainchain atoms were encompassed by density, (2) secondary structure constraints were maintained and registered with the density and (3) potential sidechain density for large, bulky sidechains were proximal to residues in the model containing corresponding sidechains. In this refinement round, torsion angles, planar peptide constraints, and Ramachandran constraints were enforced for the appropriate type of secondary structure.
For every amino acid, a rotamer search was performed allowing sidechains to be placed within corresponding sidechain density. In some instances, the “Model/Fit/Refine” tools in Coot were used to adjust the mainchain and sidechain positions to best fit the density (Fig. 1.6D).
The Ramachandran plot in Coot plots the phi–psi angles. Residues falling outside of the acceptable range are plotted in red. Clicking on these outliers, the map and model are recentered in the main Coot display. The steps 17 and 18 were performed over small stretches of residues (3–5 amino acids) containing the outliers until the residues were in acceptable or favorable conformations.
After eight iterations of steps 17–19, the Mm-cpn model had greater than 98% of all residues with acceptable Ramachandran angles (Fig. 1.6E).
Steps 11 and 12 were repeated except with a rescaled density map and the refined atomic model. A final model containing all 16 subunits was saved and deposited in the PDB.
6. DISCUSSION
While the above protocol describes a complete approach to build and refine an atomic model from a cryo-EM density map at subnanometer resolution (Fig. 1.2), individual tools can be, and most often are, used independently. Thus, it is important to know when and what tools are most appropriate for a specific problem.
Limitations in the analysis of cryo-EM density maps, due in large part to resolution, are obvious, though not prohibitive, in describing salient structural features and functions in macromolecular assemblies. The ability to analyze a cryo-EM density map hinges on the map itself; size, complexity, and quality of the density map all play critical roles in annotating structure at any resolution. Even the most experienced scientists may not be able to reliably describe features in poorly resolved regions of density maps. One would not expect to see sidechain density at 9 Å resolution; conversely, the absence of β-strand separation at 3.5 Å resolution may indicate potential problems. As such, the analysis process requires a significant investment in time and understanding the quality of the original 2D data and the reconstructed volume.
Illustrating the dependance on the resolvability of density features, de novo model building is based on establishing a sequence-to-structure correspondence using SSEs, features unique to subnanometer resolutions. This necessitates the presence of clearly identifiable SSEs in the density map, though connecting loops may be ambiguous. Density maps vary in composition, quality, and resolution making it difficult to assign a clear resolution cut-off. Model building may be easier and more reliable at near-atomic resolutions (3.5–5 Å) but still possible at lower resolutions depending on the features that are resolved in the map, as in the case of Hepatitis B (Böttcher et al., 1997; Conway et al., 1997). At higher resolutions, sidechain density can aid in the placement of Cα atoms, thereby increasing the accuracy and reliability of models. Thus, in all cases of interpreting a subnanometer resolution density map, precedence must be given to the observable features in the density map and not the stated resolution.
ACKNOWLEDGMENTS
This work is supported by grants from NIH (P41RR02250, R01GM079429, R01AI0175208) and NSF (IIS-0705644, IIS-0705474). M. R. Baker is supported by a postdoctoral training fellowship from the National Library of Medicine Training Program in Computational Biology and Biomedical Informatics provided by the Keck Center and Gulf Coast Consortia (T15LM007093).
REFERENCES
- Abeysinghe S, Ju T, Baker ML, Chiu W. Shape modeling and matching in identifying 3D protein structures. Comput. Aided Des. 2008;40:708–720. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Bowman BR, Zhou ZH, Quiocho FA, Rixon FJ, Chiu W. Architecture of the herpes simplex virus major capsid protein derived from structural bioinformatics. J. Mol. Biol. 2003;331:447–456. doi: 10.1016/s0022-2836(03)00696-x. [DOI] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Wedemeyer WJ, Rixon FJ, Baker D, Chiu W. Ab initio modeling of the herpesvirus VP26 core domain assessed by CryoEM density. PLoS Comput. Biol. 2006a;2:e146. doi: 10.1371/journal.pcbi.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Yu Z, Chiu W, Bajaj C. Automated segmentation of molecular subunits in electron cryomicroscopy density maps. J. Struct. Biol. 2006b;156:432–441. doi: 10.1016/j.jsb.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PA, Kelley LA, MacCallum RM, Sternberg MJ. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001 Suppl. 5:39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Steven AC. Macromolecular electron microscopy in the era of structural genomics. Trends Biochem. Sci. 2000;25:624–631. doi: 10.1016/s0968-0004(00)01720-5. [DOI] [PubMed] [Google Scholar]
- Booth CR, Meyer AS, Cong Y, Topf M, Sali A, Ludtke SJ, Chiu W, Frydman J. Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat. Struct. Mol. Biol. 2008;15:746–753. doi: 10.1038/nsmb.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Bradley P, Malmström L, Qian B, Schonbrun J, Chivian D, Kim DE, Meiler J, Misura KM, Baker D. Free modeling with Rosetta in CASP6. Proteins. 2005;61 Suppl 7:128–134. doi: 10.1002/prot.20729. [DOI] [PubMed] [Google Scholar]
- Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Ludtke SJ. Single particle analysis at high resolution. Methods Enzymol. 2010;482:211–236. doi: 10.1016/S0076-6879(10)82009-9. [DOI] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. 4.0-Å resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. USA. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J. Mol. Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernández JJ, Luque D, Castón JR, Carrascosa JL. Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 2008;164:170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu. Rev. Biophys. Biomol. Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Unwin NR. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14(33–8):27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: Computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Ju T, Baker ML, Chiu W. Computing a family of skeletons of volumetric models for shape description. Comput. Aided Des. 2007;39:352–360. doi: 10.1016/j.cad.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y. Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature. 1997;389:206–211. doi: 10.1038/38323. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- Kong Y, Ma J. A structural-informatics approach for mining beta-sheets: Locating sheets in intermediate-resolution density maps. J. Mol. Biol. 2003;332:399–413. doi: 10.1016/s0022-2836(03)00859-3. [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhang X, Baker TS, Ma J. A structural-informatics approach for tracing beta-sheets: Building pseudo-C(alpha) traces for beta-strands in intermediate-resolution density maps. J. Mol. Biol. 2004;339:117–130. doi: 10.1016/j.jmb.2004.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. Rotavirus architecture at subnanometer resolution. J. Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Serysheva II, Hamilton SL, Chiu W. The pore structure of the closed RyR1 channel. Structure. 2005;13:1203–1211. doi: 10.1016/j.str.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Maupetit J, Gautier R, Tufféry P. SABBAC: Online Structural Alphabet-based protein BackBone reconstruction from Alpha-Carbon trace. Nucleic Acids Res. 2006;34:W147–W151. doi: 10.1093/nar/gkl289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mizuguchi K, Go N. Comparison of spatial arrangements of secondary structural elements in proteins. Protein Eng. 1995;8:353–362. doi: 10.1093/protein/8.4.353. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- Roseman AM. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1332–1340. doi: 10.1107/s0907444900010908. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusu M, Birmanns S, Wriggers W. Biomolecular pleiomorphism probed by spatial interpolation of coarse models. Bioinformatics. 2008;24:2460–2466. doi: 10.1093/bioinformatics/btn461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva II, Hamilton SL, Chiu W, Ludtke SJ. Structure of Ca2+ release channel at 14 Å resolution. J. Mol. Biol. 2005;345:427–431. doi: 10.1016/j.jmb.2004.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl. Acad. Sci. USA. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, Miki K. Crystal structures of the group II chaperonin from Thermococcus strain KS-1: Steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J. Mol. Biol. 2004;335:1265–1278. doi: 10.1016/j.jmb.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Siebert X, Navaza J. UROX 2.0: An interactive tool for fitting atomic models into electron-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 2009;65:651–658. doi: 10.1107/S0907444909008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Navaza J, Sanejouand YH. NORMA: A tool for flexible fitting of high-resolution protein structures into low-resolution electron-microscopy-derived density maps. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1098–1100. doi: 10.1107/S090744490602244X. [DOI] [PubMed] [Google Scholar]
- Tama F, Miyashita O, Brooks CL. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J. Struct. Biol. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Tan RK, Devkota B, Harvey SC. YUP.SCX: Coaxing atomic models into medium resolution electron density maps. J. Struct. Biol. 2008;163:163–174. doi: 10.1016/j.jsb.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryomicroscopy. J. Struct. Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J. Mol. Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N. A novel three-dimensional variant of the watershed transform for segmentation of electron density maps. J. Struct. Biol. 2002;138:123–129. doi: 10.1016/s1047-8477(02)00009-6. [DOI] [PubMed] [Google Scholar]
- Volkmann N. Confidence intervals for fitting of atomic models into low-resolution densities. Acta Crystallogr. D Biol. Crystallogr. 2009;65:679–689. doi: 10.1107/S0907444909012876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J. Struct. Biol. 1999;125:176–184. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol. Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Baker ML, Jiang W, Dougherty M, Jakana J, Dong G, Lu G, Chiu W. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat. Struct. Biol. 2001;8:868–873. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]
- Zhu J, Cheng L, Fang Q, Zhou ZH, Honig B. Building and refining protein models within cryo-electron microscopy density maps based on homology modeling and multiscale structure refinement. J. Mol. Biol. 2010;397:835–851. doi: 10.1016/j.jmb.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]