Abstract
The CDKN2A gene that encodes the cell cycle inhibitor p16 shows mutations in many but not all 9p21-linked melanoma families. Most Dutch melanoma families segregate for a unique founder mutation (p16-Leiden), encoding a truncated nonfunctional p16 protein. The highly variable risk for p16-Leiden carriers to develop melanoma suggests a role for other genetic and/or environmental factors. We hypothesized that a 9p21 gene other than CDKN2A may be relevant in the remaining 9p21-linked melanoma families without p16 mutations but may also act as a risk modifier in p16-Leiden carriers. Haplotype analysis for 9p21 was performed using microsatellite markers in six p16-Leiden families originating from a founder population. p16-Leiden carriers in two families shared an unexpectedly large founder haplotype (∼20-cM) around CDKN2A, mostly in proximal direction. Melanoma-positive p16-Leiden carriers from these families showed this extensive proximal haplotype compared with melanoma-negative p16-Leiden carriers from the same families. Additional p16-Leiden families less heavily affected with melanoma showed shorter haplotypes sharing, excluding the region proximally of CDKN2A. The presence of a gene involved in melanoma susceptibility proximal of CDKN2A is corroborated by somatic deletions of 9p in tumors, which frequently do not include CDKN2A but a more proximal chromosomal area instead. Our results provide a candidate region for further gene mapping in p16-negative 9p21-linked melanoma families and guide the search for risk modifiers in melanoma development.
The familial atypical multiple mole melanoma (FAMMM) syndrome is characterized by the familial occurrence of melanoma and atypical nevi. A locus responsible for at least part of the melanoma susceptibility in FAMMM families has been mapped to 9p21 (Cannon-Albright et al. 1992). Melanoma kindreds from all over the world yield further statistical evidence for linkage to this region and display shared 9p21 haplotypes (Gruis et al. 1995b; Harland et al. 1997; Pollock et al. 1998). Deletion studies in tumor-derived cell lines resulted in the cloning from 9p21 of the MTS1 gene, which is now generally known as CDKN2A (Kamb et al. 1994; Nobori et al. 1994). Its gene product had earlier been identified as the cyclin-dependent kinase inhibitor p16 (Serrano et al. 1993). Mutations in the coding sequence of CDKN2A have been found in 40%–50% of melanoma kindreds that show linkage to 9p21 (Hussussian 1994; Walker et al. 1995). Although there is little doubt that these CDKN2A mutations are responsible for increased melanoma susceptibility, the lack of CDKN2A mutations in many of the melanoma kindreds linked to 9p21 suggests that another gene in this region may act either directly as an independent risk factor for melanoma or indirectly by influencing the expression of CDKN2A.
Somatic deletions in tumors provide further evidence for the presence of a second tumor-related gene in 9p21. Puig and coworkers (1995) found large 9p deletions in 25 of 54 primary and metastatic melanomas. Surprisingly, 4 of those 25 deletions did not include p16 itself but were located more proximally instead. Also, 100% loss of heterozygosity in the 9p region was observed in squamous cell carcinomas of the lung (Wiest et al. 1997). About half of these tumors were shown to be homozygous for a microdeletion within the area of loss of heterozygosity. Those microdeletions clustered approximately equally in two areas, one of which includes p16, whereas the other more proximal cluster may reveal the location of another tumor suppressor gene.
We hypothesized that this tentative second tumor-related gene in 9p21 may also act as a modifier of the melanoma risk conveyed by known CDKN2A mutations. To identify genetic modifiers for a known, “primary” susceptibility gene, one would ideally need to study a large group of carriers of a single mutation in that primary gene. Dutch FAMMM families provide a unique opportunity for such studies: a founder mutation defined by a 19-bp deletion in exon 2 of the CDKN2A gene (p16-Leiden) segregates in most Dutch FAMMM families (Gruis et al. 1995b). The 36% cumulative incidence for melanoma in p16-Leiden carriers illustrates the high melanoma risk associated with this mutation but also suggests that environmental and/or genetic factors act as risk modifiers. Here, we present the results of an analysis of haplotype sharing between melanoma patients and unaffected carriers from six extended FAMMM families segregating for p16-Leiden.
RESULTS
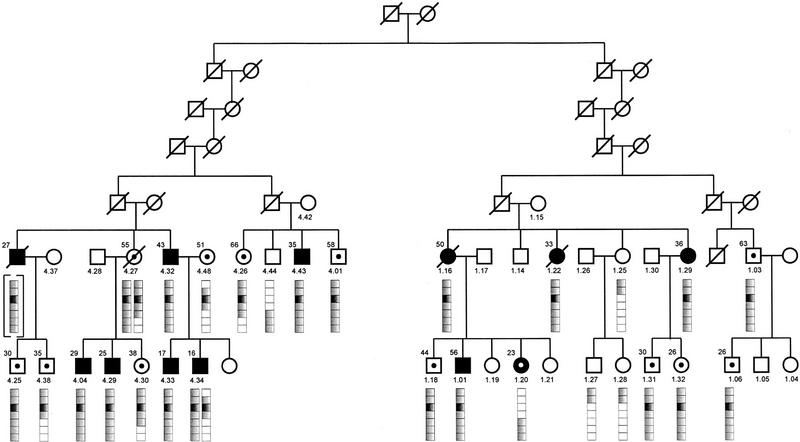
All available members of all six families from town K were tested for the presence of p16-Leiden. Figure 1 shows the segregation of p16-Leiden in families 1 and 4 and their connection through a common founder. The most recent common ancestor was identified in the late 18th century (1775), six generations before the current generation of carriers, but at least one additional connection exists between the families, two generations earlier (1707) (Hille et al. 1998). In all p16-Leiden carriers from these families, haplotypes were constructed for all genotyped microsatellite markers, and the extent of haplotype sharing was determined by direct haplotype comparison. A complicating factor in the analysis of family 4 is the fact that p16-Leiden (and part of the surrounding haplotype) was introduced two additional times into this family, via 4.48, and via the mother of 4.27, to be transmitted subsequently to 4.30. As the exact connection between these married-ins and the established common founder is not known, their haplotypes were not included in this first comparison. Of the 21 p16-Leiden carriers descending from the established common founder, 11 had developed at least one melanoma (mean age at first diagnosis, 33.4 years). The remaining ten carriers are all undergoing regular screening. The mean age of these subjects at their most recent screening was 43.3 years. A haplotype extending over at least 20 cM was found to be shared by the 11 p16-Leiden-carrying melanoma patients in families 1 and 4, who are separated by a total of 21 meioses. The probability of such extensive haplotype sharing was calculated to be ≅8%. When all unaffected carriers in these families are included in the comparison, two recombinations are observed that break up the shared haplotype, in subjects 4.38 and 4.26. As a result, the length of the segment that all unaffected carriers share with their affected carrier relatives is only 8 cM, whereas there are 10 meioses that separate these unaffected carriers from their nearest affected relative or the nearest relative that must have carried the full 20-cM haplotype. The probability that in 10 meioses a haplotype of at least 8 cM is preserved around a shared gene is ≅81%.
Figure 1.
Inheritance of the p16-Leiden allele in families 1 and 4 from a common ancestor in the late eighteenth century. Melanoma cases are shown in black. The bar underneath the family members depicts the seven-marker founder haplotype dispersed over ∼20 cM. The marker that migrates with p16-Leiden is indicated in black, and the markers that have lost the founder allele owing to recombinations in the families and in the population are depicted in white. All numbered members have been haplotyped, although only the p16-Leiden-related haplotypes are visualized. Melanoma-free carriers of the p16-Leiden allele are indicated by a black dot, whereas melanoma patient 1.20, who is no carrier of the p16-Leiden allele, is depicted by a white dot.
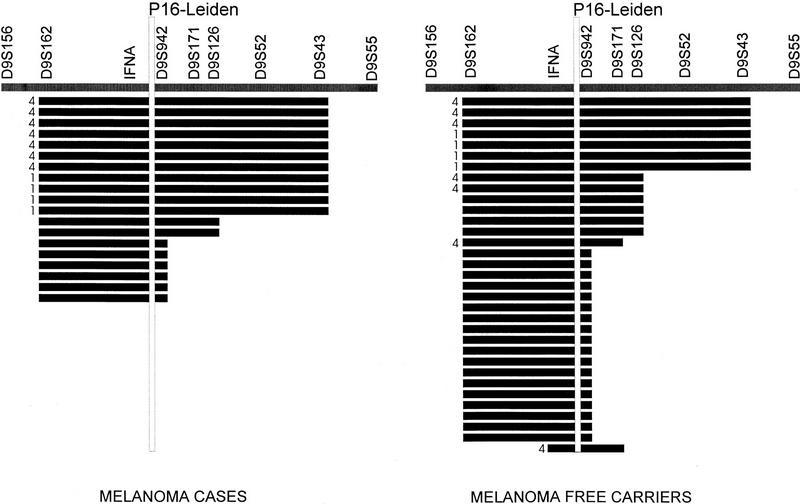
In addition we performed haplotype analysis in the four remaining p16-Leiden families (2, 3, 15, and 17) that originate from the same community as families 1 and 4 but for whom we have not been able to identify a common founder. In these four additional families, we identified 32 carriers of p16-Leiden, 8 of whom had developed melanoma. Families 2, 3, and 15 display a 5-cM remnant of the founder haplotype seen in families 1 and 4, whereas family 17 revealed haplotype sharing over a 8-cM region. Figure 2 shows all observed haplotypes in p16-Leiden carriers from all six families. A comparison between the haplotypes seen in affected carriers and those in carriers without melanoma revealed that sharing of large haplotypes proximally of p16 was concentrated in melanoma patients, whereas their unaffected carrier relatives frequently showed very little proximal sharing of the ancestral haplotype. Although it is still possible that some of the melanoma-free carriers may develop tumors, their average age of 51 years is well beyond the average age at first diagnosis (37 years) in the melanoma patients in their families. When shared haplotypes are classified as either long (including a shared allele at D9S43) or short, a comparison between melanoma (11 long haplotypes out of 19) versus unaffected carriers (7 long haplotypes out of 33) indicates a significant difference (χ2 = 7.17, P < 0.01). For a more accurate assessment of the significance of these findings, knowledge of the exact relationship between these families would be required, but this is unavailable at the moment.
Figure 2.
Haplotype sharing in the FAMMM families 1, 2, 3, 4, 15, and 17 that all originate from the same town K. Haplotypes derived from families 1 and 4 are indicated by the family number. The other haplotypes are present in families 2, 3, 15, and 17. The markers D9S156 to D9S55 cover an area of 29 cM, whereas the maximum sharing reaches from D9S162 to D9S43 and covers an area of ∼20 cM. The distances between the markers D9S162-D9S171/D9S126 and the markers D9S162-D9S942 are, respectively, 8 cM and 5 cM.
DISCUSSION
Although mutations in CDKN2A are responsible for an increased melanoma susceptibility in a large proportion of melanoma kindreds, an almost equally large group of melanoma families has been published that show cosegregating haplotypes and/or interesting lod scores for 9p21 without detectable abnormalities in the coding sequence of this gene (Hussussian et al. 1994; Walker et al. 1995). Expression mutations and germ-line deletions have occasionally been reported for CDKN2A, but it seems unlikely that such mechanisms would explain all remaining 9p21-linked families (Rizos et al. 1997; Bahuau et al. 1998). Therefore, the possibility that 9p21 harbors one or more other melanoma susceptibility genes in addition to CDKN2A should be considered.
Somatic deletions observed in melanomas and lung tumors provide independent indirect evidence for the presence of a second tumor-related gene on 9p. Four of the twenty-five observed somatic deletions do not include CDKN2A but instead concentrate in a more proximal region of <5 cM including D9S126 (Puig et al. 1995; Wiest et al. 1997). We hypothesized that variations in this tentative second tumor-related gene in 9p21 may act as risk modifiers for known mutations in CDKN2A. The majority of Dutch FAMMM families segregate for a single founder mutation in CDKN2A, a 19-bp deletion in exon 2 known as p16-Leiden. Linkage analysis in our families is consistent with the presence of a linked modifier because maximum 9p21 lod scores in an affected-only analysis are found proximal of p16 with the markers D9S171 and D9S216 (Gruis et al.1995b). We performed haplotype analysis in nine p16-Leiden families to search for systematic differences between p16-Leiden carriers with melanoma and those carriers who have remained free of melanoma. In almost all carriers, unambiguous haplotype reconstruction was possible owing to the availability of many siblings and additional relatives in these families. Therefore, the population frequency of the marker alleles that constitute the shared haplotype has little impact on our conclusions. Given that the shared haplotype in the carriers is composed of a long string of markers, it is highly unlikely that the observed sharing represents identity by state (IBS) rather than identity by descent (IBD) (Schork et al. 1998). Two distantly related families showed large haplotype sharing for 9p21 among p16-Leiden carriers. The haplotype sharing in 11 p16-positive melanoma patients from these families extended over at least 20 cM, whereas on average, one would expect these subjects to share only 8 cM around a shared gene given their degree of relationship. In contrast, nine p16-Leiden carriers free of melanoma shared only 8 cM with their affected relatives, whereas based on their degree of relationship one would expect, on average, to find a shared segment of 17 cM. In addition to this probabilistic argument, the asymmetry of the haplotype, with three-fourths of the sharing located proximal of p16 suggests that the excess haplotype sharing among patients is not simply owing to chance but that the shared haplotype contains a second locus, proximal to p16, that increases the risk for melanoma in p16-Leiden carriers. Though scarce in such a small genetic region, the observable recombinants appear to be in line with our hypothesis. The occurrence of melanoma in subject 1.20, who did not inherit p16-Leiden but only the proximal part of the founder haplotype, suggests that the tentative modifier gene, which would still be present on this part of the haplotype, may also act as an independent risk factor, although it remains quite possible that the melanoma in this individual is purely sporadic.
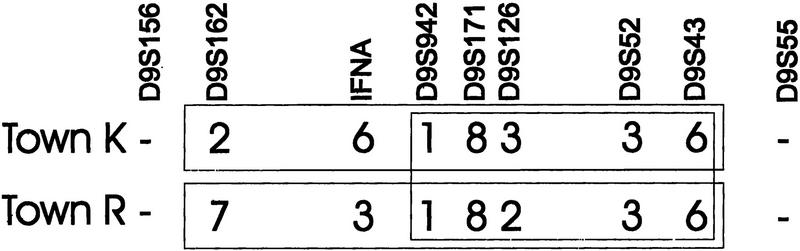
Additional p16-Leiden families were either smaller than families 1 and 4, or their exact connection to these families was unclear. The haplotype sharing between these families and families 1 and 4 is less extensive, but their shared geographic origin and the presence of the p16 deletion in a shared core haplotype are still strongly indicative of shared ancestry. As the exact degree of relationship between these families remains largely uncertain, it is not possible to determine the exact probability that the observed sharing may be owing to chance. A simple statistical comparison between carriers with and without melanoma, ignoring family relationships, yields significant results (P < 0.01) indicating that long shared haplotypes are preferentially found in carriers with melanoma. This is reflected by the summarized results in Figure 2. These joint results for all families from town K are again supportive of more sharing of the proximal part of the haplotype in p16-Leiden carriers with melanoma. This conclusion may also be phrased slightly differently: p16-Leiden-positive individuals have a higher risk to develop melanoma when they share the area between D9S126 and D9S43 with their affected relatives. The three families from town R yield results that are consistent with this conclusion: Melanoma patients from these families share between them a haplotype that extends from D9S162 to D9S43, whereas the overlap between the shared haplotype in K and the shared haplotype in R is limited to the area between CDKN2B and D9S43 (Fig. 3).
Figure 3.
Haplotype sharing is similar in size in families originating from town R and K. Because the families from R all present the same haplotype, we propose that two early events, a recombination distal of p16 and a marker mutation at D9S126, differentiate town R from K.
It may be argued that the overall risk to develop melanoma in p16-Leiden carriers will be overestimated in our study design as a consequence of ascertainment bias: p16-Leiden families without melanoma will not come to our attention. Although this is certainly true, the objective of this study was not to accurately determine the melanoma risk for p16-Leiden carriers but, rather, to determine whether there is a systematic difference in haplotype sharing between melanoma-positive and negative p16-Leiden carriers. It is difficult to see how ascertainment bias might influence the length of shared haplotypes, as the length of shared haplotypes is unknown to the investigator at the time of ascertainment.
Our results, combined with the results of somatic deletion mapping in tumors, delineate an area proximal to and including D9S126 as the most likely location for a second tumor-related gene in 9p21. Two possible candidate genes from 9p21 are CDKN2B and p14ARF (Kamb et al. 1994; Stone et al. 1995). CDKN2B encodes p15, which is functionally and structurally homologous to p16 (Hannon and Beach 1994). p14ARF is transcribed from exon 1β and exons 2 and 3 from CDKN2A but is translated in a different reading frame. Though being structurally and functionally different from p15 and p16, p14ARF can induce cell cycle arrest by interacting with p53 control (Kamijo et al. 1997, 1998; Stott et al. 1998). The extended tumor spectrum of families displaying germ-line deletions encompassing p16 and p15 or p16, p15, and p14ARF clearly adds a function to these contiguous tumor suppressor genes. However, both CDKN2B and exon 1β of p14ARF have never been found to be mutated in melanoma kindreds, and this, together with the fact that they are located somewhat more distally than our most likely area, effectively rules out these genes as candidates for the tentative second melanoma gene in 9p21. Our results should not be taken as conclusive evidence for the existence of such a gene in this region, but they may nevertheless provide a very useful hint for the further analysis of 9p21-linked families without CDKN2B mutations. Also, these results have stimulated us to initiate prospective studies in p16-Leiden carriers to determine more systematically the contribution of various possible risk factors to melanoma development.
METHODS
Family Material
Nine extended FAMMM families were ascertained through the Leiden Pigmented Lesion Clinic. Six pedigrees (1, 2, 3, 4, 15, and 17) originate from town K, and three pedigrees (6, 10, and 19) originate from town R. The population of both towns has been sedentary for several generations owing to their orthodox religion and strong family ties. Six of these families (1, 3, 4, 6, 10, and 19) were included in our previous linkage studies in view of their relatively high incidence of melanoma. Complete pedigree data and histopathological findings on families 1, 3, 4, 6, 10, and 19 have been presented elsewhere (Bergman et al. 1994).
DNA Analysis
Genomic DNA from FAMMM family members was isolated from peripheral blood leukocytes using the method of Miller (Miller et al. 1988). The families were typed for the microsatellite markers D9S156, D9S162, IFNA, D9S942, D9S171, D9S126, D9S52, D9S43, and D9S55 as described previously (Gruis et al. 1993). Genetic distances between relevant markers as obtained from reports by the Galton Lab map of Centre d’Etude du Polymorphisme Humain (CEPH) consortium data presented by Attwood et al. at the 1996 chromosome 9 workshop (www.ash.gene.ucl.ac.uk/chrg/scwg6/index.shtml) and from the genetic map from the Marshfield World Wide Web site (www.marshmed.org/genetics) are D9S162–D9S43, 20 cM; D9S162–D9S171, 8 cM; D9S162–D9S126, 10 cM. Haplotype reconstruction was performed jointly for the nine microsatellite markers and p16-Leiden, given the above map order and a location of p16-Leiden between IFNA and D9S942. The 19-bp germ-line deletion was detected after PCR with the primers A1, 5′-AGCCTTCCTTTCCGTCATGC-3′, and A2, 5′-ACCAGCGTGTCCAGGAAG-3′, after electrophoresis through a 2.5% agarose gel plus ethidium bromide (Gruis et al. 1995a).
Calculations
The probability that two distant relatives separated by n meioses share an area of at least x Morgan around a shared disease gene was calculated as by Te Meerman et al. (1995), whereas the double integration presented by those workers was replaced by an approximation via P(area ≥ x) = (1 + xn) e−xn.
Acknowledgments
We thank Leny van Mourik for technical assistance in drawing the pedigree. This study was supported by the Dutch Cancer Foundation (94-758). N.A.G. is a fellow of the Royal Netherlands Academy of Arts and Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gruis@ruly46.medfac.Leidenuniv.nl; FAX 31 71 5276075.
REFERENCES
- Bahuau M, Vidaud D, Jenkins RB, Bieche I, Kimmel DW, Assouline B, Smith JS, Alderete B, Cayuela JM, Harpey JP, Caille B, Vidaud M. Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res. 1998;58:2298–2303. [PubMed] [Google Scholar]
- Bergman W, Gruis NA, Sandkuijl LA, Frants RR. Genetics of seven Dutch familial atypical multiple mole-melanoma syndrome families: A review of linkage results including chromosomes 1 and 9. J Invest Dermatol. 1994;103:122S–125S. doi: 10.1111/1523-1747.ep12399430. [DOI] [PubMed] [Google Scholar]
- Cannon-Albright LA, Goldgar DE, Meyer LJ, Lewis CM, Anderson DE, Fountain JW, Hegi ME, Wiseman RW, Petty EM, Bale AE, et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13- p22. Science. 1992;258:1148–1152. doi: 10.1126/science.1439824. [DOI] [PubMed] [Google Scholar]
- Gruis NA, Sandkuijl LA, Weber JL, van der Zee A, Borgstein AM, Bergman W, Frants RR. Linkage analysis in Dutch familial atypical multiple mole-melanoma (FAMMM) syndrome families. Effect of naevus count. Melanoma Res. 1993;3:271–277. [PubMed] [Google Scholar]
- Gruis NA, Sandkuijl LA, van der Velden PA, Bergman W, Frants RR. CDKN2 explains part of the clinical phenotype in Dutch familial atypical multiple-mole melanoma (FAMMM) syndrome families. Melanoma Res. 1995a;5:169–177. doi: 10.1097/00008390-199506000-00005. [DOI] [PubMed] [Google Scholar]
- Gruis NA, van der Velden PA, Sandkuijl LA, Prins DE, Weaver-Feldhaus J, Kamb A, Bergman W, Frants RR. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet. 1995b;10:351–353. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Harland M, Meloni R, Gruis N, Pinney E, Brookes S, Spurr NK, Frischauf AM, Bataille V, Peters G, Cuzick J, Selby P, Bishop DT, Bishop JN. Germline mutations of the CDKN2 gene in UK melanoma families. Hum Mol Genet. 1997;6:2061–2067. doi: 10.1093/hmg/6.12.2061. [DOI] [PubMed] [Google Scholar]
- Hille ET, van Duijn E, Gruis NA, Rosendaal FR, Bergman W, Vandenbroucke JP. Excess cancer mortality in six Dutch pedigrees with the familial atypical multiple mole-melanoma syndrome from 1830 to 1994. J Invest Dermatol. 1998;110:788–792. doi: 10.1046/j.1523-1747.1998.00185.x. [DOI] [PubMed] [Google Scholar]
- Hussussian CJ, Streuwing JP, Goldstein AM, Higgins PAT, Ally DS, Sheanan MD, Clark WH, Jr, Tucker MA, Dracopoli NC. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Spurr N, Bishop T, Newton-Bishop J, Gruis N, van der Velden PA, Goldstein AM, Tucker MA, Foulkes WD, Barnhill R, Haber D, Fountain J, Hayward NK. Haplotype analysis of two recurrent CDKN2A mutations in ten melanoma families: Evidence for common founders and independent mutations. Hum Mutat. 1998;11:424–431. doi: 10.1002/(SICI)1098-1004(1998)11:6<424::AID-HUMU2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Puig S, Ruiz A, Lazaro C, Castel T, Lynch M, Palou J, Vilalta A, Weissenbach J, Mascaro JM, Estivill X. Chromosome 9p deletions in cutaneous malignant melanoma tumors: The minimal deleted region involves markers outside the p16 (CDKN2) gene. Am J Hum Genet. 1995;57:395–402. [PMC free article] [PubMed] [Google Scholar]
- Rizos H, Becker TM, Holland EA, Kefford RF, Mann GJ. Differential expression of p16INK4a and p16beta transcripts in B-lymphoblastoid cells from members of hereditary melanoma families without CDKN2A exon mutations. Oncogene. 1997;15:515–523. doi: 10.1038/sj.onc.1201217. [DOI] [PubMed] [Google Scholar]
- Schork NJ, Thiel B, St. Jean P. Linkage analysis, kinship, and the short-term evolution of chromosomes. J Exp Zool. 1998;282:133–149. [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Stone S, Jiang P, Dayananth P, Tavtigian SV, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Meerman GJ, Van der Meulen MA, Sandkuijl LA. Perspectives of identity by descent (IBD) mapping in founder populations. Clin Exp Allergy (Suppl. 2) 1995;25:97–102. doi: 10.1111/j.1365-2222.1995.tb00433.x. [DOI] [PubMed] [Google Scholar]
- Walker GJ, Hussussian CJ, Flores JF, Glendening JM, Haluska FG, Dracropoli NC, Hayward NK, Fountain JW. Mutations of the CDKN2/p16INK4 gene in Australian melanoma kindreds. Hum Mol Genet. 1995;4:1845–1852. doi: 10.1093/hmg/4.10.1845. [DOI] [PubMed] [Google Scholar]
- Wiest JS, Franklin WA, Otstot JT, Forbey K, Varella-Garcia M, Rao K, Drabkin H, Gemmill R, Ahrent S, Sidransky D, Saccomanno G, Fountain JW, Anderson MW. Identification of a novel region of homozygous deletion on chromosome 9p in squamous cell carcinoma of the lung: the location of a putative tumor suppressor gene. Cancer Res. 1997;57:1–6. [PubMed] [Google Scholar]