Abstract
A primary male autosomal linkage map of the domestic horse (Equus caballus) has been developed by segregation analysis of 140 genetic markers within eight half-sib families. The family material comprised four Standardbred trotters and four Icelandic horses, with a total of 263 offspring. The marker set included 121 microsatellite markers, eight protein polymorphisms, five RFLPs, three blood group polymorphisms, two PCR–RFLPs, and one single strand conformation polymorphism (SSCP). One hundred markers were arranged into 25 linkage groups, 22 of which could be assigned physically to 18 different chromosomes (ECA1, ECA2, ECA3, ECA4, ECA5, ECA6, ECA7, ECA9, ECA10, ECA11, ECA13, ECA15, ECA16, ECA18, ECA19, ECA21, ECA22, and ECA30). The average distance between linked markers was 12.6 cM and the longest linkage group measured 103 cM. The total map distance contained within linkage groups was 679 cM. If the distances covered outside the ends of linkage groups and by unlinked markers were included, it was estimated that the marker set covered at least 1500 cM, that is, at least 50% of the genome. A comparison of the relationship between genetic and physical distances in anchored linkage groups gave ratios of 0.5–0.8 cM per Mb of DNA. This would suggest that the total male recombinational distance in the horse is 2000 cM; this value is lower than that suggested by chiasma counts. The present map should provide an important framework for future genome mapping in the horse.
Following a general trend in modern genetics, domestic animal genome analysis has witnessed some considerable achievements over the last 5–10 years, achievements that not least can be attributed to the advent and introduction of new DNA marker technology. Significantly, reports are accumulating on the identification of genes or chromosomal regions that influence quantitative or qualitative traits of economic, agricultural, or even medical importance. Prime examples include malignant hyperthermia (MacLennan et al. 1990) and fatness (Andersson et al. 1994) in pigs, milk production (Georges et al. 1995) and double muscling (Charlier et al. 1995) in cattle, and fecundity in sheep (Montgomery et al. 1994). Identification of such loci has been mediated by the initial construction of genome maps based on genetic linkage data or physical assignments of markers to chromosomes. Primary or in some cases even second-generation linkage maps are now available for most domestic animal species, for example, cattle (Barendse et al. 1997; Kappes et al. 1997), pig (Marklund et al. 1996a; Rohrer et al. 1996), sheep (Crawford et al. 1995), goat (Vaiman et al. 1996), and dog (Mellersh et al. 1997).
In terms of genetic map information as well as of other aspects of genome analysis, one main domestic animal species has been clearly lagging behind the others—the horse. There is no primary linkage map reported for the horse. The last compilation of incidental cases of observed linkages, mainly involving protein polymorphisms, listed only 17–20 loci, few of which had been assigned to any of the 31 equine autosomes (Sandberg and Andersson 1992). The first equine microsatellite markers were derived some five years ago (Ellegren et al. 1992), but a large set of markers has not become available until recently. A few linkage groups derived from the preliminary analyses of limited numbers of microsatellites are known (e.g., Marklund et al. 1994), and initial efforts to integrate genetic and physical map data have been made (Breen et al. 1997; Godard et al. 1997). The reasons for the slow progress in equine gene mapping are several and relate mainly to the economy and practice of horse breeding (but potentially also to the fact that the horse in some cases is seen more as a companion species than as livestock). Horse breeding is associated with extensive costs and demands (personnel, stables, grazing areas), and since a large number (>100–200) of animals might be required for the construction of linkage maps, suitable pedigree might be difficult to establish. Moreover, as the horse has a gestation period of 11 months and generally produces a single foal per mating, designing large full-sib families is problematic. Thus, in practice, equine linkage mapping has to rely on family material available for other reasons than genetic studies, one typical example being large (two-generation) half-sib families present for popular stallions. Generally, such materials represent within-breed matings, that is, matings not associated with an optimized marker heterozygosity.
In an initiative to extend the knowledge of the equine genome, we have established a reference pedigree for gene mapping and we present here the construction of the first preliminary, male autosomal linkage map of the horse. The map was derived from the genotyping of 140 polymorphic markers, 100 of which were arranged into 25 linkage groups on 18 different autosomes.
RESULTS
Genetic Markers
We designed an equine reference pedigree for the purpose of gene mapping, involving eight half-sib families (four Standardbred and four Icelandic Horse stallions) and a total of 263 offspring. The pedigree was genotyped with 140 polymorphic markers, 121 of which were PCR-based microsatellites and about one-third of which had been assigned physically to chromosomes by in situ hybridization (Table 1). The markers included five new, previously undescribed restriction fragment length polymorphisms (RFLPs) that were detected using human or porcine cDNA probes, FUCA1, GLUT1, LPL, MYL1,3, and TYR; data for these polymorphisms are presented in Table 2.
Table 1.
Description of Markers Used in this Study
| Locus | Description | Primer sequences | Hoa | Phys. assign. | Reference | |
|---|---|---|---|---|---|---|
| Genes | ||||||
| Blood groups | ||||||
| EAA | erythrocyte antigen A | 50 | Bailey et al. (1979) | |||
| EAD | erythrocyte antigen D | 37.5 | Sandberg (1973) | |||
| EAQ | erythrocyte antigen Q | 37.5 | Bowling et al. (1985) | |||
| Microsatellite | ||||||
| IGF2 | insulin-like | GGAGCACAGAACATGAAAAC | 87.5 | 12q13 | H. Ellegren (unpubl.) | |
| growth factor 2 | AAATTTAATTGGCACAAACC | Raudsepp et al. (1997) | ||||
| PCR-RFLPs | ||||||
| MC1R | melanocyte-stimulating hormone receptor 1 | GATGGATCCTTCTGGGCTCCCTCAACTC | 37.5 | 3p12 | Marklund et al. (1996b) | |
| GTAGTAGGCGATGAAGAGCGTGCT | T. Raudsepp (unpubl.) | |||||
| KIT | mast cell growth factor receptor | ATTTATTCCAACTTAGCGAACTGCAGC | 25 | 3q21 | Marklund et al. (1996b) | |
| TCAGACATCTTCGTGGACAAGCAGAGG | ||||||
| Lear et al. (1997) | ||||||
| Proteins | ||||||
| A1BG | α-1-B glycoprotein | 37.5 | Juneja et al. (1987) | |||
| ALB | albumin | 50 | 3q14.3 | Andersson and Sandberg (1982),Godard et al. (1998) | ||
| CA | carbonic anhydrase | 37.5 | Sandberg et al. (1968) | |||
| ES | carboxylesterase | 50 | Andersson and Sandberg (1982) | |||
| HBA | hemoglobin A | 37.5 | 13pter | Bowling et al. (1988),Oakenfull et al. (1993) | ||
| PGD | 6-phosphogluconate | 25 | 2p12–13 | Andersson et al. (1984) | ||
| dehydrogenase | Gu et al. (1992) | |||||
| PGM | phosphoglucomutase | 37.5 | Dawson and Jaeger (1970) | |||
| TF | transferrin | 50 | Gahne (1966) | |||
| RFLPs | ||||||
| FUCA | α-L-1 fucosidase | 37.5 | this study | |||
| GLUT | glucose transporter | 12.5 | this study | |||
| type 1 | ||||||
| LPL | lipoprotein lipase | 12.5 | this study | |||
| MYL | myosin light | 75 | this study | |||
| polypeptide 1, 3 | ||||||
| TYR | tyrosinase | 12.5 | this study | |||
| SSCP | ||||||
| ELA–DRB | equine leucocyte | CTCTGCAGCACATTTCCTGGAG | 25 | 20q14 | Fraser and Bailey (1996) | |
| antigen class II | CGCCGCTGCACCAGGAA | |||||
| Ansari et al. (1988) | ||||||
| Anonymous loci | ||||||
| A-14 | microsatellite | CAGCTGGGTGACACAGAGAG | 75 | 2q14–21 | Marti et al. (1998) | |
| GTCATCACTACTCCCTACAC | ||||||
| A-17 | microsatellite | GTGGAGAGATAAAAGAAGATCC | 50 | 26q13–14 | Marti et al. (1998) | |
| GGCCACAAGGAATGAACACAC | ||||||
| ASB1 | microsatellite | AGCAGAAACCCACTCAAGCC | 75 | Breen et al. (1997) | ||
| GCATAATACCCTCAAGGTC | ||||||
| ASB2 | microsatellite | CCTTCCGTAGTTTAAGCTTCTG | 87.5 | 15q21.3–23 | Breen et al. (1997) | |
| CACAACTGAGTTCTCTGATAGG | ||||||
| ASB3 | microsatellite | AATTCATCTCAGTGCTCTACCAGC | 87.5 | 4q12–13 | Breen et al. (1997) | |
| TTCATTTTCTACATGCACTACAGC | ||||||
| ASB4 | microsatellite | TAAATTGTAAAAGCTGGAGCCG | 100 | 9q16–18 | Breen et al. (1997) | |
| GCAAATAGTAGTTAAGTCCTC | ||||||
| ASB5 | microsatellite | TCGAGGAGCTCATGACCTGG | 75 | 9q16–18 | Breen et al. (1997) | |
| TTGTACAACTCTCCACCATAGC | ||||||
| ASB6 | microsatellite | GGCACAGATGTTAGCTCAGC | 75 | 10p13 | Breen et al. (1997) | |
| ATGGAACCAGCCTGGATTGC | ||||||
| ASB7 | microsatellite | CTGGAAATTACAGTGGTCTTCTGG | 62.5 | 19q14–16 | Breen et al. (1997)b | |
| AGGTTTTCAGGGGCTTGCGAAGC | ||||||
| ASB8 | microsatellite | GACAACGTGGCAGCTCACTGCC | 37.5 | 1q16–17 | Breen et al. (1997) | |
| GCAAGTAAGCCATATGTGCATGCG | ||||||
| ASB9 | microsatellite | GTGCGCATGTATGTGCGTGCC | 75 | 10q21–23 | Breen et al. (1997) | |
| ATTTCCACAAGGGACATGAGG | ||||||
| ASB10 | microsatellite | GTTGTCTAGGTGCAGAATCTGG | 75 | Breen et al. (1997) | ||
| GTTATGTCTCCCCTTTCTCTACC | ||||||
| ASB11 | microsatellite | CCACCTATGTGTTCAGTTCACC | 50 | 19q21–22 | Breen et al. (1997)b | |
| GCACCAATGTTTATAGACTCCC | ||||||
| ASB12 | microsatellite | TCAGCAATAGAAGCCAGCTCC | 75 | 1q12–13 | Breen et al. (1997) | |
| TCCTATGGAGGTGACCTTCCC | ||||||
| ASB13 | microsatellite | CTCTGAAAGAGCAGGATTGG | 37.5 | 2q14.3–21.2 | Breen et al. (1997)b | |
| GTCTTCTAAGTGGTAAGAGCC | ||||||
| ASB14 | microsatellite | CTCCATGAATTCTCGCAGGTTGG | 25 | 6q21 | Breen et al. (1997) | |
| CCATGGGCCATATGCACACTGC | ||||||
| ASB15 | microsatellite | GTCCCAAAGGGACTCAGGAAGG | 37.5 | 15q21 | Breen et al. (1997) | |
| TGGATGCCAGTGCATAGACAG | ||||||
| ASB17 | microsatellite | GAGGGCGGTACCTTTGTACC | 62.5 | 2p14–15 | Breen et al. (1997) | |
| ACCAGTCAGGATCTCCACCG | ||||||
| ASB18 | microsatellite | TGCAGACAAAGCTGGACACTC | 50 | Breen et al. (1997) | ||
| CTGCTGAGAAAGCTTCTGC | ||||||
| ASB19 | microsatellite | GAGTTGGAGCTCAAGTCTGTC | 62.5 | 15q21.3–23 | Breen et al. (1997) | |
| GTTTAGCAACTACAGCGTAGG | ||||||
| ASB22 | microsatellite | GAGGAATGTGAAATACAGGAGG | 75 | 4q21 | Breen et al. (1997) | |
| TTTGTGGTCTTCCGTGCACC | ||||||
| ASB23 | microsatellite | GAGGTTTGTAATTGGAATG | 62.5 | 3q22 | Irvin et al. (1998) | |
| GAGAAGTCATTTTTAACACCT | Lear et al. (1998) | |||||
| ASB35 | microsatellite | ATGCATGAGCAGAGTGTTCTTCC | 62.5 | M. Breen et al. | ||
| TAGTACTTCTCTCTTAATATAAGC | (unpubl.) | |||||
| ASB36 | microsatellite | GAACATGTAGTGTTTACTCTGCC | 50 | M. Breen et al. | ||
| GAAGGTTTGTTGGGTCTTACAAGG | (unpubl.) | |||||
| ASB37 | microsatellite | CCTGCAACTTTTTCCCAGCC | 12.5 | 13q11–12 | Irvin et al. (1998) | |
| GGCAGATGTTAGCTCATGGC | Lear et al. (1998) | |||||
| ASB38 | microsatellite | TGGGGTTGCCTTGGTTACC | 62.5 | Irvin et al. (1998) | ||
| TCAGAGGATGAGGCACAGC | ||||||
| B-8 | microsatellite | TCCTCAGTCCTTTCTCATGC | 62.5 | 15q14–21 | Marti et al. (1998) | |
| AGCTGAAGGCAATCTGTACC | ||||||
| D-8 | microsatellite | TTTTTGTGTCTCAGGAGTGTG | 62.5 | 11p12–13 | Marti et al. (1998) | |
| AGTCTGATGGTGGAGGAAGG | ||||||
| ECA-2 | microsatellite | TTCCCTCCCATGGTTATTTTTC | 100 | (1q2.1)c | Sakagami et al. (1995) | |
| TCTCTACTTTCATATACATTTGG | ||||||
| ECA-3 | microsatellite | GGTTCACACAGGAGTCAGGGA | 50 | (2p1.3–4)c | Tozaki et al. (1995) | |
| CCTTCTGGTTTGCCTCGTCTC | ||||||
| HLM3 | microsatellite | GAAGGTAGAAAAGGAGGGCTAGAAC | 12.5 | Vega-Pla et al. (1996) | ||
| TCTAGAGGACCATTCTCTGGGCTGTG | ||||||
| HMB1 | microsatellite | GTGTGTATGCTTCCCAACCCTT | 50 | Binns et al. (1995) | ||
| GTTATAAAGCACTATGATCTCA | ||||||
| HMB2 | microsatellite | GTGCCACCACCTCTGTGATT | 50 | Binns et al. (1995) | ||
| TGGAGAAGGATCTTGGGCTC | ||||||
| HMB3 | microsatellite | CAAACATCAGTTAAGAGTGA | 75 | Binns et al. (1995) | ||
| CTCTAATCCAGCAGTGTTCA | ||||||
| HMB4 | microsatellite | AACCGCCTGAGCAAGGAAGT | 62.5 | Binns et al. (1995) | ||
| GCTCCCAGAGAGTTTACCCT | ||||||
| HMB5 | microsatellite | ACGGACACATCCCTGCCTGC | 75 | Binns et al. (1995) | ||
| GCAGGCTAAGGAGGCTCAGC | ||||||
| HMB6 | microsatellite | GAAGATGTCCGCTTTGATAT | 75 | Binns et al. (1995) | ||
| CACTGGCACATCCAGATTTG | ||||||
| HMS1 | microsatellite | CATCACTCTTCATGTCTGCTTGG | 62.5 | Guérin et al. (1994) | ||
| TTGACATAAATGCTTATCCTATGGC | ||||||
| HMS2 | microsatellite | ACGGTGGCAACTGCCAAGGAAG | 62.5 | Guérin et al. (1994) | ||
| CTTGCAGTCGAATGTGTATTAAATG | ||||||
| HMS3 | microsatellite | CCAACTCTTTGTCACATAACAAGA | 62.5 | Guérin et al. (1994) | ||
| CAATCCTCACTTTTTCACTTTGTT | ||||||
| HMS5 | microsatellite | TAGTGTATCCGTCAGAGTTCAAAG | 62.5 | Guérin et al. (1994) | ||
| GCAAGGAAGTCAGACTCCTGGA | ||||||
| HMS6 | microsatellite | GAAGCTGCCAGTATTCAACCATTG | 75 | Guérin et al. (1994) | ||
| CTCCATCTTGTGAAGTGTAACTCA | ||||||
| HMS7 | microsatellite | CAGGAAACTCATGTTGATACCATC | 75 | Guérin et al. (1994) | ||
| TGTTGTTGAAACATACCTTGACTGT | ||||||
| HMS18 | microsatellite | CAACAATGAAAATTTGTCCTGTGC | 62.5 | Godard et al. (1997) | ||
| GTAAATGAGTAGACAATCATGAGG | ||||||
| HMS19 | microsatellite | CTAACCAGCACAGAATGAATGGC | 25 | 4q21 | Godard et al. (1997, 1998) | |
| TAAAAGAACAGTGGAGAGTAAAGTG | ||||||
| HMS20 | microsatellite | TGGGAGAGGTACCTGAAATGTAC | 100 | Guérin and Bertaud (1996) | ||
| GTTGCTATAAAAAATTGTCTCCCTAC | ||||||
| HMS23 | microsatellite | GATCCAATATTGTAAACCCCGCC | 37.5 | Godard et al. (1997) | ||
| CCTTCATAACCCTTATTGCAGCC | ||||||
| HMS45 | microsatellite | TGTTACAGGTATTGGTAAACTGTGC | 75 | Godard et al. (1997) | ||
| GGAACAAGAAGAAATCACTAATGTC | ||||||
| HMS46 | microsatellite | GTCTCAGCCAAAAGGTATTCAAGC | 50 | Godard et al. (1997) | ||
| TGGCACCAATATAGGTCACCTGG | ||||||
| HMS47 | microsatellite | CCTGCTGAGGACCTTGGAAGCT | 25 | 22q19 | Godard et al. (1997, 1998) | |
| ATGTATTTTCAAGTCTAATATCTGCC | ||||||
| HTG2 | microsatellite | GATTGGCAACAGATGTTAACTCGG | 12.5 | Ellegren et al. (1992) | ||
| CCCCATGAGAACTAACAATGTTAG | ||||||
| HTG3 | microsatellite | TAACCTGGGTGCAAAGCCACCCAT | 62.5 | Ellegren et al. (1992) | ||
| TCAGGGCCAATCTTCCTCAC | ||||||
| HTG4 | microsatellite | CTATCTCAGTCTTCATTGCAGGAC | 100 | Ellegren et al. (1992) | ||
| CTCCCTCCCTCCCTCTGTTCTC | ||||||
| HTG5 | microsatellite | TGCTAAGCCTCAGCACATACA | 50 | Ellegren et al. (1992) | ||
| TGGAAATAAGGTTAGCAGGGATGC | ||||||
| HTG6 | microsatellite | CCTGCTTGGAGGCTGTGATAAGAT | 37.5 | 15q26–27 | Ellegren et al. (1992) | |
| GTTCACTGAATGTCAAATTCTGCT | Godard et al. (1998) | |||||
| HTG7 | microsatellite | CCTGAAGCAGAACATCCCTCCTTG | 25 | Marklund et al. (1994) | ||
| ATAAAGTGTCTGGGCAGAGCTGCT | ||||||
| HTG8 | microsatellite | CAGGCCGTAGATGACTACCAATGA | 62.5 | Marklund et al. (1994) | ||
| TTTTCAGAGTTAATTGGTATCACA | ||||||
| HTG9 | microsatellite | TGTGGGAAGAGTGTCAATAGCTGT | 62.5 | 4q21.3 | Marklund et al. (1994) | |
| AGGCATCTGGTTTGCTGCAATTTC | ||||||
| Godard et al. (1998) | ||||||
| HTG10 | microsatellite | CAATTCCCGCCCCACCCCCGGCA | 87.5 | Marklund et al. (1994) | ||
| TTTTTATTCTGATCTGTCACATTT | ||||||
| HTG11 | microsatellite | CAATGATGGTACTTTGCATATTAA | 37.5 | Marklund et al. (1994) | ||
| ATCGGCATGCACACTCATAGGTAG | ||||||
| HTG12 | microsatellite | CACTAGAGTCAGGGGGGGTGGGCT | 12.5 | Marklund et al. (1994) | ||
| TTGGAGTACTCTTTCTCCCTTCCC | ||||||
| HTG13 | microsatellite | TTAGCACGGGGAGATCGGATCCTG | 75 | Marklund et al. (1994) | ||
| GGTCTCCCTCTCCATTCACCCTGC | ||||||
| HTG14 | microsatellite | CCAGTCTAAGTTTGTTGGCTAGAA | 62.5 | Marklund et al. (1994) | ||
| CAAAGGTGAGTGATGGATGGAAGC | ||||||
| HTG15 | microsatellite | TCTTGATGGCAGAGCCAGGATTTG | 37.5 | Marklund et al. (1994) | ||
| AATGTCACCATGCGGCACATGACT | ||||||
| HTG17 | microsatellite | GCTATCCCTCCTGAGTCTTA | 87.5 | Lindgren et al. (1998) | ||
| AGGTAATTTGAAATAAAATACAC | ||||||
| I-12 | microsatellite | AACTAAGCACGTCATACAAG | 37.5 | 19q12–14 | Marti et al. (1998) | |
| CTTGTAGTTTTCGTTGTATAGC | ||||||
| I-18 | microsatellite | CAACAAAGATGTTGCAAGGG | 50 | 16q23–25 | Marti et al. (1998) | |
| TGTGCCTCTTGTCTCTTAGG | ||||||
| LEX2 | microsatellite | AAAAGGAAGACTGGCGACAG | 12.5 | Coogle et al. (1996a) | ||
| GGTGGGGGAAAGAATGGT | ||||||
| LEX4 | microsatellite | AATAGCAAATCTCCCACTTCA | 25 | Coogle et al. (1996a) | ||
| GTCCTCACAACCTCATCATAA | ||||||
| LEX5 | microsatellite | AAGGCAATGCTTATCAAATGC | 50 | Coogle et al. (1996a) | ||
| TTACCCGCAGTGACTTCTATT | ||||||
| LEX7 | microsatellite | GGTAGGGCTCTGGGATGA | 50 | Coogle et al. (1996a) | ||
| AACACTGGGGAAAAGTCAG | ||||||
| LEX8 | microsatellite | AAACTGTCACAACGGTTAGGAC | 37.5 | Coogle et al. (1996a) | ||
| CGAAAAAGCCACTTGAGGTC | ||||||
| LEX9 | microsatellite | AAAGCCGTAAGATTGGGACA | 75 | Coogle et al. (1996a) | ||
| TCCATTGTGAGGGTGTAACA | ||||||
| LEX11 | microsatellite | ATTCCCAGTGAAGTATTGCCA | 37.5 | Coogle et al. (1996a) | ||
| AGAGATGGGTACCTGGGATTC | ||||||
| LEX14 | microsatellite | CCTTACTCACTGGGGAATAAA | 87.5 | Coogle et al. (1996a) | ||
| AGACTGAACACCTAACTATGA | ||||||
| LEX15 | microsatellite | GCATTCCCATCATCACAT | 37.5 | Coogle et al. (1996b) | ||
| CCTGCCTTGCCTCTTTCT | ||||||
| LEX16 | microsatellite | GTGGGGCCGGTATAGTGATTG | 50 | Coogle et al. (1996b) | ||
| ACCCTAACTGATAACTGATAGA | ||||||
| LEX17 | microsatellite | CCTGCCCAAGAAGAACTCAGA | 100 | Coogle et al. (1996b) | ||
| AGCAGTGTATTTTTGAAACAT | ||||||
| LEX18 | microsatellite | TTTCATCACTTTCTGCTTCC | 25 | Coogle et al. (1996b) | ||
| TTCTCTTCCTTTGCTCATCCT | ||||||
| LEX19 | microsatellite | TTCCCTTTTCCTCACATCCT | 87.5 | Coogle et al. (1996b) | ||
| TTTTAGGTTCATCTATGTTGTTGC | ||||||
| LEX20 | microsatellite | GGAATAGGTGGGGGTCTGTT | 37.5 | Coogle et al. (1996b) | ||
| AGGGTACTAGCCAAGTGACTGC | ||||||
| LEX21 | microsatellite | GTAGGCTTTCTGCCAAAAT | 62.5 | Coogle et al. (1996b) | ||
| TGAGGGGAGTCATAAAAA | ||||||
| LEX22 | microsatellite | AACATATCCATCGCCTCACA | 25 | Coogle et al. (1996b) | ||
| TGCAAATTCACTGAGAGTGG | ||||||
| LEX23 | microsatellite | GGATGAAACAGGGAAGGAAA | 62.5 | Coogle et al. (1996b) | ||
| CCAACGGATTCATGAAAGCTA | ||||||
| LEX25 | microsatellite | CAATCGTGGCCCGGTAAC | 50 | Coogle et al. (1996c) | ||
| TTCACTCCAATCCTCAGTCA | ||||||
| LEX27 | microsatellite | ACCACTGGGAAACTGTGTAA | 25 | Coogle et al. (1996c) | ||
| GCCCAGAATCCGAACC | ||||||
| LEX29 | microsatellite | TGGGGTGTCACTGCTTCTC | 62.5 | Coogle et al. (1996c) | ||
| ACTGAGGGCCAGGTTTCTAA | ||||||
| LEX30 | microsatellite | GGAGGGTGCAAGGTGCTA | 37.5 | Coogle et al. (1996c) | ||
| GGCAGGTCAGAAGGGACA | ||||||
| LEX31 | microsatellite | CCCATTAAGAACTTTTCATCCTG | 87.5 | Coogle et al. (1996c) | ||
| GGCAAGCCCCACAAAATTAT | ||||||
| LEX32 | microsatellite | CGTAGTAGGGTTTTGGGTCC | 50 | Coogle et al. (1996c) | ||
| TTGCGTTTCAATTTTTAATGAC | ||||||
| LEX33 | microsatellite | TTTAATCAAAGGATTCAGTTG | 75 | Coogle et al. (1996c) | ||
| TTTCTCTTCAGGTGTCCTC | ||||||
| LEX34 | microsatellite | GCGGAGGTAAGAAGTGGTAG | 50 | Coogle et al. (1997) | ||
| GGCCTAAGATGAGGGTGAA | ||||||
| LEX35 | microsatellite | CCCAGCATATCAAAGATGTT | 75 | Coogle et al. (1997) | ||
| GCTCAGTGTACTTCAAGCAG | ||||||
| LEX37 | microsatellite | GGATTCCTCAACCTCCTAAA | 25 | Coogle et al. (1997) | ||
| AGGGATAAGTGACCACCAC | ||||||
| LEX38 | microsatellite | CTGCATTCCCATCATCACAT | 37.5 | Coogle et al. (1997) | ||
| TGCCTTGCCTCTTTCTGTTTA | ||||||
| LEX39 | microsatellite | CCTCTGTCCCCACTACTCTC | 37.5 | Coogle et al. (1997) | ||
| TTGATCTCCACTCCCAATG | ||||||
| LEX40 | microsatellite | TTTGGCCGTTAGTCGTGT | 37.5 | Coogle et al. (1997) | ||
| GACAAATCGGAAAGTTGGAA | ||||||
| MPZ002 | microsatellite | GATCCCCCCTATTTTATATACAG | 25 | Breen et al. (1994) | ||
| AGGTTCTCATTCTACCTACAAGG | ||||||
| SGCV1 | microsatellite | AGTCACCACCACTCACCTTGT | 37.5 | ?d | Godard et al. (1997) | |
| CCAACACAGGATACGGATGA | ||||||
| SGCV3 | microsatellite | CCTTGTGGTGAGTTTTCCTCTT | 37.5 | ?d | Godard et al. (1997) | |
| CTGCAAAGCTCTGAAGGTC | ||||||
| SGCV4 | microsatellite | CGACGCCTCCTCCTAAAC | 37.5 | 23q19 | Godard et al. (1997) | |
| CAGCTGTGTGCCTTTGATTAT | ||||||
| SGCV6 | microsatellite | GGGCCTGGTTTTCCTTCTAA | 62.5 | 15q24 | Godard et al. (1997) | |
| GCATTTGTGGCCTGTGTCATA | ||||||
| SGCV7 | microsatellite | GAATTTGAATGTATCTATTCTGAATG | 50 | 18q21 | Godard et al. (1997) | |
| GTGAGTTTTCAAGCTGGCATATTC | ||||||
| SGCV8 | microsatellite | GAGTTCATTCTTTTTCGTGGCTG | 37.5 | 19 | Godard et al. (1997) | |
| GGAAACACCCTAAGTGTCCCTTG | ||||||
| SGCV10 | microsatellite | CATCCATCCTTTCCAGCTCGATATTC | 62.5 | 12p13 | Godard et al. (1997) | |
| CAAGACCGTAACTCAGGAGCCC | ||||||
| SGCV13 | microsatellite | GGACTAAAGCCCAACCATCCAGC | 75 | 11q12 | Godard et al. (1997) | |
| CTCACCAGTAAGGGGTTATGGGGC | ||||||
| SGCV14 | microsatellite | CCCCAGTGGTTCCATTTAGATGT | 75 | 21q13 | Godard et al. (1997) | |
| GGGGAGAGCATTTTGGTGA | ||||||
| SGCV16 | microsatellite | AATTCTCAAATGGTTCAGTGA | 37.5 | 21q13 | Godard et al. (1997) | |
| CTCCCTCCCTTCCTTCTA | ||||||
| SGCV17 | microsatellite | GGCCCAACGTCTATAGAAAGATGT | 37.5 | ?d | Godard et al. (1997) | |
| CCCCCAAATGGCTATTTTCTAA | ||||||
| SGCV18 | microsatellite | TGGGGAAGAGGGATTCAT | 62.5 | 3p13-14 | Godard et al. (1997) | |
| AAATGCCAAGCCTATCTATGC | ||||||
| SGCV19 | microsatellite | GCCCCCACCTGCTCCACC | 62.5 | 22q19 | Godard et al. (1997) | |
| GGGGCAAAGTGGAAATCC | ||||||
| SGCV23 | microsatellite | GGCTTAAGATATGGGTGAGTAAGG | 87.5 | 4q27 | Godard et al. (1997) | |
| GCCCACCCTCTTACTTTTCTCAA | ||||||
| SGCV24 | microsatellite | CTACCATTGAAGAGGGGTGGC | 50 | 11p12 | Godard et al. (1997) | |
| GAAACGAGCAGGAAGTGAATCTCC | ||||||
| SGCV25 | microsatellite | GCCCATATTAGTAGGACTGTG | 37.5 | 1q14 | Godard et al. (1997) | |
| GGCCATATTCAGCAGAGCT | ||||||
| SGCV28 | microsatellite | CTGTGGCAGCTGTCATCTTGG | 25 | Godard et al. (1997) | ||
| CCCAATTCCAGCCCAGCTTGC | ||||||
| SGCV30 | microsatellite | ACTGGAGGGGTGAAACAGATTCAGA | 75 | 10qter | Godard et al. (1997, 1998) | |
| GGAAGGGAGGTCATCAGAA | ||||||
| SGCV32 | microsatellite | TGTTCCAAAATGGAGGGTGAGCC | 62.5 | ?d | Godard et al. (1997) | |
| CCACAGGCTCTTAAAACCAGAAGC | ||||||
| VHL20 | microsatellite | CAAGTCCTCTTACTTGAAGACTAG | 87.5 | van Haeringen et al. (1994) | ||
| AACTCAGGGAGAATCTTCCTCAG | ||||||
| VIAS-H34 | microsatellite | GTATCAGCTTAACAGCTTTCTTTAAATG | 37.5 | Ewen and Matthews (1994a) | ||
| CTCCCGTCTCCTCTCTTGTTC | ||||||
| VIAS-H39 | microsatellite | AATGTGATTATAGCAGATAGGGTT | 37.5 | Ewen and Matthews (1994b) | ||
| CTATCCAATCTTCACAATCATGTA | ||||||
Observed heterozygosity among the eight sires in the reference pedigree.
The assignment provided here is based on new experiments, which indicates that these clones are chimeric (M. Breen, unpubl.). Within the chimeric clones, the largest pieces of DNA map to the locations indicated in Breen et al. (1997). However, the sequence surrounding the microsatellite loci have now been confirmed to map to the locations given here.
Chromosome band identification was done using an old idiogram nomenclature.
Physical assignment does not fit with linkage data.
Table 2.
Data for New RFLP Markers
| Locus | Enzyme | Alleles | Polymorphic fragments (kb) | Observed heterozygosity | Reference |
|---|---|---|---|---|---|
| FUCA1 | TaqI | A | 1.3 | 0.38 | Fukushima et al. (1985) |
| B | 1.4 | ||||
| GLUT1 | TaqI | A | 1.6 | 0.13 | Mueckler et al. (1985) |
| B | 1.9 | ||||
| LPL | TaqI | A | 3.5 + 5.4 | 0.13 | Harbitz et al. (1992) |
| B | 8.5 | Gu et al. (1992) | |||
| MYL1, 3 | MspI | A | 2.8 | 0.75 | Seiden et al. (1987) |
| B | 3.3 | ||||
| TYR | TaqI | A | 2.6 | 0.25 | Barton et al. (1988) |
| B | 3.4 |
Marker heterozygosity varied extensively between loci, from one out of the eight sires being heterozygote to all being so. The average number of heterozygous sires was 4.3, which corresponds to an observed heterozygosity (Ho) of 53%. In general, microsatellites were more variable than the other markers in this study (Ho = 56% vs. 35%). Two important consequences of only the sires and their offspring being included in the study were that we only measured male recombination fractions and could not deduce the transmission of paternal alleles for all offspring. In a heterozygous offspring showing the same two alleles as its father, knowledge of the genotype of its mother is required for following the inheritance of the sire’s allele. The average number of fully informative offspring per marker was 89, ranging from 9 to 235.
Linkage Analysis
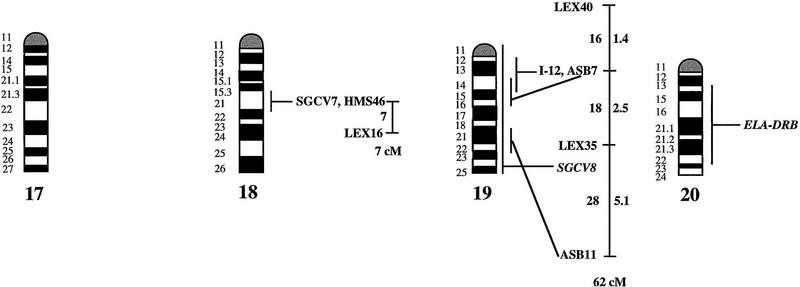
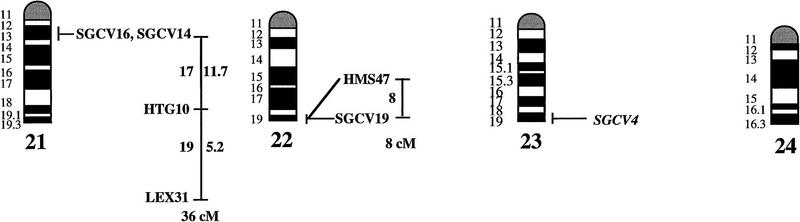
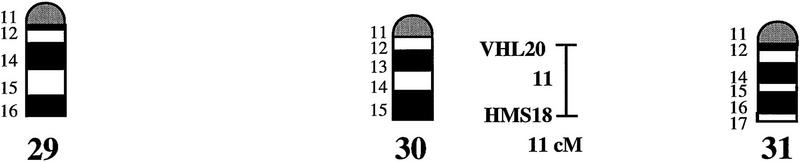
An overview of the linkage data is given in Figure 1, where the equine idiogram (Bowling et al. 1997a) is shown together with the established linkage groups. One hundred of the 140 markers (72%) showed linkage to at least one other marker. The average pair-wise recombination fraction between linked markers, using an lod score of three as threshold level for significant linkage, was 9.7% ± 0.7 s.e., with 26% as the longest interval for which linkage was detected (ASB6–ASB9). By multipoint analysis we could establish 25 linkage groups. Twenty-two of these were assigned to 18 different chromosomes, generally caused by one or more of the markers included in a linkage group being mapped physically by in situ hybridization (see legend to Fig. 1). Chromosomes covered by linkage groups were ECA1, ECA2, ECA3, ECA4, ECA5, ECA6, ECA7, ECA9, ECA10, ECA11, ECA13, ECA15, ECA16, ECA18, ECA19, ECA21, ECA22, and ECA30. For four additional chromosomes (ECA12, ECA20, ECA23, and ECA26), a physically anchored genetic marker was included in our material but showed no linkage to other markers. Only two metacentric (ECA8 and ECA12) and two of the larger acrocentric chromosomes (ECA14 and ECA17) were not tagged by physically assigned linkage groups.
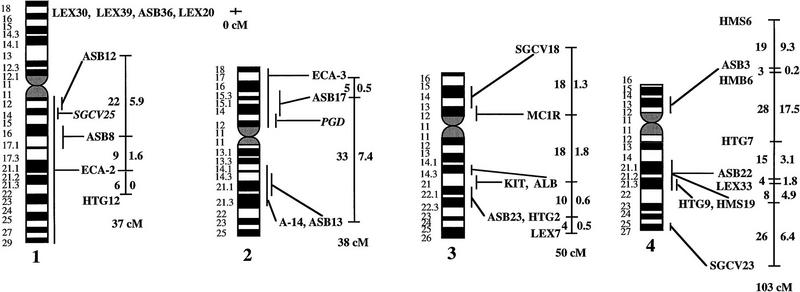
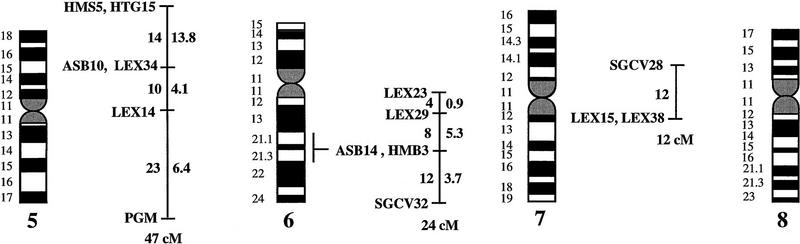
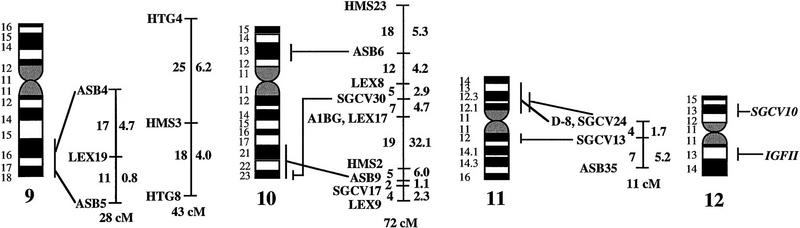
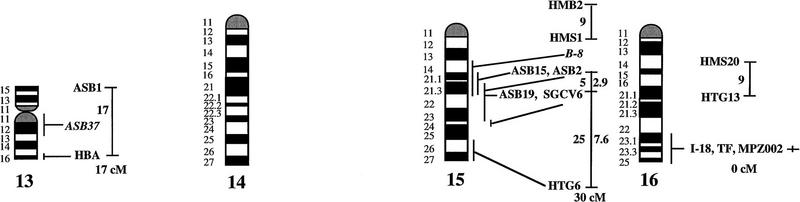
Figure 1.
A preliminary male autosomal linkage map of the equine genome. The map depicts all established linkage groups anchored to chromosomes, as well as unassigned linkage groups (bottom). Chromosomal assignments of linkage groups are based on one or more of the markers being physically mapped by in situ hybridization (vertical bars close to chromosomes). The only exceptions are the LEX30–LEX39–MPZ027–LEX20 linkage group on ECA1, the HMS5–HTG15–ASB10–LEX34–LEX14–PGM linkage group on ECA5, the SGCV28–LEX15–LEX38 linkage group on ECA7, the HTG4–HMS3–HTG8 linkage group on ECA9, and HMS20–HTG13 on ECA16, which chromosomal assignments are based on synteny data (Shiue et al. 1998; see also Bailey et al. 1997; Godard et al. 1997). The assignment of the HMB2-HMS1 group to ECA15 is based on a previously observed linkage between one of these markers and physically anchored markers on this chromosome (Godard et al. 1997). The assignment of the group on ECA30 is based on a defined aneuploidy (Bowling et al. 1997b). Values to the left of main vertical bars represent genetic distances between markers expressed as multipoint Kosambi cM, and values to the right are the log 10 odds against reversed order of adjacent markers. Loci shown at the same vertical position had 0% recombination. Below each linkage group is indicated its total multipoint length. In situ mapped markers genotyped in the family material but not showing linkage are depicted in italics on the map according to their band assignments; their exclusion from linkage groups is evident from the absence of a horizontal line in the vertical bar connecting linked markers. Three markers (ASB13 on ECA2, MC1R on ECA3, and ASB11 on ECA19) known from in situ hybridization to reside within established linkage groups were included in multipoint analyses although only showing a maximum two-point lod score of between 2 and 3.
Linkage groups ranged between 0 and 103 cM (multipoint distances in Kosambi cM), the longest residing on ECA4, and the number of markers within groups between 2 and 10. The order of markers within linkage groups could in several cases not be resolved with confidence (i.e., the odds against reversed order of adjacent markers being lower than 1000:1), and the precise order of markers in such groups should therefore be regarded as tentative. For the purpose of this study, however, we consider it important to report even tentative orders in light of the previous absence of map information for most horse chromosomes. The sum of the length of all linkage groups was 679 cM, with an average distance between adjacent markers of 12.6 cM. Clearly, a much greater total genetic length is revealed if one takes into account the flanking distances covered by end markers in linkage groups, and the distances covered by the 40 unlinked markers. Using the mean number of informative meioses per marker (89), we can estimate that, on average, our data set allow linkage between two random markers spaced up to 15 cM apart to be detected with an lod score criterion of 3. Thereby assuming that each of the 2 × 25 = 50 end markers on average cover 15 flanking cMs, the total map length would be about 1425 cM. Furthermore, with the addition of unlinked markers, it is reasonable that the marker set covers well above 1500 cM. These can only be seen as very rough estimates as, for instance, some end markers will be close to telomeres already.
The linkage groups on ECA3, ECA4, and ECA15 contained five in situ mapped markers, the one on ECA2 had four, whereas ECA1, ECA9, ECA10, ECA11, ECA19, and ECA22 had either two or three physically anchored markers. In most of these cases this allowed determining the orientation of the linkage group along the respective chromosomes. Data from chromosomes with more than one physical tag also allowed a rough analysis of the relationship between genetic and physical distances in the equine genome. Using the approach described in Ellegren et al. (1994), we analyzed this by measuring the physical distance between the most distant anchored markers within linkage groups, expressed as the relative proportion of the genome covered by these markers (measured with a ruler on the karyotype, from the midpoints of in situ assignments), and compared this with the recombinational distance between these markers. A minimum length of linkage groups to be considered was set to 20 cM. Six groups fulfilled these criteria and gave estimates of 0.70 cM/Mb (ASB12–ASB8 on ECA1), 0.50 (ASB17–ASB13 on ECA2), 0.54 (SGCV18–ASB23 on ECA3), 0.84 (ASB3–SGCV23 on ECA4), 0.73 (ASB6–ASB9 on ECA10), 0.60 (ASB15–HTG6 on ECA15), and 0.83 (I-12–ASB11 on ECA19), the mean being 0.68 cM/Mb ± 0.05 s.e.
DISCUSSION
This study constitutes the most comprehensive mapping effort so far for the horse genome to date, and is the first to present a preliminary male autosomal linkage map for this species. The number of genetically mapped markers, ∼100, far exceeds the sum of that included in earlier overviews (e.g., Sandberg and Andersson 1992) and recent linkage studies (Marklund et al. 1994; Breen et al. 1997; Godard et al. 1997). The majority of the markers genetically mapped in the present study thus represent new linkage assignments. Moreover, for most of the 18 autosomes tagged by linkage groups (i.e., ECA1, ECA2, ECA5, ECA6, ECA7, ECA9, ECA11, ECA13, ECA15, ECA16, ECA18, ECA19, ECA21, and ECA22), the present data either represent the first linkage groups assigned to these chromosomes, or the first to involve more than a single pair of markers.
The total male map distance residing within linkage groups was 679 cM, and we estimate that the map covers well above 1500 cM when distances covered by end markers and by unlinked markers are taken into account. It is hard to deduce how large a fraction of the genome is thereby covered, notably because we do not know the total recombinational distance (genetic length) of the equine genome. Total, sex-average distances for other mammalian species range from ∼1600 cM (mouse; Davisson and Roderick 1989) to 3500 cM or even higher (human; Weissenbach et al. 1992). In at least some species the male recombination rate is considerably less than that in females (Morton 1991; Ellegren et al. 1994), but this might not be a ubiquitous phenomenon among mammals as suggested by data for cattle and sheep (Crawford et al. 1995; Barendse et al. 1997). The only clue to the genetic length of the equine genome comes from the analysis of meiotic chromosomes (Scott and Long 1980). The number of per cell chiasma in late diplotene or diakinesis among stallions was counted to 54.4 ± 1.8, which is comparable to that found in sheep (Chapman and Bruere 1977; Long 1978), but is higher than that in pig (Burt and Bell 1987), cattle, and goat (Logue 1977). The observed number of chiasma in horse would suggest a total male genetic distance of 2720 cM (whereas it would be 2000–2500 cM in pig, cattle, and goat). It is, however, difficult to properly assess genetic lengths from chiasma counts, and also differences between species, as such estimates depend on the precise meiotic stage at which cells are analyzed, something which may vary between studies. As a minimum estimate, it seems reasonable that our map covers at least 50% of the equine genome.
In a sense, we found it somewhat surprising that the proportion of genotyped markers showing linkage was not higher (140 markers genotyped, 100 linked). Earlier studies of other livestock species have generally noted a higher proportion of linked markers at the corresponding stage of map development (e.g., Ellegren et al. 1994). Although the discrepancy may potentially relate to a difference in the total genetic length of genomes, it seems evident that this mainly reflects an inherent problem in equine genome mapping. The possibility of detecting linkage between markers residing on the same chromosomes will depend obviously on the number of informative meioses shared between the markers. This, in turn, will be a function of marker heterozygosity in the segregating generation and the number of offspring in which transmission can be followed. Because horse gene mapping for reproductive, biological, and practical reasons generally will have to rely on half-sib families (embryo transfer is not yet used in horse), the number of potentially informative meioses will only be half that obtained in analyses of the same number of offspring from full-sib families. This difference is further accentuated if dams are excluded from the family material being genotyped, as was the case in this study, given that it prohibits tracing paternal allele transmission in offspring with the same genotype as their fathers. Furthermore, as marker heterozygosity in the segregating F1 generation in mapping pedigrees of other species is often maximized through crosses between genetically divergent parental lines (Beattie 1994), the fact that the economy and practice of horse breeding generally do not allow crosses between divergent breeds implies that a lower proportion of markers will be in heterozygote state in the segregating generation. It could be noted finally that for a majority of the markers used in this study there is no information available on if the degree of genetic variability differs between breeds, so it cannot be judged if linkage would generally be more easily detected within some breeds than within others.
Some of the linkage assignments made in this study were not in agreement with previously reported map data. The assignments of SGCV17 and SGCV32 to linkage groups on ECA10 and ECA6, respectively (supported by lod scores of 9.98 for SGCV17 and of 7.72 for SGCV32), contradict both their reported FISH mapping to ECA9 (Godard et al. 1997). By somatic cell hybrid mapping, Shiue et al. (1998) similarly placed SGCV17 on ECA10, giving support to our linkage data. New FISH experiments with the SGCV17 and SGCV32 cosmids indicate that they may indeed map to ECA10 and ECA6, respectively (G. Guérin, pers. comm.). Another discrepancy was our observation of close linkage between SGCV1 and SGCV3 (0% recombination, lod score 11.74). These two markers have been FISH mapped to ECA13 and ECA19, respectively (Godard et al. 1997). Peculiarly, SGCV1 and SGCV3 gave identical microsatellite amplification profiles among unrelated individuals as well as in families (homo/heterozygosity, relative positions of alleles, segregation, etc.), and it appears that they may in fact represent the same locus, their previous disparate in situ assignment being because of human error (G. Guérin, pers. comm.). Yet another discrepancy was the precise location of the marker SGCV30. Physical mapping places it terminal on ECA10q (Godard et al. 1997), however, our linkage data suggests that it is closer to the centromere. All these physical assignments of SGCV markers were done with cosmids and it is possible that the cosmid library contained a significant proportion of chimeric clones, as observed for other equine cosmid libraries (M. Breen, pers. comm.). A few cases of linkage between two markers associated with lod score values slightly above three were likely to represent chance events rather than true linkage. LEX14 showed 25% recombination to SGCV30 on ECA10 at lod score 3.01, however, synteny mapping places LEX14 on ECA5. Similarly, linkages between HMS20 and SGCV18 on ECA3 (24% recombination, lod score 3.13), and between HTG5 and HMS23 on ECA10 (11% recombination, lod score 3.69) may be chance events as HMS20 maps to ECA16 and HTG5 to ECA20 in hybrid panel analysis (Shiue et al. 1998). These three linkages have therefore been omitted from the compilation in Figure 1.
Heterologous chromosome painting (ZOO-FISH) has delineated the overall homologies between the human and equine genomes on the chromosomal level (Raudsepp et al. 1996). In general, chromosomes tend to be well conserved with, for instance, the majority of equine chromosomes corresponding to single human chromosomes (though not necessarily the reverse—a consequence of the horse karyotype containing more chromosomes than the human). Whereas this gross-level information provides an important starting point in the transfer of map information between species, more detailed information on, for example, the positions of genes and gene orders on chromosomes is required for fine-tuned analysis. Two new gene assignments were made in this study, phosphoglucomutase (PGM) to ECA5 and transferrin (TF) to ECA16. Human PGM maps to HSA1p31. According to ZOO-FISH (Raudsepp et al. 1996), HSA1 corresponds to three different horse chromosomes, ECA2p, ECA5, and ECA30, but it is not known which parts of the human chromosome are homologous to each of the three horse chromosomes. Our mapping of PGM now shows that ECA5 is homologous to at least parts of the p arm of HSA1. As the linkage group on ECA5 is not oriented, we cannot deduce how the HSA1 conservation is arranged along ECA5. The assignment of the TF locus to ECA16 is in agreement with ZOO-FISH data as human TF maps to HSA3q21 and HSA3 corresponds to ECA16 and ECA19. The HSA3q21 band is relatively close to the centromere and given the rather distal location of TF on ECA16, it is possible that the entire q arm of HSA3 corresponds to ECA16, but is orientated reversely. If so, the HSA3–ECA19 homology would involve the p arm of HSA3. Of course, internal rearrangements may occur, but ZOO-FISH data suggest that such events have been rare following the split of the human and equine lineages (Raudsepp et al. 1996).
Only a few mutations causing disease or affecting other important traits have been identified yet in the horse, that is, the adult skeletal muscle sodium channel α subunit gene (SCN4A) associated with hyperkalemic periodic paralysis (HYPP; Rudolph et al. 1992), the catalytic subunit of the DNA-dependent protein kinase (DNA–PKCS) associated with severe combined immune deficiency (SCID; Shin et al. 1997), the endothelin receptor B (EDNRB) associated with overo lethal white foal syndrome (OLWS; Santschi et al. 1998), and the melanocyte-stimulating hormone receptor (MC1R) associated with the extension (E) chestnut coat color (Marklund et al. 1996b). All of these mutation identifications have been based on a comparative candidate gene approach, using information from other species in which a similar phenotype and a causative gene has been identified already. With the linkage map presented here, it will now become feasible to make genome scans for traits of unknown genetic background in the horse.
In what way should future equine genome mapping go? A first step will obviously have to be extending the present linkage map to reach a nearly complete genome coverage, and to construct a framework map based on highly informative markers ordered with confidence. It is likely that this goal could be reached in the near future by merging maps presently under development in different laboratories, for example, through the international Equine Gene Mapping Workshop collaboration, and by the selected analysis of markers known from synteny mapping to reside on chromosomes with poor coverage (Shiue et al. 1998). In the latter perspective, isolation of markers from chromosome-specific libraries will also be an important tool. During the process of map expansion, there is as well a need for extensive integration of physically anchored markers. The availability of equine BAC libraries (e.g., Godard et al. 1998) will allow the identification of large-insert clones containing genetically mapped markers, and the subsequent physical mapping of such clones with FISH. Furthermore, a large number of coding markers should be integrated into the linkage map to facilitate comparative approaches in the search for candidate genes. Recent progress with expressed sequence tags (ESTs) mapping in domestic animals (e.g., Fridolfsson et al. 1997) suggests that this can greatly increase the possibility of exploiting human gene map information across species (e.g., Hasler-Rapacz et al. 1998). As a second step, the development of equine radiation hybrid (RH) panels would greatly facilitate more fine-tuned mapping.
METHODS
Reference Families
A Swedish reference pedigree for equine gene mapping has been established in the form of eight half-sib families comprising sires and offspring. Four families are composed of Standardbred (S) trotters and four of Icelandic (I) horses. The number of offspring in the respective families is 25 (S), 27 (S), 31 (S), 31 (I), 31 (I), 31 (I), 40 (S), and 47 (I), totaling 263. Because the number of mares is the same as the number of offspring and given that the informativeness of these mares in linkage analysis would be limited, mares were not included in the reference material. As a consequence, only the recombination fractions obtained through male meiosis are followed in this material. Furthermore, this means that X chromosome linkages were not covered by our data.
Genetic Markers and Genotyping
A list of genetic markers genotyped in this study is given in Table 1 and includes 121 microsatellites, eight protein polymorphisms, five RFLPs, three blood group polymorphisms, two PCR–RFLPs, and one single-strand conformational polymorphism (SSCP), in total 140 markers. The vast majority of microsatellites was of the (CA)n dinucleotide repeat type, 45 of which had been assigned physically by in situ hybridization (Table 1).
Markers were genotyped essentially as reported in the original references provided in Table 1. Briefly, PCR amplification of microsatellite loci was performed in a MJ Research (PTC-100) thermal cycler in 10-μl reactions containing 0.25 units of AmpliTaq DNA polymerase (Perkin-Elmer), 200 μm dNTPs, 1.5 mm MgCl2, 50 mm KCl, 10 mm Tris-HCl (pH 8.3), 0.001% (wt/vol) gelatin, 100 ng of genomic DNA, and 1–3 pmoles of each primer. 5′ end labeling of one primer in each primer pair was carried out in 25-μl reactions using 0.1 μCi of [γ32P] per pmole primer, 10 units of T4 polynucleotide kinase (New England Biolabs, Beverly, MA), 70 mm Tris-HCl (pH 7.6), 10 mm MgCl2, and 5 mm DTT. The standard PCR profile consisted of one cycle of 94°C for 3 min, 58°C for 30 sec, and 72°C for 1 min, followed by 29 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. After the final cycle a prolonged extension step of 10 min was included. The PCR products were mixed with loading buffer (95% formamide, 0.05% xylene cyanol FF, 0.05% bromophenol blue, 0.02 m EDTA) and electrophoresed for 1–2 hr in 6% denaturating polyacrylamide gels (Sequagel XR, National Diagnostics, Atlanta, GA). Subsequently, gels were soaked in 10% acetic acid, dried at 70°C, and exposed to autoradiographic films overnight.
Five heterologous mammalian cDNA probes, fucosidase 1 (FUCA1), glucose transporter 1 (GLUT1), lipoprotein lipase (LPL), myosin light chain 1 and 3 (MYL1,3), and tyrosinase (TYR), were used for RFLP analysis (Table 2). Fifteen micrograms of genomic DNA was digested with either MspI, PvuII, or TaqI (Promega, Madison, WI), separated in 0.9% agarose gels, and transferred to Hybond N+ membranes (Amersham Pharmacia Biotech, Uppsala, Sweden). Hybridization was made in 0.26 m Na2HPO4, 7% SDS, 5% dextran sulfate, 1% bovine serum albumin, and 0.2 mg/ml salmon sperm DNA at 65°C, using probes labeled with [α32P]dCTP by nick translation. Membranes were washed at a final stringency of 0.2× SSC at 60°C and exposed to autoradiographic films for 2–6 days.
Linkage Analysis
Linkage between markers was analyzed using the program CRIMAP, version 2.4 (Green et al. 1990). First, the TWOPOINT option of CRIMAP was used to detect pair-wise linkages. An lod score threshold of three was set as criterion for significant linkage. For multipoint analysis of larger linkage groups we used the option BUILD to select markers to be used as a framework for the continuing ordering of additional markers; the order of selected markers was supported by an lod score of three or higher. The option ALL was used subsequently to incorporate the rest of the markers, one at a time. The FLIPS and FIXED options were used finally for evaluating the statistical support of the proposed order and the distance between markers, respectively. All multipoint distances are expressed as Kosambi cM. For smaller linkage groups (≤5 markers) we started with the ALL option and then ran the FLIPS2 option. Genotype data were checked for typing errors using the CHROMPIC option, as a means to identify unlikely double recombinants.
Acknowledgments
The technical assistance of Sigridur Björnsdottir, Lena Oscarsson, and Eva Pettersson is acknowledged. Leif Andersson is acknowledged for support during the project. Financial support was obtained from the Swedish Research Council for Agriculture and Forestry, the Erik Philip-Sörensens Foundation, the Trygger foundation, and The Royal Swedish Academy of Agriculture and Forestry.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Gabriella.Lindgren@bmc.uu.se; FAX 46-18-504461.
REFERENCES
- Andersson L, Sandberg K. A linkage group composed of three coat colour genes and three serum protein loci in horses. J Hered. 1982;73:91–94. [PubMed] [Google Scholar]
- Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, Andersson K, Andersson-Eklund L, Edfors-Lilja I, Fredholm M, Hansson I, Håkansson J, Lundström K. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science. 1994;263:1771–1774. doi: 10.1126/science.8134840. [DOI] [PubMed] [Google Scholar]
- Ansari HA, Hediger R, Fries R, Stranzinger G. Immunogenetics 28: 362–364. 1988. Chromosomal localization of the major histocompatibility complex of the horse (ELA) by in situ hybridization. [DOI] [PubMed] [Google Scholar]
- Bailey E, Reid RC, Skow LC, Mathiason K, Lear TL, McGuire TC. Linkage of the gene for equine combined immunodeficiency disease to microsatellite markers HTG8 and HTG4; synteny and FISH mapping to ECA9. Anim Genet. 1997;28:268–273. doi: 10.1111/j.1365-2052.1997.00152.x. [DOI] [PubMed] [Google Scholar]
- Bailey E, Stormont C, Suzuki Y, Trommershausen Smith A. Linkage of loci controlling alloantigens on red blood cells and lymphocytes in the horse. Science. 1979;204:1317–1319. doi: 10.1126/science.451540. [DOI] [PubMed] [Google Scholar]
- Barendse W, Vaiman D, Kemp SJ, Sugimoto Y, Armitage SM, Williams JL, Sun HS, Eggen A, Agaba M, Aleysin SA, et al. A medium-density genetic linkage map of the bovine genome. Mamm Genome. 1997;8:21–28. doi: 10.1007/s003359900340. [DOI] [PubMed] [Google Scholar]
- Barton DE, Kwon BS, Francke U. Human tyrosinase gene, mapped to chromosome 11 (q14-q21), defines second region of homology with mouse chromosome 7. Genomics. 1988;3:17–24. doi: 10.1016/0888-7543(88)90153-x. [DOI] [PubMed] [Google Scholar]
- Beattie CW. Livestock genome maps. Trends Genet. 1994;10:334–338. doi: 10.1016/0168-9525(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Binns MM, Holmes NG, Holliman A, Scott AM. The identification of polymorphic microsatellite loci in the horse and their use in thoroughbred parentage testing. Br Vet J. 1995;151:9–15. doi: 10.1016/s0007-1935(05)80057-0. [DOI] [PubMed] [Google Scholar]
- Bowling AT, Clark RS. Blood group and protein polymorphism gene frequencies for seven breeds of horses in the United States. Anim Blood Groups Biochem Genet. 1985;16:93–108. doi: 10.1111/j.1365-2052.1985.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Bowling AT, Scott AM, Flint J, Clegg JB. Novel alpha haemoglobin haplotypes in horses. Anim Genet. 1988;19:87–101. doi: 10.1111/j.1365-2052.1988.tb00795.x. [DOI] [PubMed] [Google Scholar]
- Bowling AT, Breen M, Chowdhary BP, Hirota K, Lear T, Millon LV, Leon FAP, Rausepp T, Stranzinger G. International System for Cytogenetics Nomenclature of the domestic Horse. Chromosome Res. 1997a;5:433–443. doi: 10.1023/a:1018408811881. [DOI] [PubMed] [Google Scholar]
- Bowling AT, Millon LV, Dileanis S. Physical mapping of genetic markers to chromosome 30 using a trisomic horse and evidence for maternal origin of the extra chromosome. Chromosome Res. 1997b;5:429–431. doi: 10.1023/a:1018456727811. [DOI] [PubMed] [Google Scholar]
- Breen M, Downs P, Irvin Z, Bell K. Six equine dinucleotide repeats: Microsatellites MPZ002, 3, 4, 5, 6 and 7. Anim Genet. 1994;25:124. doi: 10.1111/j.1365-2052.1994.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Breen M, Lindgren G, Binns MM, Norman J, Irvin Z, Bell K, Sandberg K, Ellegren H. Genetical and physical assignments of equine microsatellites-first integration of anchored markers in horse genome mapping. Mamm Genome. 1997;8:267–273. doi: 10.1007/s003359900407. [DOI] [PubMed] [Google Scholar]
- Burt A, Bell G. Mammalian chiasma frequencies as a test of two theories of recombination. Nature. 1987;326:803–805. doi: 10.1038/326803a0. [DOI] [PubMed] [Google Scholar]
- Chapman HM, Bruere A. Chromosome morphology during meiosis of normal and Robertsonian translocation-carrying rams (Ovis aries) Can J Genet Cytol. 1977;19:93–102. doi: 10.1139/g77-011. [DOI] [PubMed] [Google Scholar]
- Charlier C, Coppieters W, Farnir F, Grobet L, Leroy PL, Michaux C, Mni M, Schwers A, Vanmanshoven P, Hanset R, Georges M. The mh gene causing double-muscling in cattle maps to bovine chromosome 2. Mamm Genome. 1995;6:788–792. doi: 10.1007/BF00539005. [DOI] [PubMed] [Google Scholar]
- Coogle L, Bailey E, Reid R, Russ M. Equine dinucleotide repeat polymorphisms at loci LEX002, -003, -004, -005, -007, -008, -009, -010, -011, -013 and -014. Anim Genet. 1996a;27:126–127. [PubMed] [Google Scholar]
- Coogle L, Reid R, Bailey E. Equine dinucleotide repeat loci LEX015-024. Anim Genet. 1996b;27:217–218. [PubMed] [Google Scholar]
- ————— Equine dinucleotide repeat loci from LEX025 to LEX033. Anim Genet. 1996c;27:289–290. doi: 10.1111/j.1365-2052.1996.tb00500.x. [DOI] [PubMed] [Google Scholar]
- ————— Equine dinucleotide repeat loci LEX034-LEX048. Anim Genet. 1997;28:308–322. [PubMed] [Google Scholar]
- Crawford AM, Dodds KG, Ede AJ, Pierson CA, Montgomery GW, Garmonsway HG, Beattie AE, Davies K, Maddox JF, Kappes SW, et al. An autosomal genetic linkage map of the sheep genome. Genetics. 1995;140:703–724. doi: 10.1093/genetics/140.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT, Roderick TH. Linkage map. In: Lyon MF, Searle AG, editors. Genetic variants and strains of the laboratory mouse. 2nd ed. Stuttgart, Germany: Gustav Fischer Verlag; 1989. pp. 416–427. [Google Scholar]
- Dawson DM, Jaeger S. Heterogeneity of phosphoglucomutase. Biochem Genet. 1970;4:1–9. doi: 10.1007/BF00484014. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Johansson M, Sandberg K, Andersson L. Cloning of highly polymorphic microsatellites in the horse. Anim Genet. 1992;23:133–142. doi: 10.1111/j.1365-2052.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Chowdhary BP, Johansson M, Marklund L, Fredholm M, Gustavsson I, Andersson L. A primary linkage map of the porcine genome reveals a low rate of genetic recombination. Genetics. 1994;137:1089–1100. doi: 10.1093/genetics/137.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen KR, Matthews ME. VIAS-H17 and VIAS-H34: Two new polymorphic equine microsatellite loci. Anim Genet. 1994a;25:63. [PubMed] [Google Scholar]
- ————— VIAS-H39, an equine tetranucleotide microsatellite repeat polymorphism. Anim Genet. 1994b;25:433. doi: 10.1111/j.1365-2052.1994.tb00542.x. [DOI] [PubMed] [Google Scholar]
- Fraser DG, Bailey E. Demonstration of three DRB loci in a domestic horse family. Immunogenetics. 1996;44:441–445. doi: 10.1007/BF02602805. [DOI] [PubMed] [Google Scholar]
- Fridolfsson A-K, Hori T, Winterø A-K, Fredholm M, Yerle M, Robic A, Andersson L, Ellegren H. Expansion of the pig comparative map by expressed sequence tags (EST) mapping. Mamm Genome. 1997;8:907–912. doi: 10.1007/s003359900609. [DOI] [PubMed] [Google Scholar]
- Fukushima H, de Wet JR, O’Brien SJ. Molecular cloning of a cDNA for human α-L-fucosidase. Proc Natl Acad Sci. 1985;82:1262–1265. doi: 10.1073/pnas.82.4.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahne B. Studies on the inheritance of electrophoretic forms of transferrins, albumins, prealbumins and plasma esterases of horses. Genetics. 1966;53:681–694. doi: 10.1093/genetics/53.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges M, Nielsen D, Mackinnon M, Mishra A, Okimoto R, Pasquino AT, Sargeant LS, Sorensen A, Steele MR, Zhao X, Womack JE, Hoeschele I. Mapping quantitative trait loci controlling milk production in dairy cattle by exploiting progeny testing. Genetics. 1995;139:907–920. doi: 10.1093/genetics/139.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godard S, Vaiman D, Oustry A, Nocart M, Bertaud M, Guzylack S, Mériaux JC, Cribiu EP, Guérin G. Characterization, genetic and physical mapping analysis of 36 horse plasmid and cosmid-derived microsatellites. Mamm Genome. 1997;8:745–750. doi: 10.1007/s003359900558. [DOI] [PubMed] [Google Scholar]
- Godard S, Schibler L, Oustry A, Cribiu EP, Guérin G. Construction of a horse BAC library and cytogenetical assignment of 20 type I and type II markers. Mamm Genome. 1998;9:633–637. doi: 10.1007/s003359900835. [DOI] [PubMed] [Google Scholar]
- Green P, Falls KA, Crooks S. Documentation for CRI-MAP, version 2.4. St. Louis, MO: Washington University School of Medicine; 1990. [Google Scholar]
- Gu F, Harbitz I, Chowdhary BP, Chaudhary R, Gustavsson I. Localization of the 6-phosphogluconate dehydrogenase (PGD) gene in horses by in situ hybridization. Hereditas. 1992;117:93–95. doi: 10.1111/j.1601-5223.1992.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Gu F, Harbitz I, Chowdhary BP, Davies W, Gustavsson I. Mapping of the porcine lipoprotein lipase (LPL) gene to chromosome 14q12-q14 bands by in situ hybridisation. Cytogenet Cell Genet. 1992;59:63–64. doi: 10.1159/000133201. [DOI] [PubMed] [Google Scholar]
- Guérin G, Bertaud M. Characterization of two polymorphic horse microsatellites: HMS15 and HMS20. Anim Genet. 1996;27:123. [PubMed] [Google Scholar]
- Guérin G, Bertaud M, Amigues Y. Characterization of seven new horse microsatellites: HMS1, HMS2, HMS3, HMS5, HMS6, HMS7 and HMS8. Anim Genet. 1994;25:62. [PubMed] [Google Scholar]
- Harbitz I, Kristensen T, Davies W. Isolation and sequencing of porcine lipoprotein lipase cDNA and its use in multiallelic restriction fragment length polymorphism detection. Anim Genet. 1992;23:517–522. doi: 10.1111/j.1365-2052.1992.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Hasler-Rapacz J, Ellegren H, Fridolfsson A-K, Kirkpatrick B, Kirk S, Andersson L, Rapacz J. Identification of a mutation in the low density lipoprotein receptor gene associated with recessive familial hypercholesterolemia in swine. Am J Med Genet. 1998;76:379–386. [PubMed] [Google Scholar]
- Irvin Z, Giffard J, Brandon R, Breen M, Bell K. Equine dinucleotide repeat polymorphisms at loci ASB21, 23, 25 and 37-43. Anim Genet. 1998;29:67. [PubMed] [Google Scholar]
- Juneja RK, Gahne B, Stratil A. Polymorphic plasma postalbumins of some domestic animals (pig PO2, horse Xk, and dog Pa proteins) identified as homologous to human plasma alpha 1B-glycoprotein. Anim Genet. 1987;18:119–124. doi: 10.1111/j.1365-2052.1987.tb00750.x. [DOI] [PubMed] [Google Scholar]
- Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TP, Lopez-Corrales NL, Beattie CW. A second-generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- Lear TL, Irwin Z, Brandon R, Bell K, Mathiason K, Bailey E. Proceedings, plant and animal genome VI. 1998. Physical assignment of microsatellite markers to horse chromosomes using FISH; p. 162. . San Diego, CA. [Google Scholar]
- Lear TL, Adams MH, McDowell KJ, Sullivan ND, Coogle L, Ferguson E, Jr, Chambers TM, Bailey E. Proceedings of the 10th North American colloquium on gene mapping and cytogenetics in human and domestic species. 1997. Chromosomal location of the genes for ESR, ETS2, GOT2, KIT, MX1 and PGR in the horse, Equus caballus; p. 24. . Apalachicola, FL. [Google Scholar]
- Lindgren, G., H. Persson, and H. Ellegren. 1998. Five equine dinucleotide microsatellite loci HTG17, HTG20, HTG21, HTG28 and HTG31. Anim. Genet. (in press). [DOI] [PubMed]
- Long SE. Chiasma counts and non-disjunction frequencies in a normal ram and in rams carrying the Massey I (t1) Robertsonian translocation. J Reprod Fertil. 1978;53:353–356. doi: 10.1530/jrf.0.0530353. [DOI] [PubMed] [Google Scholar]
- Logue DN. Meiosis in the domestic ruminants with particular reference to Robertsonian translocations. Ann Genet Sel Anim. 1977;9:493–507. doi: 10.1186/1297-9686-9-4-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan DH, Duff C, Zorzato F, Fujii J, Philips M, Korneluk RG, Frodis W, Britt BA, Worton RG. Ryanodine receptor gene is a candidate for predisposition to malignant hyperthermia. Nature. 1990;343:559–561. doi: 10.1038/343559a0. [DOI] [PubMed] [Google Scholar]
- Marklund S, Ellegren H, Eriksson S, Sandberg K, Andersson L. Parentage testing and linkage analysis in the horse using a set of highly polymorphic microsatellites. Anim Genet. 1994;25:19–23. [PubMed] [Google Scholar]
- Marklund L, Johansson Moller M, Høyheim B, Davies W, Fredholm M, Juneja RK, Mariani P, Coppieters W, Ellegren H, Andersson L. A comprehensive linkage map of the pig based on a wild pig–Large White intercross. Anim Genet. 1996a;27:255–269. doi: 10.1111/j.1365-2052.1996.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Marklund L, Johansson Moller M, Sandberg K, Andersson L. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm Genome. 1996b;7:895–899. doi: 10.1007/s003359900264. [DOI] [PubMed] [Google Scholar]
- Marti E, Breen M, Fischer P, Swinburne J, Binns MM. Six new cosmid derived and physically mapped equine dinucleotide repeat microsatellites. Anim Genet. 1998;29:236–238. doi: 10.1046/j.1365-2052.1998.00236.x. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Langston AA, Acland GM, Fleming MA, Ray K, Wiegand NA, Francisco LV, Gibbs M, Aguirre GD, Ostrander EA. A linkage map of the canine genome. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Lord EA, Penty JM, Dodds KG, Broad TE, Cambridge L, Sunden SLF, Stone RT, Crawford A. The Booroola fecundity (FecB) gene maps to sheep chromosome 6. Genomics. 1994;22:148–153. doi: 10.1006/geno.1994.1355. [DOI] [PubMed] [Google Scholar]
- Morton NE. Parameters of the human genome. Proc Natl Acad Sci. 1991;88:7474–7476. doi: 10.1073/pnas.88.17.7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, Allard WJ, Lienhard GE, Lodish HF. Sequence and structure of a human glucose transporter. Science. 1985;229:941–945. doi: 10.1126/science.3839598. [DOI] [PubMed] [Google Scholar]
- Oakenfull EA, Buckle VJ, Clegg JB. Localization of the horse (Equus caballus) α-globin gene complex to chromosome 13 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1993;62:136–138. doi: 10.1159/000133456. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Frönicke L, Scherthan H, Gustavsson I, Chowdhary BP. Zoo-FISH delineates conserved chromosomal segment in horse and man. Chromosome Res. 1996;4:218–225. doi: 10.1007/BF02254963. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Otte K, Rozell B, Chowdhary BP. FISH mapping of the IGF2 gene in horse and donkey—detection of homoeology with HSA11. Mamm Genome. 1997;8:569–572. doi: 10.1007/s003359900505. [DOI] [PubMed] [Google Scholar]
- Rohrer GA, Alexander LJ, Hu Z, Smith TP, Keele JW, Beattie CW. A comprehensive map of the porcine genome. Genome Res. 1996;6:371–391. doi: 10.1101/gr.6.5.371. [DOI] [PubMed] [Google Scholar]
- Rudolph JA, Spier SJ, Byrns G, Rojas CV, Bernoco D, Hoffman EP. Periodic paralysis in Quarter horses: A sodium channel mutation disseminated by selective breeding. Nat Genet. 1992;2:144–147. doi: 10.1038/ng1092-144. [DOI] [PubMed] [Google Scholar]
- Sakagami M, Tozaki T, Mashima S, Hirota K, Mukoyama H. Equine parentage testing by microsatellite locus at chromosome 1q2.1. Anim Genet. 1995;26:123–124. doi: 10.1111/j.1365-2052.1995.tb02647.x. [DOI] [PubMed] [Google Scholar]
- Sandberg K. Genetic polymorphism in carbonic anhydrase from horse erythrocytes. Hereditas. 1968;60:411–412. [Google Scholar]
- ————— The D blood group system of the horse. Anim Blood Groups Biochem Genet. 1973;4:193–205. doi: 10.1111/j.1365-2052.1973.tb01300.x. [DOI] [PubMed] [Google Scholar]
- Sandberg K, Andersson L. Horse (Equus caballus) In: O’Brien SJ, editor. Genetic maps. 6th ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 4.276–4.278. [Google Scholar]
- Santschi EM, Purdy AK, Valberg SJ, Vrotsos PD, Kaese H, Mickelson JR. Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mamm Genome. 1998;9:306–309. doi: 10.1007/s003359900754. [DOI] [PubMed] [Google Scholar]
- Scott IS, Long SE. An examination of chromosomes in the stallion (Equus caballus) during meiosis. Cytogenet Cell Genet. 1980;26:7–13. doi: 10.1159/000131415. [DOI] [PubMed] [Google Scholar]
- Seidel U, Bober E, Winter B, Lenz S, Lohse P, Arnold HH. The complete nucleotide sequences of cDNA clones coding for human myosin light chains 1 and 3. Nucleic Acids Res. 1987;15:4989. doi: 10.1093/nar/15.12.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EK, Perryman LE, Meek K. A kinase-negative mutation of DNA-PKCS in equine SCID results in defective coding and signal joint formation. J Immunol. 1997;158:3565–3569. [PubMed] [Google Scholar]
- Shiue, Y., L.A. Bickel, A.R. Caetano, L.V. Millon, R.S. Clark, M.L. Eggleston, R. Michelmore, E. Bailey, G. Guérin, S. Godard, J.R. Mickelson, S.J. Valberg, J.D. Murray, and A.T. Bowling. 1998. A synteny map of the horse genome comprised of 240 microsatellite and RAPD markers. Anim. Genet. (in press). [DOI] [PubMed]
- Tozaki T, Sakagami M, Mashima S, Hirota K, Mukoyama H. ECA-3: Equine (CA) repeat polymorphism at chromosome 2p1.3-4. Anim Genet. 1995;26:283. doi: 10.1111/j.1365-2052.1995.tb03265.x. [DOI] [PubMed] [Google Scholar]
- Vaiman D, Schibler L, Ourgeois F, Oustry A, Amigues Y, Cribiu EP. A genetic linkage map of the male goat genome. Genetics. 1996;144:279–305. doi: 10.1093/genetics/144.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haeringen H, Bowling AT, Stott ML, Lenstra JA, Zwaagstra KA. A highly polymorphic horse microsatellite locus: VHL20. Anim Genet. 1994;25:207. doi: 10.1111/j.1365-2052.1994.tb00129.x. [DOI] [PubMed] [Google Scholar]
- Vega-Pla JL, Garrido JJ, Dorado G, de Andrés-Cara DF. Three new polymorphic equine microsatellites: HLM2, HLM3, HLM5. Anim Genet. 1996;27:215. doi: 10.1111/j.1365-2052.1996.tb00961.x. [DOI] [PubMed] [Google Scholar]
- Weissenbach J, Gyapay G, Dib C, Vignal A, Morisette J, Millasseau P, Vaysseix G, Lathrop M. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]