Abstract
The modification of proteins with ubiquitin chains can change their localization, activity and/or stability. Although ubiquitylation requires the concerted action of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s), it is the E2s that have recently emerged as key mediators of chain assembly. These enzymes are able to govern the switch from ubiquitin chain initiation to elongation, regulate the processivity of chain formation and establish the topology of assembled chains, thereby determining the consequences of ubiquitylation for the modified proteins.
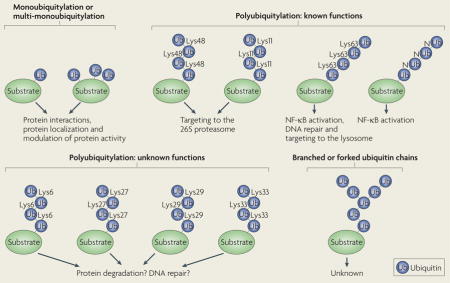
Ubiquitylation is a powerful mechanism for regulating most aspects of cell physiology1,2. Substrate proteins can be modified with a single ubiquitin on one (mono-ubiquitylation) or multiple (multi-monoubiquitylation) sites (BOX 1). Alternatively, the substrates can be modified with a chain of ubiquitin molecules (polyubiquitylation), which are linked through one of the seven Lys residues or the amino terminus of ubiquitin3–6. Ubiquitin chains containing branches (two ubiquitin molecules linked to a single ubiquitin within a chain) or a mixture of different linkages exist, but the physiological relevance of these modifications remains unclear7–9.
Box 1. The ubiquitin code.
Ubiquitin is usually attached to the ε-amino group of Lys residues in substrates (see the figure). The transfer of a single ubiquitin to one (monoubiquitylation) or multiple (multi-monoubiquitylation) sites can recruit binding partners, inhibit interactions, change protein localizations or modulate protein activities1,2. Ubiquitin itself contains seven Lys residues, which can function as acceptor sites for another ubiquitin moiety during the assembly of ubiquitin chains. In addition, the amino terminus of a substrate-linked ubiquitin (see the figure; N) can serve as an acceptor for the formation of linear ubiquitin chains. Depending on the connection between li nked ubiquitin molecules, ubiquitin chains can differ in structure and function. Lys48- and Lys11-linked ubiquitin chains target proteins for degradation by the 26S proteasome. Lys63-linked chains usually mediate the recruitment of binding partners, which can lead to activation of nuclear factor-κB (NF-κB), orchestration of different steps during DNA repair or targeting of the modified protein to the lysosome. Other ubiquitin chains, such as Lys6- or Lys29-linked chains, have been detected in vitro or in vivo, but substrates or enzymes responsible for their assembly are poorly defined. Branched or forked ubiquitin chains result from the attachment of two ubiquitin molecules to two different Lys residues in a ubiquitin that is already linked to a substrate, but their significance in cellular regulation has not yet been established.
Substrates modified with ubiquitin are recognized in cells by a cohort of proteins containing ubiquitin-binding motifs such as the ubiquitin-interacting motif (UIM) and ubiquitin-associated (UBA) domains10,11. Many of these ubiquitin-binding ‘decoder’ proteins preferentially associate with a distinct conjugate, such as monoubiquitin or a ubiquitin chain of a specific linkage. These proteins also bind to downstream effectors of signalling pathways and thereby couple ubiquitylation to the desired biological outcome. For example, proteins recognizing Lys48-linked ubiquitin chains escort substrates to the 26S proteasome for degradation12–14, whereas those binding Lys63-linked chains mediate the activation of the transcription factor nuclear factor-κB (NF-κB) or orchestrate different steps in the DNA repair programme5,15–18. The interplay between the different forms of ubiquitylation and their recognition by a plethora of binding partners defines the highly complex ‘ubiquitin code’ (BOX 1).
Specific ubiquitylation of the thousands of human substrates depends on the sequential action of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). The human genome encodes 2 E1s, at least 38 E2s and 600–1,000 E3s19,20. E1s use ATP to generate a thioester bond between the Cys at their active site and the carboxyl terminus of ubiquitin. This ubiquitin is then transferred to the Cys residue in the active site of an E2, which in turn cooperates with two different classes of E3s to modify the substrates (FIG. 1a). For the ~60 HECT family E3s, ubiquitin is shuttled from an E2 to a Cys in the HECT domain of the E3 before being attached to a substrate21. Most E3s, however, contain a RING domain or a structurally related U-box and act as matchmakers, bringing a substrate and a charged E2 together and activating the E2 to ligate ubiquitin to a Lys in the substrate3.
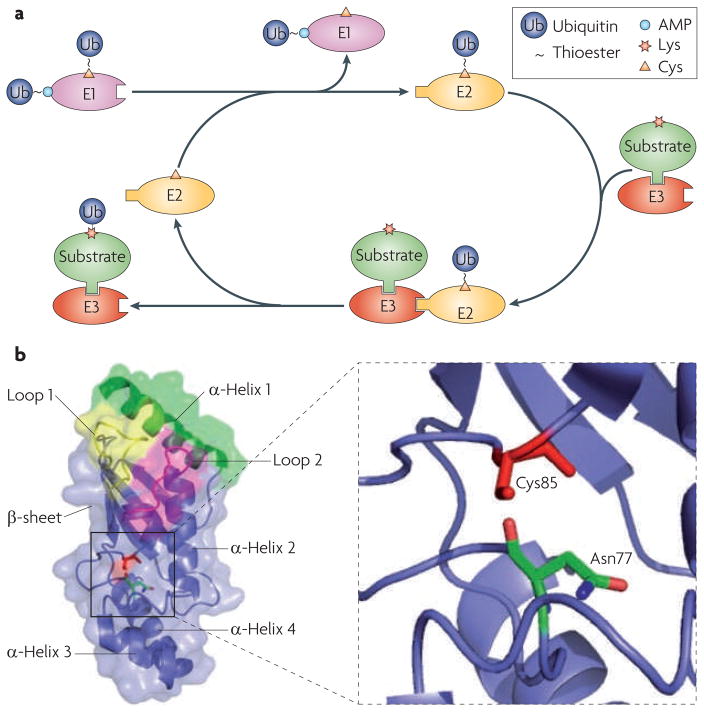
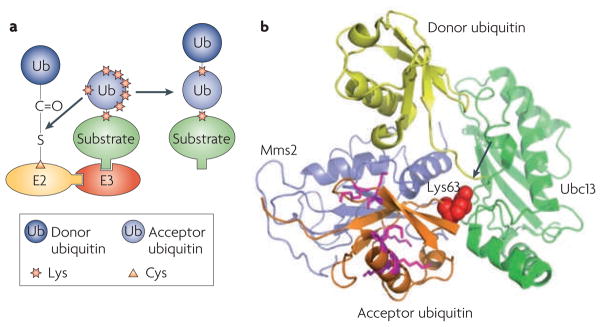
Figure 1. Ubiquitylation from an e2 perspective.
a. Schematic overview of ubiquitylation. A ubiquitin-conjugating enzyme (E2) first interacts with the ubiquitin-activating enzyme (E1) that has been loaded with two ubiquitin molecules (one at its adenylation domain as an adenylate (~AMP) and the other linked to a Cys at its active site as a thioester). The activated ubiquitin is transferred to the Cys in the E2 active site. The E2 has to dissociate from the E1 before it engages with a cognate ubiquitin ligase (E3), which recruits substrates. Once ubiquitin has been transferred to the substrate, the E2 dissociates from the E3, allowing it to be recharged with ubiquitin for the next round of transfers. b. Structure of the core ubiquitin-conjugating (UBC) domain of the E2 UBE2D224. The UBC core is comprised of four α-helices and an antiparallel β-sheet formed by four β–strands. The E1-interacting α-helix 1 is shown in green, and the two E3-interacting loops are shown in yellow (loop 1) and magenta (loop 2). The close-up view shows the relative spatial positions of the side chains of the active-site Cys85 and Asn77.
Because of their capacity to recruit specific substrates, much of the initial work in the ubiquitin field focused on E3s, and E2s were often considered as ‘ubiquitin carriers’ with auxiliary roles. However, many recent studies have revealed that E2s have an active role in determining the length and topology of ubiquitin chains and the processivity of the chain assembly reaction. Here, we integrate these findings into a model of E2 function, underscoring the importance of E2s for chain assembly and determining the cellular consequences of ubiquitylation.
Interactions of E2s during chain assembly
The E2 family comprises 13 genes in Saccharomyces cerevisiae and at least 38 genes in humans (BOX 2; see Supplementary information S1 (table)). Human E2s can be classified into 17 subfamilies based on phylogenetic analyses22. Active E2s possess a core ubiquitin-conjugating (UBC) domain, which contains the catalytic Cys residue and interacts with E1s. Ubiquitin E2 variant (UEV) proteins also have a UBC domain but lack an active-site Cys residue. The UBC domains from different E2s have a high degree of sequence homology and adopt similar structures comprised of four α-helices, an anti-parallel β-sheet formed by four strands, and a short 310-helix23–25. The highly conserved active-site Cys is located in a shallow groove formed by a short loop connecting α-helix 2 with α-helix 3 and a long loop proximal to the active site (FIG. 1b).
Box 2. The E2 nomenclature.
The nomenclature of ubiquitin-conjugating enzymes (E2s) is currently confusing. When the first E2 genes were cloned researchers mostly used the form E2-nK (where n denotes the molecular weight of the E2) and UBCn in yeast or UBCHn in humans (where n corresponds to the order of discovery). Other E2s were labelled following their discovery in genetic or proteomic screens, without a reference to their E2 function, for example Huntingtin-interacting protein 2 (HIP2; also known as E2-25K, UBCH1 and UBE2K). As a result, E2s from different organisms bearing the same number are often not functionally related, and most E2s have multiple names. To unify the nomenclature of mammalian E2s, we therefore suggest that a system based on the bioinformatics-driven identification of all predicted human E2s is used. This system uses the form UBE2Xn, where the combination of letter ‘X’ and number ‘n’ specifies different E2s (see Supplementary information S1 (table)).
Interaction of E2s with E1s
The first important task of a ubiquitin E2 is to ensure that it receives ubiquitin, but not related ubiquitin-like modifiers (UBLs), on its active site. UBLs are structurally similar to ubiquitin and they are also conjugated to substrate Lys residues with the aid of specific E1s, E2s and E3s. The modification of proteins with UBLs usually modulates protein localization or activation1. Ubiquitin and UBLs can compete for the same Lys in substrates to trigger different reactions. For example, monoubiquitylated proliferating cell nuclear antigen promotes translesion synthesis DNA repair, whereas modification of the same Lys residue with the UBL small ubiquitin-like modifier (SUMO) blocks recombination between sister chromatids17.
Although E1s and E2s for UBLs have structures similar to those of the corresponding enzymes for ubiquitin, E2s for ubiquitin specifically interact with the two E1s of the ubiquitin pathway19. In general, E2s bind their cognate E1s with significant affinity only if the E1 is carrying their modifier19,26. A series of impressive structural analyses revealed that charging of an E1 with ubiquitin or a UBL triggers conformational changes in the E1, which exposes cryptic E2-binding sites and allows the formation of the proper E1–E2 complex27–29. During these rearrangements, a negatively charged groove within a ubiquitin fold domain (UFD) in the E1 becomes available for recognition by two highly conserved Lys residues present in α-helix 1 of all ubiquitin E2s but absent in E2s for the UBLs small ubiquitin- like modifier and neuronal precursor cell-expressed developmentally downregulated protein 8 (NEDD8) (FIG. 2a). Subtle and not yet fully understood differences in the UFDs of the two human ubiquitin E1s, UBE1 (also known as UBA1) and UBE1L2 (also known as UBA6), allow ubiquitin E2s to discriminate between them30. Sequences outside the UBC domain of E2s also contribute to the specificity of E1 binding. The N-terminal extension of the NEDD8 E2 UBE2M, for example, stabilizes the interaction between UBE2M and its E1, and at the same time reduces its affinity for the ubiquitin E1 (REFS 31,32). Some ubiquitin E2s, such as UBE2C, contain similar N-terminal appendices, which decrease their affinity for their E1 and thereby reduce the efficiency by which ubiquitin is transferred to their active site32–34.
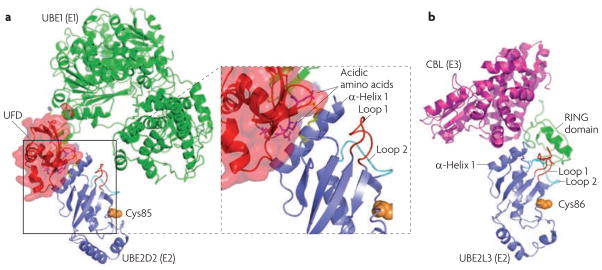
Figure 2. Structural representation of e2 interactions.
a. Structural model of the ubiquitin-conjugating enzyme (E2)–ubiquitin-activating enzyme (E1) interaction. The example shown is the E2 UBE2D2 in complex with the E1 UBE1 (also| known as UBA1) (Protein Data Bank (PDB) code 3CMM)29. The structure of UBE2M (the neuronal precursor cell-expressed developmentally downregulated protein 8 (NEDD8) E2) in complex with the ubiquitin fold domain (UFD) of the corresponding E1 is used as a reference (PDB code 1Y8X)55. The enlarged view shows that the UFD contains a groove formed by acidic amino acids, which are proposed to interact with the side chains of two Lys residues (yellow) in α-helix 1 of the E2. b. The structure of the E2–ubiquitin ligase (E3) interaction. The example shown is the E2 UBE2L3 in complex with the RING E3 casitas B-lineage lymphoma (CBL) (PDB code 1FBV)44. Note that the E2 has overlapping binding sites on α-helix 1 for the E1 and the E3. The two loops that contact the RING domain and the catalytic Cys86 are also shown.
Interactions of E2s with E3s
After being charged with ubiquitin, E2s engage E3s to catalyse substrate ubiquitylation. A single E2 can interact with several different E3s — this is seen most dramatically with members of the UBE2D family of E2s. Although the analysis of E2–E3 interactions in vitro is straightforward, determining the physiological E2–E3 pairs, especially in human cells, is more difficult. Well characterized physiological E2–E3 pairs include the yeast Skp–cullin–F-box protein (SCF) and the E2 cell division cycle 34 (Cdc34)35; the human anaphase-promoting complex or cyclosome (APC/C) and the E2s UBE2C and UBE2S33,36; the human TNF receptor-associated factor 6 (TRAF6) and the heterodimeric E2 UBE2N–UBE2V137; the endoplasmic reticulum (ER)-resident E3 gp78 and the E2 UBE2G238,39; the E3 radiation sensitive protein 18 (RAD18) and UBE2A, which function during DNA repair40,41; and fanconi anemia group L protein (FANCL) and UBE2T, which monoubiquitylate fanconi anaemia complementation group D2 protein (FANCD2) during DNA repair42,43. Because an E2’s choice of E3 can influence the outcome of substrate ubiquitylation, developing means to understand the rules that govern E2–E3 pairing will be an important avenue of future research.
All E2s characterized so far recognize E3s through the L1 and L2 loops and the N-terminal α-helix 1 on the E2 surface. Slight sequence variations in these motifs contribute to the specificity of E3 binding (FIG. 2b). For example, the E2 UBE2L3 uses Pro62 and Phe63 in the L1 loop, Pro97 and Ala98 in the L2 loop, and Arg5 and Arg15 in α-helix 1 to interact with the E3 casitas B-lineage lymphoma (CBL), a proto-oncogene that negatively regulates receptor Tyr kinase signalling pathways44. By contrast, the relevant E2 residues for the interaction between UBE2D2 and the E3 CNOT4 (CCR4–NOT transcription complex, subunit 4) are Asp59 and Lys63 in the L1 loop, Ser94 and Ala96 in the L2 loop, and Lys4 in α-helix 1 (REF. 45). When a single E2 interacts with multiple E3s, the E2 residues involved in E3 recognition are not necessarily the same: UBE2N uses Arg6 and Lys10 to recognize residues upstream of the RING domain of the E3 TRAF6, whereas Arg7 and Lys10 of UBE2N mediate its contact to the U-box of C terminus of HSC70 interacting protein (CHIP; also known as STUB1)46,47 (FIG. 3a). Similar to the association with E1s, additional components unique to E2s can further increase the specificity of E3 binding. The yeast E2 Rad6 interacts with a specific motif in Rad18 independently of the RING domain48, and Cdc34 uses a negatively charged C-terminal tail to bind to its E3, SCF49,50. Likewise, human UBE2G2 uses Leu163 and Leu165 to bind a second motif, the G2BR motif, in the E3 gp78, which significantly increases the affinity of this E2–E3 interaction39,51,52 (FIG. 3b). Interestingly, if a G2BR peptide is added in trans, the affinity of UBE2G2 for the RING domain of gp78 is increased52, indicating that different binding sites can cooperate to achieve the optimal affinity of an E2 for its E3.
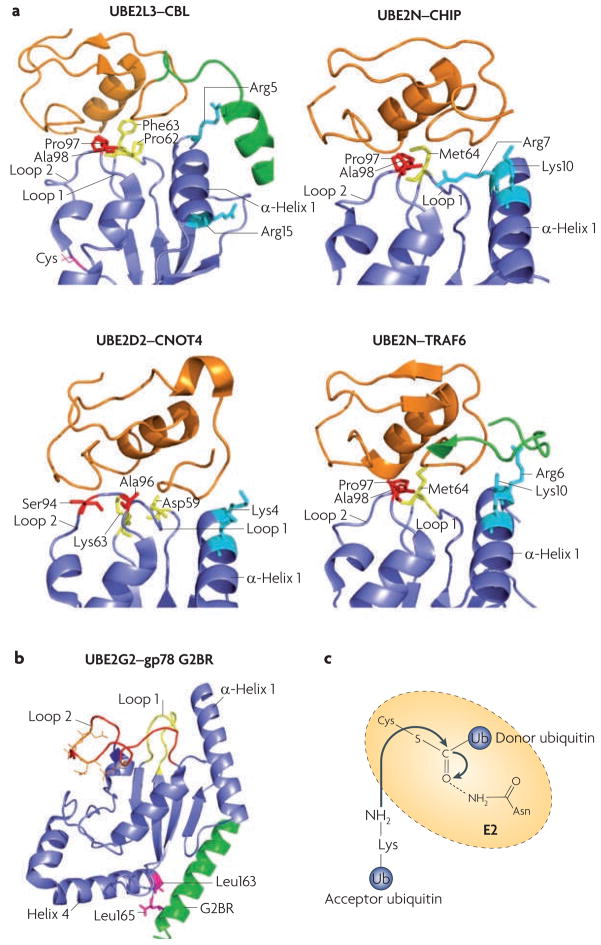
Figure 3. E2–E3 interaction specificity and the proposed mechanism of E2 catalysis.
a. Comparison of different ubiquitin-conjugating enzyme (E2)–ubiquitin ligase (E3) interactions: UBE2L3–casitas B-lineage lymphoma (CBL) (Protein Data Bank (PDB) code 1FBV44), UBE2N–carboxy terminus of HSC70 interacting protein (CHIP; also known as STUB1) (PDB code 2C2V46), UBE2D2–CNOT4 (CCR4–NOT transcription complex, subunit 4) (PDB code 1UR645) and UBE2N–TNF receptor associated factor 6 (TRAF6) (PDB code 3HCT47). All E2s use loop 1, loop 2 and α-helix 1 to interact with E3s. The side chains of the residues involved in E3 recognition are colour-coded (residues in loop 1 are yellow, in loop 2 are red and in α-helix 1 are cyan). Note that for CBL and TRAF6, E3 elements outside the RING domain (shown in green) participate in E2 binding. The CNOT4–UE2D2 structure is an NMR model, whereas the others were solved by crystallography. The RING finger (or the U-Box for CHIP) is in orange, and the cognate E2s are in blue. b. An example structure of a RING-independent E2–E3 interaction: that between UBE2G2 (blue) and the G2BR peptide from gp78 (also known as AMFR; green) (PBD code 3FSH)51. The E3 peptide interacts with two Leu residues of the E2, at a position remote from the predicted RING binding site (loop 1, loop 2 and α-helix 1). Acidic residues in the UBE2G2 acidic loop are in orange. c. A catalysis model for isopeptide bond formation during ubiquitin chain synthesis. The side chain of a conserved Asn residue in the E2 is proposed to interact with the active-site Cys (carrying the donor ubiquitin), which stabilizes the oxyanion transition state of the nucleophilic attack by the Lys residue of the acceptor ubiquitin.
The interactions between E2s and E3s are usually weak, with dissociation constants in the micromolar range. UBE2N, for example, binds the E3 TRAF6 with a dissociation constant of 1.2 μM, and the affinity of UBE2N for the isolated RING domain of TRAF6 is even lower, with a dissociation constant of 2 mM47. Although this weak interaction with RING or HECT domains takes place on an E2 surface distant from the active site, the binding of an E3 is required for full E2 activity. Indeed, E3s substantially increase the rate of ubiquitin discharge from E2 active sites24,52,53. It is believed that E3s induce conformational changes in E2s that position a crucial Asn near the active site to stabilize an oxyanion intermediate in the transition state24,54 (FIGS. 1b, 3c). A highly conserved hydrophobic residue (Ile88 in UBE2D2) was suggested to mediate the communication between E3 binding and the E2 active site24, but further structural work is required to fully understand the mechanism underlying the allosteric activation of E2s.
The low affinity of E2s for E3s is probably advantageous for ubiquitin chain formation because E2s use overlapping surfaces for interacting with E1s and E3s55,56 (FIG. 2a,b). Therefore, an E2 cannot be recharged by an E1 while bound to a cognate E3, but instead has to commit to several rounds of E3 binding and dissociation during ubiquitin chain assembly. Once dissociated, recharging of E2s seems to take place rapidly as free E2s are mostly loaded with ubiquitin at steady state in cells30,57. As discussed above, some E2s increase their capacity to stimulate chain formation by recognizing additional sites on their E3s, distinct from the RING or HECT domains. It is tempting to speculate that such E3-binding sites could allow an E2 to disengage from the RING domain for recharging while remaining associated with an E3. Alternatively, the RING-independent binding sites might increase the kinetics of an E2 rebinding to an E3 after being charged by an E1.
Interaction of E2s with cofactors
In addition to cycling between E1s and E3s, some E2s bind cofactors that influence their localization, activity or specificity. This is best understood for UEV proteins. UEV proteins contain a UBC domain but lack a catalytic Cys residue58. Instead of having catalytic activity of their own, they bind an active E2 and regulate its activity or linkage specificity (discussed below). Another well-characterized E2 cofactor is the yeast coupling of ubiquitin conjugation to ER degradation protein 1 (Cue1), a transmembrane protein of the ER that uses a C-terminal motif to recruit the E2 Ubc7 (REFS 59,60). The association with Cue1 also increases the ubiquitylation activity of Ubc7 (REFS 60,61) and protects Ubc7 from autoubiquitylation and proteasomal degradation62. These properties allow Cue1 to focus Ubc7 activity on substrates at the ER membrane and, accordingly, Cue1 is required for Ubc7-dependent ER-associated degradation (ERAD)59.
Whereas the association of E2s with UEV proteins or Cue1 occurs independently of the E2s’ activity, some E2s bind cofactors only when charged with ubiquitin. An intriguing example of this type of interaction is the coupled monoubiquitylation that is observed for many regulators of endocytosis63,64. During endocytosis, proteins such as epidermal growth factor receptor pathway substrate 15 (Eps15) use a diverse set of ubiquitin-binding domains (UBDs) to interact with ubiquitylated endocytic cargos. These UBDs also recognize ubiquitin attached to the Cys at the active site of an E2 (REF. 64), and the resulting association with the E2 leads to the robust monoubiquitylation of the UBD-containing protein. It is not yet fully understood whether the interaction with UBD-containing proteins provides a means of regulating the activity of E2s. In a variation on this theme, the E2s UBE2E2 and UBE2E3 associate with the nuclear transport receptor importin 11 only when they are charged with ubiquitin65. This interaction results in the selective nuclear translocation of charged, and presumably active, E2s65. Together, these observations reveal an intriguing set of specific, dynamic and tightly regulated interactions for the small E2s, placing them at a central position within the ubiquitylation cascade.
Roles of E2s during ubiquitin chain assembly
The complexity of the interactions carried out by E2s contradicts their early image as simple carriers of activated ubiquitin. Indeed, as we discuss below, it is now evident that E2s can determine the linkage specificity and length of ubiquitin chains and can strongly influence the processivity of chain formation.
Switching from ubiquitin chain initiation to elongation
The assembly of ubiquitin chains is usually initiated by the transfer of the first ubiquitin to a Lys on a substrate. Subsequently, the E2–E3 pair switches to chain elongation, during which additional ubiquitin molecules are attached to the substrate-linked ubiquitin. The decision of whether a Lys residue in the substrate or in the ubiquitin will receive the next ubiquitin is often made by the E2, and E2s with dedicated roles in ubiquitin chain initiation and elongation have recently been described.
In an example of the division of labour of E2s between ubiquitin chain initiation and elongation, the yeast APC/C uses Ubc4 to modify Lys residues in substrates and Ubc1 to extend Lys48-linked ubiquitin chains66 (FIG. 4a). Similarly, the human Ubc4 homologues of the UBE2D family prefer to modify Lys residues in substrates when incubated with APC/C in vitro, but the importance of UBE2D for APC/C activity in cells is unclear8,34,67. In the case of the heterodimeric E3 breast cancer type 1 susceptibility protein (BRCA1)–BRCA1-associated RING domain 1 (BARD1), several E2s, including UBE2W and UBE2E2, function in ubiquitin chain initiation, whereas the heterodimer UBE2N–UBE2V1 and UBE2K specifically promote chain elongation68.
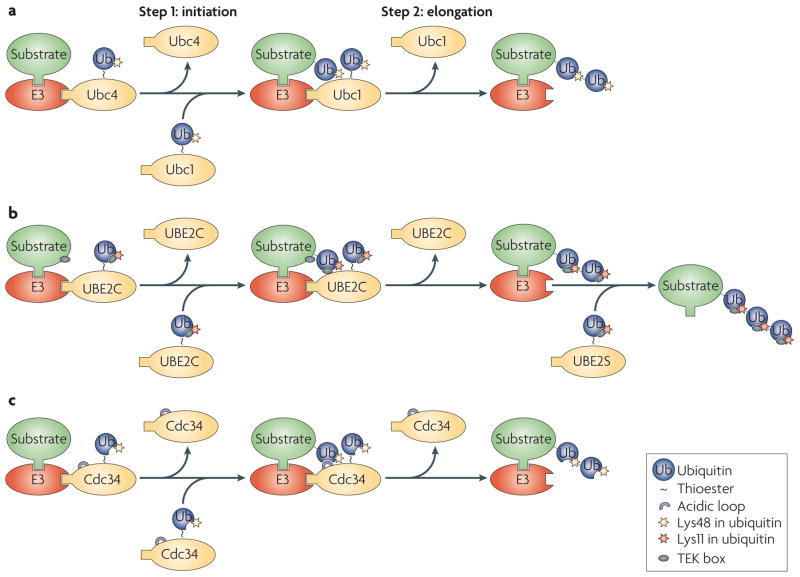
Figure 4. Mechanisms for ubiquitin chain initiation and elongation.
a. The yeast anaphase-promoting complex or cyclosome (APC/C) uses the ubiquitin-conjugating enzyme (E2) Ubc4 to initiate ubiquitylation, and a different E2, Ubc1, to elongate Lys48-linked ubiquitin chains. b. UBE2C recognizes a TEK box in the substrate to initiate ubiquitylation. Once a ubiquitin moiety has been added to the substrate, a similar TEK box present in ubiquitin takes over to promote elongation of the Lys11-linked ubiquitin chain. The efficient extension of Lys11-linked ubiquitin chains requires a second E2, UBE2S. c. The yeast E2 cell division cycle 34 (Cdc34) can initiate and elongate ubiquitin chains. Efficient chain elongation requires the acidic loop of Cdc34, which might orient the attacking Lys group relative to the E2 active site (charged with the ubiquitin thioester) to facilitate the formation of ubiquitin–ubiquitin linkage. E3, ubiquitin ligase.
Some E2s involved in ubiquitin chain initiation, especially those of the UBE2D family, lack specificity for a Lys residue in the substrate, which allows them to initiate chain formation on a diverse set of substrates for multiple E3s8. By contrast, other E2s are more selective in promoting ubiquitin chain initiation and potentially recognize substrate residues in proximity to the modified Lys. An example of a selective type of E2 is UBE2T, which ubiquitylates specific Lys residues in its substrate FANCD2 but lacks any ubiquitin chain extension activity and does not cooperate with chain-elongating E2s43. UBE2T thereby catalyses the monoubiquitylation of FANCD2.
E2s involved in ubiquitin chain elongation often depend on the prior attachment of the first ubiquitin to a Lys residue in the substrate. Both the Lys63-specific chain-elongating E2 UBE2N–UBE2V1 (yeast Ubc13–Mms2) and the Lys11-specific chain-elongating E2 UBE2S lack the capability for ubiquitin chain initiation36,68–70. Accordingly, formation of Lys63-linked ubiquitin chains on yeast proliferating cell nuclear antigen during post-replicative DNA repair is initiated by the E2 Rad6, before Ubc13–Mms2 can extend the chains17. In a similar manner, the E3 TRAF6 probably uses UBE2D to initiate and UBE2N–UBE2V1 to elongate Lys63-linked ubiquitin chains during activation of the transcription factor NF-κB69. Most E2s involved in ubiquitin chain elongation interact with the substrate-attached ubiquitin, which results in the striking specificity of UBE2S for Lys11-linked chains, UBE2K for Lys48-linked chains and UBE2N–UBE2V1 for Lys63-linked chains58,71,72.
In contrast to a strict separation of ubiquitin chain initiation and elongation steps, some initiating E2s extend short ubiquitin chains, before the elongating E2 takes over, thereby increasing the rate of ubiquitin chain formation. An example is the E2 UBE2C, which initiates the formation of Lys11-linked ubiquitin chains on the human E3 APC/C, promoting the degradation of a large family of substrates during the short time span of mitosis73. This reaction is strongly promoted by a Lys-rich region in the substrates, the TEK box, which is located ~20 residues downstream of the APC/C-binding D box or KEN box67. A similar TEK box motif is found in ubiquitin close to Lys11 (Lys6, Leu8 or Thr9 of ubiquitin), which strongly promotes the formation of short Lys11-linked ubiquitin chains by UBE2C (FIG. 4b). These chains are subsequently elongated by the Lys11-specific chain-elongating E2 UBE2S36. Similar to mutation of Lys11 or the TEK box in ubiquitin, the co-depletion of UBE2C and UBE2S abrogates APC/C activity in cells, implying that the cooperation between these particular chain-initiating and chain-elongating E2s is essential for the human APC/C.
Finally, a few E2s catalyse both the initiation and the elongation of specific ubiquitin chains. The yeast E2 Cdc34, for example, cooperates with the E3 SCF to add Lys48-linked ubiquitin chains to the cell cycle inhibitor subunit inhibitor of cyclin-dependent kinase 1 (Sic1), triggering Sic1 degradation and entry of cells into S phase74. As Sic1 does not seem to interact with Cdc34, ubiquitin chain initiation by Cdc34 results from stochastic collisions between Sic1 and charged Cdc34 and, therefore, lacks strict specificity for Lys residues in Sic1 (REFS 53,75,76). As a result, the transfer of the first ubiquitin to Lys residues in Sic1 is slow. By contrast, chain elongation by Cdc34 is rapid and ubiquitin is only attached to Lys48 of an attached ubiquitin. The efficient elongation of the ubiquitin chain requires an interaction between an acidic loop located near the active site in Cdc34 and the substrate-attached ubiquitin to orient the attacking ε-amino group of Lys48 (REFS 53,77) (FIG. 4c). In support of this notion, Cdc34 lacking its acidic loop is defective in ubiquitin chain elongation, although it can effectively transfer the first ubiquitin to the SCF substrates Sic1 and IκBα53,77. Moreover, if two residues close to Lys48 (Ile44 and Gly47) are mutated in the acceptor ubiquitin, Cdc34 is unable to catalyse the formation of Lys48-linked ubiquitin dimers in an in vitro reaction that recapitulates the chain extension process53,77. Together, these observations demonstrate that the choice of E2 can be an effective mechanism to switch between ubiquitin chain initiation and the formation of specific ubiquitin chains in a regulated manner.
Controlling the processivity of ubiquitin chain formation
The substrate receptors of the 26S proteasome recognize ubiquitylated proteins only after a chain containing at least four ubiquitin molecules has been attached78. In the competitive environment of a cell, assembling long ubiquitin chains is not a trivial task, and it is limited by the availability of chain-elongating E2s, competition for a finite pool of E3s and opposition by deubiquitinases (DUBs; also known as deubiquitylating or deubiquitinating enzymes)79. A crucial factor in determining the probability of assembling a ubiquitin chain of sufficient length is the processivity of the chain formation reaction73,75,80.
The processivity of ubiquitylation is defined as the number of ubiquitin molecules transferred to a growing chain during a single round of substrate association with an E3 (REF. 80). It can be determined by the affinity of a substrate for its E3 (that is, how long the substrate remains bound to the E3 and therefore able to receive ubiquitin) and by the rate of ubiquitin transfer catalysed by the E2 (that is, how fast ubiquitin is transferred during the time a substrate is bound to an E3). The higher the processivity of chain assembly, the greater the likelihood that a substrate will receive a ubiquitin chain that is long enough to be recognized by the substrate receptors of the 26S protea-some. The processivity of a ubiquitylation reaction can have dramatic effects on the biological consequences of the reaction. For example, differences in the processivity of chain formation on APC/C substrates can determine the timing of their degradation during mitosis, which is required for cell cycle progression67,73.
Only a few E3s bind to their substrates with nanomolar affinity and, in most cases, the affinity for substrates is too low to allow the purification of stable E3–substrate complexes from cells. However, E3s are still able to ubiquitylate substrates with high processivity. One example is APC/C, which can rapidly form ubiquitin chains on securin even though securin is hardly ever detected in an APC/C complex73. What then allows APC/C to catalyse chain formation on securin so rapidly that it can occur in a single binding event? A partial answer to this question comes from the discovery of TEK boxes in securin and ubiquitin, as described above67. Consecutive recognition of these motifs by UBE2C allows ubiquitin chain initiation and elongation to occur in quick succession. The human APC/C also uses a dedicated chain-elongating E2, UBE2S, which further enhances processivity36.
An alternative mechanism underlying processive ubiquitin chain formation has been suggested for the E3 complex BRCA1–BARD1, which regulates DNA repair during interphase and spindle formation during mitosis81,82. Using E2s of the UBE2D family, the BRCA1–BARD1 complex can catalyse its own ubiquitylation, which occurs with high processivity in vitro. This reaction requires a non-covalent interaction between ubiquitin and residues of the β-sheet of UBE2D83. This β-sheet is on the back of UBE2D, opposite the catalytic centre, and is unlikely to be used for aligning a Lys residue in the bound ubiquitin with the UBE2D active site. Instead, the non-covalent interaction between UBE2D and ubiquitin could lead to an oligomeric assembly of UBE2D charged with ubiquitin at the active site83. This can congregate active E2s close to substrates and E3s and thereby increase the processivity of ubiquitylation.
Some E2s have resorted to even more dramatic measures to improve the processivity of ubiquitin chain formation. For example, the mammalian E2 UBE2G2 and its yeast homologue, Ubc7, pre-assemble ubiquitin chains on their active sites both in vivo and in vitro61,62,84,85. These chains can then be transferred en bloc to the substrate, increasing processivity. In mammals, the pre-assembly of ubiquitin chains on the active site of UBE2G2 requires a conserved acidic loop close to the catalytic Cys (FIG. 3b). It also depends on the stable interaction of UBE2G2 with its oligomeric E3, gp78, which brings multiple ubiquitin-charged UBE2G2 molecules into close proximity. The formation of large E2–E3 hetero-oligomers reduces the reliance on E2 recharging for ubiquitin chain synthesis, as ubiquitin can be transferred from one E2 active site to a ubiquitin at a second active site to form active site-linked ubiquitin chains51,84. This mechanism of ubiquitin chain assembly might be particularly important for the rapid elimination of toxic UBE2G2 substrates, such as misfolded and aggregation-prone proteins emerging from the ER en route to the cytoplasmic 26S proteasome. In conclusion, E2s use several distinct mechanisms to catalyse ubiquitin chain formation with high processivity, which are tightly connected to the physiological roles of these enzymes.
Selecting the correct linkage
Connecting ubiquitin molecules in a defined manner by modifying specific Lys residues in ubiquitin is another intrinsic property of many E2s. Early studies showed that E2s can synthesize ubiquitin chains of a distinct linkage even in the absence of an E3 (REFS 71,86–88). For example, UBE2S catalyses the formation of Lys11-linked ubiquitin chains; UBE2K, UBE2R1 and UBE2G2 assemble Lys48-linked chains; and the UBE2N–UBE2V1 complex links ubiquitin molecules through Lys63. The linkage specificity of these E2s is not altered when their cognate E3s are present. The preference for a specific Lys in ubiquitin probably results from the E2 orienting the acceptor ubiquitin in a way that exposes the favoured Lys to its active site (charged with the donor ubiquitin)25,53 (FIG. 5a). Structural studies dissecting the mechanism of Lys63-linked ubiquitin chain formation by the yeast Ubc13–Mms2 heterodimer have supported this model25,58. In Ubc13–Mms2, the donor ubiquitin is covalently linked to the catalytic Cys in Ubc13, whereas the acceptor ubiquitin is non-covalently bound to the back of the UEV protein Mms2. The interaction with Mms2 positions the acceptor ubiquitin relative to Ubc13 so that Lys63, but no other Lys residue, can attack the thioester bond between the donor ubiquitin and Ubc13 (REF. 25) (FIG. 5b).
Figure 5. Model of ubiquitin chain linkage selection.
a. A model for ubiquitin chain linkage selection by ubiquitin-conjugating enzymes (E2s). The E2 orients the acceptor ubiquitin in a way that exposes only the favoured Lys residue to its active site (charged with the donor ubiquitin), leading to the formation of ubiquitin chains of a specific linkage. b. Structural illustration of ubiquitin chain linkage selection by the E2 heterodimer Ubc13–Mms2 (Protein Data Bank code 2GMI)25. In the structure of the Ubc13~Ub-Mms2 complex (where ~ represents a thioester bond), an acceptor ubiquitin from a neighbouring complex in the crystal makes contact with Mms2 in such a way that only its Lys63 is aligned with the thioester (arrow) that links Ubc13 to the donor ubiquitin.
Variations on this theme also apply to E2s that form ubiquitin chains of different linkages. For example, the formation of Lys48-linked chains by yeast Cdc34 and human UBE2G2 seems to involve interactions between an acidic loop in these E2s and the incoming acceptor ubiquitin53,84. Mutations in the acidic loop inhibit ubiquitin chain formation by both Cdc34 and UBE2G2 and turn Cdc34 into a promiscuous E2 without specificity for Lys48. It is tempting to speculate that the interaction between these E2s and ubiquitin aligns Lys48 for efficient ubiquitin transfer. Likewise, UBE2K contains a unique C-terminal extension that interacts non-covalently with ubiquitin and is crucial for assembling Lys48-linked ubiquitin chains with high processivity66,72,89. However, for the yeast UBE2K homologue, Ubc1, the specificity for Lys48-linked ubiquitin chains is determined by its catalytic UBC domain, independently of its UBA domain66. How UBE2S establishes its striking specificity for Lys11-linked ubiquitin chains is not yet understood.
Unlike the E2s mentioned above, members of the UBE2D family of E2s do not confer linkage specificity and instead synthesize ubiquitin chains of all possible linkages in vitro. The UBE2D proteins are the smallest E2s, containing only the UBC core. UBE2D can bind ubiquitin non-covalently on its back, but this is unlikely to present a specific Lys residue of the bound ubiquitin to the active site of UBE2D on the opposite end of the protein83. As a result, the transfer of the donor ubiquitin to Lys residues in the acceptor ubiquitin might be a stochastic event, and linkage selection can be influenced by an E3 instead of UBE2D. It is also possible that UBE2D in cells might primarily function in ubiquitin chain initiation, rather than elongation.
Although E2s determine most of the linkage specificity for RING E3s, they might be less important for selecting the correct linkage of HECT E3s. HECT E3s can be regarded as E2–E3 ‘hybrids’, which are charged with ubiquitin on a Cys at their active site while directly binding to substrates. It is conceivable that HECT E3s not only position a substrate, but also help orient specific Lys residues in the acceptor ubiquitin to promote formation of a distinct ubiquitin chain. In support of this hypothesis, the usually nonspecific UBE2D1 preferentially assembles Lys29- and Lys48-linked ubiquitin chains in conjunction with the HECT E3 UBE3C (also known as KIAA10)90.
E2s: conductors of chain assembly
Taking these observations into account, it is evident that E2s lie at the heart of ubiquitylation by regulating ubiquitin chain formation on several levels. First, an E2 strongly influences the selection of the correct modifier, ubiquitin, and a suitable E3. The UBC domain provides the E2 with a structural framework to communicate with the correct E1 to pick up ubiquitin, but not UBLs, and to engage with an E3. The E2 uses overlapping binding sites to associate with the E1 and E3s, making ubiquitin chain assembly a highly dynamic reaction.
The E2 then helps to determine the length of the attached ubiquitin chain. Some E2s preferentially transfer ubiquitin to a Lys in the substrate to initiate ubiquitin chain formation, whereas others are powerful chain-elongating factors. Although a few E2s can perform both tasks, we suggest that in many cases the collaboration between chain-initiating and chain-elongating E2s is crucial for the rapid assembly of ubiquitin chains. Chain-initiating E2s are probably less selective in modifying specific Lys residues in substrates than chain-elongating E2s, which often extend only a given type of ubiquitin chain. Regulating the availability of initiating and elongating E2s will directly affect the outcome of the ubiquitin chain formation reaction.
E2s also regulate the processivity of chain formation. They have evolved several strategies to increase processivity, including the recognition of substrate motifs for rapid ubiquitin chain initiation, binding of E3s using RING-independent sites to increase affinity, oligomerization of charged E2s and preassembly of ubiquitin chains on their active sites followed by en bloc transfer. We propose that the E2 properties that determine processivity can be regulated to fine-tune ubiquitin chain formation in response to changes in signalling or cell physiology.
Finally, many E2s have the capacity to determine ubiquitin chain topology. This probably requires a non-covalent interaction between an E2 and the acceptor ubiquitin, which exposes a specific Lys on the acceptor ubiquitin to the active site of the E2 charged with the donor ubiquitin. Depending on the recognized surface of ubiquitin, chains of different linkage will be assembled and the modified proteins will be assigned different fates. By determining the specificity for the correct modifier, the length and topology of ubiquitin chains and the processivity of chain assembly, E2s have a major role in determining the outcome of ubiquitylation and the consequences for the modified protein.
Perspectives
Dissecting the complexity and dynamics of the interactions that underlie the distinct activities of E2s will help us understand how the ubiquitin code is written. A central question in the field is how the limited number of E2s pair up with almost a thousand putative E3s in a specific and often tightly regulated manner. Despite structural and biochemical analyses of E2–E3 interactions91,92, we are unable to predict which E3 cooperates with a given E2 and we are surprisingly ignorant about physiological E2–E3 pairs. Indeed, for most mammalian E2s we still have very limited knowledge about their function in cells. These E2s can be responsible for the assembly of ubiquitin chains of different linkages, including those with as yet undetermined biological functions.
Another key challenge is to understand how E2s are activated by E3s to transfer ubiquitin to a target protein, a process that probably occurs by an allosteric mechanism that is not yet known. Crystallography has provided fascinating glimpses into the world of ubiquitin, and the structure of an E2–E3 complex bound to both a donor and an acceptor ubiquitin will be crucial to understand the complex relationship between E2s and E3s during the transfer reaction. Finally, although progress has been made33,36,62,93, we still know little about how E2 activity is regulated in cells.
Without doubt, shedding more light on E2s will improve our understanding of protein homeostasis and cellular signalling. Aberrations in the ubiquitin system are linked to many diseases, including neuro-degeneration and cancer. Hopefully, our accumulating mechanistic insight will aid in the development of effective therapeutics that modulate the activity of specific E2s. Such drugs might join the proteasome inhibitor Velcade (also known as Bortezomib)94, which has been approved for treatment of multiple myeloma and mantle cell lymphoma, or inhibitors of E1s, which have been demonstrated to possess anti-cancer activities in mouse models95.
Supplementary Material
Acknowledgments
We thank the members of our laboratories for many stimulating discussions. We are grateful to J. Schaletzky for discussions and critically reading the manuscript and C. Wolberger (Johns Hopkins University, Maryland, USA) for providing the coordinates of the Mms2-bound ubiquitin. The work in our laboratories is funded by a National Institutes of Health Director’s New Innovator Award (M.R.), RO1 5R01GM083064-02 (M.R.), a March of Dimes grant (M.R.), and the intramural research programme of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (Y.Y.). M.R. is a Pew Scholar.
Glossary
- Ubiquitin-interacting motif
A small motif that mediates the interaction of a protein with the hydrophobic patch of ubiquitin around Ile44
- Ubiquitin-associated (UBA) domain
A protein domain that forms a three-helix bundle and interacts with hydrophobic regions of ubiquitin
- 26S proteasome
A multisubunit protease that degrades proteins with attached ubiquitin chains. It contains a barrel-like 20S proteolytic core particle that houses the active sites and a 19S regulatory particle that governs substrate recognition and entry into the 20S core particle
- HECT domain
A domain of ~40 kDa (350 amino acids) that is found at the C terminus of HECT E3s. It contains a catalytic Cys residue that accepts ubiquitin from an E2 to form a ubiquitin thioester intermediate before transferring the ubiquitin to substrates
- RING domain
A domain that is present in most E3s and is defined by the consensus sequence CX2CX(9–39) CX(1–3)HX(2–3)C/HX2CX(4–48)CX2C (where X means any amino acid). It coordinates two structural zinc cations
- Ubiquitin-conjugating (UBC) domain
A conserved core domain of ~150 residues that is found in all E2s, including those for UBLs. It contains the catalytic Cys residue of E2s
- 310-helix
A type of secondary protein structure in which the amino acids are in a right-handed helical arrangement. The hydrogen bonds are formed between the NH group of an amino acid and the CO group of the amino acid three residues earlier (as opposed to four residues earlier in an α-helix)
- Ubiquitin fold domain
A domain found in E1s that mediates binding to an E2 and forms a similar structure to ubiquitin
- TEK box
A Lys-rich region in APC/C substrates downstream of initial APC/C recognition sites (such as the D box or KEN box), which promotes initiation of ubiquitin chain formation
- D box
The amino acid sequence RXXL(X)nN (where X means any amino acid), which mediates binding of an APC/C substrate to the co-activators Cdc20 and Cdh1 and potentially also to subunits of the core APC/C
- KEN box
The amino acid sequence KEN(X)nP (where X means any amino acid), which mediates binding of APC/C substrates to the co-activator Cdh1
Footnotes
Contributor Information
Yihong Ye, Email: yihongy@mail.nih.gov.
Michael Rape, Email: mrape@berkeley.edu.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nature Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 6.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. Demonstrates the existence and importance of non-canonical ubiquitin chains in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HT, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 8.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nature Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin binding domains — from structures to functions. Nature Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nature Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 18.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 21.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 22.Michelle C, Voure’h P, Mignon L, Andres CR. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J Mol Evol. 2009;68:616–628. doi: 10.1007/s00239-009-9225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Hwang WC, Basavappa R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. J Biol Chem. 2002;277:21913–21921. doi: 10.1074/jbc.M109398200. [DOI] [PubMed] [Google Scholar]
- 24.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. Suggests that RING E3s mediate the allosteric activation of E2s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2–Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. Together with reference 58, reveals the mechanism of linkage-specific ubiquitin chain formation, in this case for Lys63-linked chains. [DOI] [PubMed] [Google Scholar]
- 26.Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988;263:13268–13275. [PubMed] [Google Scholar]
- 27.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang DT, et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. An elegant structural study that defines the molecular basis underlying the communication between E1s and E2s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 30.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- 31.Huang DT, et al. A unique E1–E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nature Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nature Struct Mol Biol. 2008;15:280–287. doi: 10.1038/nsmb.1387. Provides striking insight into the role of N- or C-terminal appendices of E2s in determining E2 specificity. [DOI] [PubMed] [Google Scholar]
- 33.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 34.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitinligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 36.Williamson A, et al. Identification of a physiological E2 module for the human anaphase promoting complex. Proc Natl Acad Sci USA. 2009 Oct 12; doi: 10.1073/pnas.0907887106. Defines the molecular mechanism underlying the formation of Lys11-linked ubiquitin chains by APC/C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng L, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 38.Fang S, et al. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B, et al. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 41.Xin H, et al. The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res. 2000;28:2847–2854. doi: 10.1093/nar/28.14.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. First structural study to illustrate the interaction of an E2 with a RING E3. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Zhang M, et al. Chaperoned ubiquitylation — crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 47.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nature Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailly V, Prakash S, Prakash L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol Cell Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolman CJ, Toth J, Gonda DK. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, et al. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das R, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitinligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. A thorough dissection of ubiquitin chain formation by Cdc34 that led to the concept that chain initiation and elongation are carried out by distinct mechanisms. [DOI] [PubMed] [Google Scholar]
- 54.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. A biochemical study providing crucial insight into the mechanism of E2-dependent ubiquitin transfer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang DT, et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 56.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nature Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 57.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 58.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 59.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 60.Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM. A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J Cell Sci. 2009;122:1374–1381. doi: 10.1242/jcs.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nature Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- 63.Polo S, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 64.Hoeller D, et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Plafker SM, Plafker KS, Weissman AM, Macara IG. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J Cell Biol. 2004;167:649–659. doi: 10.1083/jcb.200406001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. An elegant study demonstrating the role of distinct ubiquitin chain-initiating E2s and ubiquitin chain-elongating E2s for the yeast APC/C. [DOI] [PubMed] [Google Scholar]
- 67.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. Identifies the first role for Lys11-linked ubiquitin chains in controlling cell cycle progression and provides a mechanism by which an E2 and an E3 can cooperate in efficient ubiquitin chain initiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nature Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 69.Petroski MD, et al. Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. J Biol Chem. 2007;282:29936–29945. doi: 10.1074/jbc.M703911200. [DOI] [PubMed] [Google Scholar]
- 70.Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- 71.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 72.Haldeman MT, Xia G, Kasperek EM, Pickart CM. Structure and function of ubiquitin conjugating enzyme E2–25K: the tail is a core-dependent activity element. Biochemistry. 1997;36:10526–10537. doi: 10.1021/bi970750u. [DOI] [PubMed] [Google Scholar]
- 73.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 74.Verma R, Feldman RM, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gazdoiu S, Yamoah K, Wu K, Pan ZQ. Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol Cell Biol. 2007;27:7041–7052. doi: 10.1128/MCB.00812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 80.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 81.Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 82.Joukov V, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 83.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 84.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. Together with references 51 and 62, shows that ubiquitin chains can be preassembled on a catalytic Cys of E2 and subsequently be transferred en bloc to a substrate protein. [DOI] [PubMed] [Google Scholar]
- 85.Cao J, et al. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 2007;6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 86.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 87.Van Nocker S, Vierstra RD. Cloning and characterization of a 20-kDa ubiquitin carrier protein from wheat that catalyzes multiubiquitin chain formation in vitro. Proc Natl Acad Sci USA. 1991;88:10297–10301. doi: 10.1073/pnas.88.22.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haas AL, Reback PB, Chau V. Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. Comparison to their putative rabbit 266, homologs, E220K and E232K. J Biol Chem. 1991:5104–5112. [PubMed] [Google Scholar]
- 89.Merkley N, Shaw GS. Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J Biol Chem. 2004;279:47139–47147. doi: 10.1074/jbc.M409576200. [DOI] [PubMed] [Google Scholar]
- 90.Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez-Noel G, Muller U, Harbers K. Identification of molecular determinants required for interaction of ubiquitin-conjugating enzymes and RING finger proteins. Eur J Biochem. 2001;268:5912–5919. doi: 10.1046/j.0014-2956.2001.02541.x. [DOI] [PubMed] [Google Scholar]
- 92.Markson G, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009 Jun 23; doi: 10.1101/ gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gazdoiu S, et al. Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc Natl Acad Sci USA. 2005;102:15053–15058. doi: 10.1073/pnas.0507646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 95.Yang Y, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.