Abstract
We address the problem of efficiently aligning a transcribed and spliced DNA sequence with a genomic sequence containing that gene, allowing for introns in the genomic sequence and a relatively small number of sequencing errors. A freely available computer program, described herein, solves the problem for a 100-kb genomic sequence in a few seconds on a workstation.
With large amounts of both expressed and genomic DNA sequence data being made available, it is becoming more common to align the two. We have written a computer program, called sim4, to perform such alignments very efficiently and accurately, under the assumption that the differences between the two sequences are limited to (1) introns in the genomic sequence, and (2) sequencing errors (in either sequence).
The next section describes use of sim4 in a production setting. Then, the tool’s accuracy is assessed using simulated data, into which “sequencing errors” are introduced using a random number generator. Next, we report some experimental data obtained by aligning human mRNAs with the homologous genomic sequence from the mouse. This application is somewhat outside sim4’s intended scope, as evolutionary differences such as long insertions need to be handled, but useful results are frequently produced. We then illustrate how the capabilities of sim4 can be incorporated into larger tools and software packages and finish with a brief description of sim4’s algorithmic approach.
The program can be obtained by anonymous ftp from globin.cse.psu.edu or over the World Wide Web from http://globin.cse.psu.edu/.
The BDGP: cDNA vs. Genomic Alignments
The Berkeley Drosophila Genome Project (BDGP) is a consortium whose goal is to determine the complete DNA sequence of the euchromatic genome of the fruit fly Drosophila melanogaster and to develop experimental and computational tools to probe its biological significance (Rubin 1996). It includes a large-scale sequencing project, together with both biological and computational annotation projects, the results of which are curated by experienced Drosophila biologists. This work is available on the World Wide Web at http://fruitfly.berkeley.edu/.
Among genomic annotations, the location of genes in the genomic sequence is of great interest to both biologists and computer scientists. An accurate and well-curated transcript map helps biologists understand mutational effects and the regulation of gene expression, and it gives computational biologists a powerful data set for training and evaluating algorithms. Like other large-scale genome projects (Eddy 1994; Cherry et al. 1998) the BDGP provides both computational predictions and experimental results. Computational results come from a collection of gene finders, including Genie (Reese et al. 1997) and dGrail (Xu and Uberbacher 1997), each of which has different strengths and weaknesses. Experimental annotations are based on sequence data from a variety of EST and full-length cDNA sequencing projects. These cDNA sequences have been positioned on the genomic sequence using a variety of tools [primarily Blast (Altschul et al. 1990)] with substantial manual intervention. Increasing quantities of data have made this technique unworkable, necessitating a specialized tool for aligning cDNA and genomic sequences. sim4 fills this need by quickly aligning a cDNA sequence to its parent genomic sequence with sufficient accuracy to require minimal manual editing.
Validating sim4’s Alignments
To evaluate sim4’s alignments on a set of genes with known structures, we started with a curated set of genomic GenBank sequences for multiexon Drosophila genes that was developed to train the Drosophila version of Genie (Reese et al. 1997). GenBank entries for these sequences include feature annotations that describe both mRNA and coding sequence (CDS) subsequences within each entry. The mRNA features most closely approximate our experimental data, but the CDS entries are more carefully curated, so they were the basis for our experiments. The data set contains 202 GenBank flatfile entries, 184 of which have usable CDS features with a total of 681 exons. Of these 184 entries, 156 have 2–5 exons, 21 have 6–9 exons, and 7 have 11–15 exons. The average exon length is 425 bases. There are 64 exons with <50 bases, 441 exons with lengths between 50 and 499 bases, 107 exons with lengths between 500 and 999 bases, and 69 exons with lengths between 1000 and 5000 bases. A number of entries have unusually small exons (16 entries have exons <10 bases, 15 have exons between 11 and 20 bases, 13 have exons between 21 and 30 bases). In many cases, this is an artifact of only using the CDS portion of the transcript; some of the initial and final exons only contribute a few bases to the coding sequence.
We extracted each CDS sequence from its parent sequence and used sim4 to align it to the original GenBank sequence. sim4’s performance was measured by how closely the intron–exon boundaries corresponded to the GenBank CDS annotation. The reported error is the number of bases misidentified for each exon. For example, if an exon is known to occur at the location 345–465 but sim4 reported an exon of 340–460, the error would be 10.
If sim4 is told that a pair of sequences contain very few errors and are very similar (by setting the N = 1 option) it will make extra efforts to accurately handle small exons at the beginning and ends of the alignments. We examined sim4’s performance both without and with this optimization.
Without using optimizations for high-quality sequences, sim4 generated alignments that exactly matched the GenBank sequence annotations for 166 of these sequences. Of the remaining 18 alignments, 11 had errors in the range of 1–10 bases, 6 had errors in the range of 11–20 bases, and 1 had an error of 25 bases. All of these erroneous alignments resulted from a common mistake, namely that sim4 failed to align small initial or final exons in their proper locations and frequently included some or all of their bases in a neighboring exon, doubly penalizing the mistake. Many of these small exons are an artifact of our using the CDS feature sequences in our experiments. In these sequences, only a small portion of the initial or final exon is part of the CDS; the remainder is part of the the 5′- or 3′-untranslated region (UTR). Although we decided that the mRNA feature entries are not curated well enough to be used as the basis of our experiments, we did use them to test this explanation for sim4’s difficulties. Of the 18 GenBank entries for the problem sequences, 9 have mRNA features (DMU52952 has 3 mRNA features with different inital exons, none of which are included in its CDS feature). In each of these nine cases, sim4 produced correct alignments between the mRNA subsequence and the genomic sequence.
Using the optimizations for handling small exons in high-quality sequences, sim4 was able to correctly align 172 of the sequences (the 166 sequences that were identified by the unoptimized run plus 6 sequences that the unoptimized run had aligned incorrectly). Of the 12 erroneous alignments, 9 had difficulties that were similar to those described above. Two of the alignments had a previously unobserved type of error: Each had a small exon that was perfectly aligned to an incorrect location. The final erroneous sequence missed a small “internal” exon, distributing some of its bases among the neighboring exons.
It might be possible to recognize this mistake in a sim4 postprocessor, as the alignments have less than perfect similarity, with the mismatches occuring at the ends of the sequences. Tuning with a splice site predictor, as described in Reese et al. (1997) could help increase the accuracy of the prediction.
Comparisons with Similar Tools
We know of three other tools that are designed to align spliced sequences (mRNA, cDNA, ESTs) to the corresponding genomic sequences. The goal of this section is to compare sim4 with these other tools. Gelfand et al. (1996) describe a tool for identifying genes in genomic sequence using alignments of spliced sequences. Birney and Durbin (1997) have developed a set of tools for automatically generating alignment programs based on a high-level description of the desired dynamic programming recurrence. Their tool kit contains an example program, est2gen, which aligns EST and genomic sequences. Richard Mott’s tool (Mott 1997) est_genome uses a carefully crafted implementation of a linear space dynamic programming recurrence to optimally align spliced sequences to their genomic counterparts.
We were unable to acquire an implementation of the ideas presented by Gelfand et al. (1996). est2gen produces only graphic displays of the alignments that it generates, and it does not explicitly generate a list of exons and their positions on the sequences. A visual inspection of its alignments showed that although it usually aligned several of the major exons, it frequently omitted exons and its alignments usually included a great deal of noise around exon boundaries. These characteristics made it too unwieldy to explicitly score the alignments using the scheme discussed above, but we did include it in our running time comparison. Like sim4, est_genome produces an output format that is easily parsed and that explicitly describes the exon–intron structure of the alignment it found.
Table 1 presents the results of our comparisons. We used est2gen, est_genome, and sim4 to align 184 sequences from the Drosophila training set, as described above (Reese et al. 1997). (To successfully align these sequences it was necessary to increase an upper bound on the amount of memory that est_genome could allocate.) All alignments were performed on a dual 266-MHz Intel Pentium II system with 128 Mb of RAM running RedHat Linux 5.0.
Table 1.
Comparison of Running Times and Accuracy of Programs That Align Spliced Sequences to their Genomic Counterparts
| Time (sec)/seq. | Errors per alignment | ||||
|---|---|---|---|---|---|
| 0 | 1–10 | 11–20 | 21–35 | ||
| est2gen | 156 | — | — | — | — |
| est_genome | 20 | 143 | 21 | 13 | 7 |
| sim4, N = 0 | 0.06 | 166 | 11 | 6 | 1 |
| sim4, N = 1 | 0.06 | 172 | 6 | 5 | 1 |
Running times are the average time per sequence in the 184-sequence Drosophila test set. Errors were scored as described in Validating sim4’s Alignments. As discussed in Comparisons with Similar Tools, est2gen results could not be scored but were less accurate than est_genome or sim4.
Both sim4 and est_genome had some trouble handling small exons. As discussed above in Validating sim4’s Alignments, very small exons occur as an artifact in our experimental data set. Generally, sim4 had trouble only with exons that are so small (10–20 bases) that they are very unlikely to occur in real sequences. When run with its default settings, the exons that est_genome had difficulty positioning were slightly larger, though still unlikely to occur in real data. It also had a pronounced tendency to include an extra base at the beginning of the first exon, which can be seen in the large number of alignments in the 1–10 base error category. It should be possible to increase the likelihood that est_genome will correctly handle small exons by tuning its parameters, although there is also an increased risk of spurious alignments (Mott 1997).
Assigning cDNAs to a Genomic Clone
The Adh region of the Drosophila genome has been the object of intense genetic and biochemical scrutiny for many years. Because of the wealth of available information, the BDGP has been using it in a pilot study for its annotation project. It is one of the foci of the large-scale sequencing project, and much of our cDNA sequencing has been concentrated on transcripts from this region. As part of the annotation project we have identified 27 cDNA sequences in GenBank that are from the Adh region and have been assigned to particular P1 clones. We used these sequences to determine if sim4 would be able to detect the correct location of a cDNA sequence in our pool of genomic sequence.
Each of the 27 cDNA sequences was compared to the current collection of 3120 contigs from our P1 clones, covering the Adh region as well as other regions of the genome, for a total of 84,240 alignments. Selecting the alignments that included >90% of the cDNA’s sequence and were >90% similar over all of the exons gave a single alignment for each of 21 of the sequences. All of the six sequences missed by this simple screening rule were easily accounted for.
Four of the cDNAs spanned multiple P1 clones. Their alignment to an individual clone accounted for <90% of their length, though they were very similar.
One of the cDNA sequence annotations refered to a related, but incorrect, GenBank entry. This incorrect entry is not in a region for which we have genomic sequence, so sim4 was correct when it was unable to assign it to a P1 clone. Using the correct GenBank sequence for the gene results in a three-exon match with 100% identity using 100% of the clone.
The final cDNA had two difficulties. First, it is only partially contained in our collection of genomic clones and the clone ends in the middle of a large intron. Second, there are some substantial differences between the genomic and cDNA sequences that are probably due to differences in the parent Drosophila strains or to sequencing errors. sim4’s alignment to the correct P1 clone found three exons, which were 100%, 94%, and 88% similar, respectively. The mismatches were all clustered in multibase deletions.
Our simple screen could easily be augmented to pass alignments that have strong similarity and that have exons that run off of the end of a clone. With some tuning it might also be possible to pass correct alignments that have weaker similarities and/or introns that run off of the ends of the clones. Because relaxing the filter too much might result in a high false-positive rate, it may be necessary to manually intervene in these cases.
Tests on Simulated Data
To further assess the accuracy of sim4, we extracted mRNAs from 16 genes in a 222,930-bp genomic sequence from human Chromosome 12p13 (Ansari-Lari et al. 1997; GenBank accession no. HSU47924) based on the annotated exon boundaries. Using a random number generator, nucleotide substitutions were introduced an average of twice as frequently as either (single-nucleotide) insertions or deletions. We modeled two kinds of data—ESTs and full-length mRNAs.
ESTs were simulated by randomly selecting 500 bp from the mRNA and introducing errors at rates of 1%, 3%, and 5%. The results cited in Table 2 indicate that even with ESTs, sim4 should usually give the correct alignment. For full-length mRNAs, we measured performance with error rates of 0.1% and 1%. sim4 failed to correctly identify the boundaries of a short (6 nucleotides) internal exon in the hBAP gene. The 6 nucleotides were instead distributed at the ends of the adjacent exons. Even so, the experiment’s results, summarized in Table 3, suggest that with highly accurate full-length cDNA sequences, sim4’s alignment should be completely correct the vast majority of the time.
Table 2.
Accuracy of sim4 with Simulated ESTs
| Rate (%) | 1 | 2 | 3 |
|---|---|---|---|
| 1 | 0.15 | 0.23 | 89.5 |
| 3 | 0.20 | 0.35 | 81.7 |
| 5 | 0.26 | 0.47 | 75.7 |
The numbered columns record the following data. (1) The percentage of nucleotides in putative sim4 exons that are not in the true mRNA. For instance, at a rate of 1% simulated errors, each 500-bp simulated EST had an average of five errors. The 0.15% false-positive rate means that an average of 500 × 0.0015 = 0.75 predicted nucleotides were not in the true mRNA. (2) The percentage of nucleotides in the true mRNA but not in putative sim4 exons. (3) The percentage of splice junctions that were determined exactly.
Table 3.
Performance of sim4 with Simulated Full-Length mRNAs
| Rate (%) | 1 | 2 | 3 |
|---|---|---|---|
| 0.1 | 0.031 | 0.031 | 98.3 |
| 1 | 0.059 | 0.060 | 94.7 |
Columns are as in Fig. 2. The default setting for sequence accuracy (N = 0) was used.
Cross-Species Alignments
sim4 is intended to produce a correct alignment that accounts for introns and for sequencing errors. It is not designed to deal properly with evolutionary mutations, such as multinucleotide insertions and deletions. To get a better feel for the rate at which sim4’s accuracy degrades with evolutionary divergence, we measured its effectiveness at aligning the 16 human mRNAs discussed in the previous section with the orthologous genomic sequence from the mouse, which is available as GenBank accession numbers AC002393 and AC002397 (Ansari-Lari et al. 1998).
Of the 16 genes, 13 are more highly conserved than the average of 84.6% nucleotide identity reported in a survey of 1196 human/mouse orthologs by Makalowski et al. (1996). The only gene that is substantially less conserved than this average, CD4, is associated with the immune system, which is frequently the case with highly divergent genes.
Table 4, column 4, reports how much of each mRNA was aligned by sim4, and column 5 shows how much of each protein-coding region was aligned. We also compared the positions of exon boundaries with the positions determined by sim4’s putative exons. Column 6 gives the number of nucleotides that were aligned to non-mRNA regions of the mouse, as a percentage of the mRNA’s length. Each time an exon boundary was misplaced by, for example k nucleotides, k was added to this amount, and in one case (gene A-2) an erroneous exon of length 8 was predicted. Thus, we are assuming that the mouse mRNA preserves the human splice junctions.
Table 4.
Performance of sim4 When Aligning Human mRNAs with the Orthologous Mouse Genomic Sequence
| Gene | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| ISOT | 91.9 | 98.6 | 20 (20) | 100 | 100 | 0.00 (0/3115) |
| HSENO-2 | 91.7 | 99.1 | 12 (13) | 83.0 | 100 | 1.23 (28/2274) |
| hBAP | 91.3 | 100 | 10 (9) | 100 | 100 | 0.89 (11/1240) |
| GNB3 | 89.8 | 98.2 | 11 (10) | 75.0 | 100 | 0.10 (2/1922) |
| A-2 | 89.5 | 96.8 | 5 (7) | 94.0 | 98.5 | 1.78 (27/1515) |
| HSPTP1CG | 89.2 | 96.1 | 15 (14) | 80.1 | 96.8 | 1.87 (38/2033) |
| HUMDRPLA | 88.9 | 95.1 | 10 (10) | 98.4 | 99.2 | 0.12 (5/4341) |
| C10 | 88.7 | 97.6 | 3 (3) | 78.2 | 100 | 0.00 (0/519) |
| TPI | 88.7 | 96.0 | 7 (7) | 52.9 | 100 | 0.00 (0/1843) |
| C3f | 88.6 | 93.2 | 12 (12) | 100 | 100 | 0.32 (6/1856) |
| C9 | 87.4 | 92.0 | 2 (2) | 98.5 | 100 | 0.11 (1/877) |
| B | 86.4 | 90.9 | 14 (14) | 74.2 | 93.3 | 1.17 (25/2129) |
| C2f | 85.0 | 92.2 | 6 (6) | 72.8 | 100 | 0.23 (2/886) |
| B7 | 81.6 | 86.6 | 7 (5) | 64.2 | 64.4 | 0.33 (4/1208) |
| C8 | 81.5 | 75.9 | 6 (6) | 71.0 | 98.4 | 0.48 (6/1247) |
| CD4 | 70.2 | 62.9 | 10 (7) | 21.0 | 47.3 | 0.59 (18/3051) |
Genes, as named in the left-most column, are taken from data reported by Ansari-Lari et al. (1997, 1998) and sorted according to nucleotide identity. The numbered columns record the following data: (1) Percentage of nucleotide identity between the human and mouse sequences in the protein-coding region. (2) Percentage of amino acid similarity. (Data in columns 1 and 2 are taken from Ansari-Lari et al. 1998.) (3) Number of exons in the gene and, in parentheses, number of putative exons found by sim4. (4) Percentage of the entire mRNA aligned by sim4. (5) Percentage of the protein-coding region aligned by sim4. (6) Percentage of nucleotides aligned by sim4 to positions not in the true mouse mRNA (assuming preservation of splice junctions).
Two trends are evident from the data presented in Table 4. First, sim4 is frequently much more effective at aligning protein-coding regions than for the UTRs at the ends of the mRNA. For instance, for 9 of the 16 genes, sim4 was 100% accurate in the coding regions, whereas 100% accuracy for the entire gene was attained in only three cases. This reflects the fact that a gene’s 5′ and 3′ UTR are usually much less well conserved than the coding region (Makalowski et al. 1996). Second, typically <1% of the nucleotides in sim4’s putative exons were not in the true mRNA, even in cases where sim4 was unable to find the gene accurately.
Other Uses of Sim4
The approach implemented in sim4 may be fruitfully integrated into a variety of sequence analysis packages, as illustrated here. One natural use of these methods is for comparing a genomic sequence with an EST database. That problem was addressed earlier by Huang et al. (1997), using other computational methods.
To explore the use of sim4’s algorithm for this potential application, we built a prototype program, called blEST, that can quickly identify near-identity matches between a genomic sequence and an EST database. After masking interspersed repeats (e.g., Alus) and low-complexity regions in the genomic sequence, blEST extracts from the database all ESTs that share a 32-bp exact match with the genomic sequence. The resulting ESTs are then compared with the unmasked genomic sequence using a variant of sim4 that reports only those ESTs that meet certain (adjustable) conditions, such as (1) the putative identified exons must cover at least 70% of the database sequence, and (2) the overall identity within those exons must be at least 95%. Although the running time depends on the number of matching ESTs, we found it to average ∼1 min/100 kb of genomic sequence on a 200-MHz workstation, when comparing a human genomic sequence with all human ESTs in the dbEST database (Boguski et al. 1993). However, the loss of effectiveness caused by restricting attention to only very strong matches (e.g., at least 95% identity) remains to be evaluated before this approach can be recommended for general use.
Typically, results from database searches are combined with other sources of information to reach certain conclusions. In particular, a major use of ESTs is for identifying genes in a sequenced genomic region (e.g., Smith et al. 1996; Ansari-Lari et al. 1997; Flint et al. 1997; Ruddy et al. 1997). Several groups have found that the information provided by ESTs substantially enhances the results of gene-prediction programs, such as GRAIL (Uberbacher et al. 1996).
We recently began to explore the predictive power of combining human/mouse sequence comparison with other tools to identify genes (Ansari-Lari et al. 1998). A goal is to produce a system that can automatically analyze orthologous human and mouse genomic sequence data at, for example, a rate of 100 kb in a few minutes (i.e., in a small multiple of the time taken to identify repeats) and that presents the results in a readily understood graphic format.
A number of approaches and software tools have been developed by various groups to provide a graphic summary of sequence positions that match ESTs. At one extreme are programs (e.g., Harris 1997; Ansari-Lari et al. 1998) that do not distinguish regions that match only one EST from regions with multiple matches. At the other extreme, the program PowerBLAST (Zhang and Madden 1997) shows each match, complete with the identification of positions where sequences disagree. An innovative approach of Smith et al. (1996) uses colors and a kind of “projected three-dimensional” display to indicate how many ESTs match in a given region, as well as the strengths of those matches.
There is a strong rationale for at least giving some indication of how many ESTs match the genomic sequence in a given region. A number of investigators have observed that genomic regions aligning with several ESTs are more likely to contain a gene than if only one EST aligns. For instance, in tests using genomic sequence data with well-characterized gene content, and at stringencies comparable to those used by blEST, essentially every EST cluster detected a gene, whereas only 70% of singleton aligning ESTs did so (Fig. 2C in Bailey et al. 1998). Moreover, an indication of the number of hits may provide at least a weak indication of expression levels for each gene.
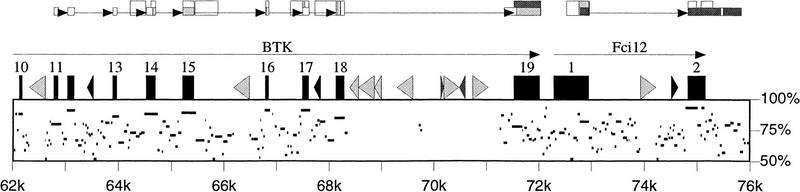
Figure 1 shows part of a pip (percent identity plot; Hardison et al. 1997) of a human/mouse alignment in the BTK region (Oeltjen et al. 1997), that has been automatically annotated using the output of blEST. Note the singleton human ESTs containing portions of introns 13, 15, 16, and 17 of BTK and the mouse EST extending slightly upstream of exon 19. Also note that the introns estimated by blEST and the indication of EST redundancy accurately identify the true exons.
Figure 1.
Graphic representation of a genomic alignment together with EST matches computed by blEST. Human genes and interspersed repeats are drawn along the top of the box, with horizontal lines inside the box indicating the human positions and percent identity of gap-free portions of an alignment with the mouse genomic sequence. Above these are representations of the matches between the genomic sequences and the dbEST database (Boguski et al. 1993). The taller boxes show human ESTs matching the human sequence; and the shorter boxes show mouse ESTs matching the mouse sequence (with position on the human sequence deduced from the alignment). Shading of boxes indicates one match (white), two to three matches (light gray), or four to nine matches (dark gray). Thus, there are at least four mouse ESTs extending beyond the annotated end of the Fci12 gene, suggesting a longer 3′ UTR in the mouse. Arrows connecting EST boxes indicate introns identified by blEST using the sim4 strategy.
METHODS
In the approach described here, an expressed sequence is aligned with a genomic sequence in the following steps.
Determine high-scoring segment pairs (HSPs). An HSP is just a high-scoring gap-free alignment of regions of the two sequences, such as computed by the blast program (Altschul et al. 1990). sim4 detects exact matches of length 12 and extends them in both directions with a score of 1 for a match and −5 for a mismatch, stopping when extensions no longer increase the score. Code to locate HSPs in a pair of long DNA sequence was borrowed from a program described by Schwartz et al. (1991).
Select a set of HSPs that could represent a gene. A dynamic programming algorithm selects a best chain of the HSPs subject to the constraint that (a) their starting positions in the expressed sequence are in increasing order, and (b) the diagonals of consecutive HSPs are either nearly the same or differ by enough to be a plausible intron. HSP scores are multiplied by 100 and reduced by the differences between diagonals of consecutive HSPs to determine a score for a chain.
Find exon boundaries. When consecutive “exon cores” (each given by a collection of HSPs on nearly the same diagonal in the gene model) overlap, the ends are trimmed in an attempt to find an intron matching either GT…AG or CT…AC. (It might be worthwhile to consider more sophisticated rules for splice junctions, e.g., those used by Burge and Karlin (1997), but we have not done so.) If the cores do not overlap, they are extended toward one another using a “greedy” strategy (Miller and Myers 1985) until they meet at a row of the dynamic programming matrix, and that row is then adjusted, if necessary, to satisfy the above intron consensus signals. If the extension procedure fails, the region between the two adjacent exon cores is searched for HSPs at a reduced stringency (starting with exact matches of length 8). Similarly, the first and last exon cores are extended toward the ends of the expressed sequence, first by a greedy approach, and then, if necessary, by a reduced stringency search for HSPs. We added an option for handling highly accurate expressed sequence data (the N = 1 option mentioned in Table 1). The program looks for very small first or last exons whose splice-signal orientation is consistent with that of other introns.
Determine the alignment. The alignment for each exon (whose boundaries in each of the sequences are determined by the previous step) is computed by the method of Chao et al. (1997).
Acknowledgments
We thank Jinghui Zhang for suggesting that we write sim4, Sima Misra for her help with Drosophila genome information, Martin Reese for sharing his Drosophila training set, and Michael Ashburner for sharing his set of genes in the Adh region. This work was supported in part by a grant from the National Human Genome Research Institute to G.M.R. and by grant R01 LM05110 from the National Library of Medicine to W.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL webb@cse.psu.edu; FAX (814) 865-3176.
REFERENCES
- Altschul S, Gish W, Miller W, Myers E, Lipman D. A basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ansari-Lari MA, Shen Y, Muzny DM, Lee W, Gibbs RA. Large-scale sequencing in human chromosome 12p13: Experimental and computational gene structure determination. Genome Res. 1997;7:268–280. doi: 10.1101/gr.7.3.268. [DOI] [PubMed] [Google Scholar]
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, Gibbs RA. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region on mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Bailey LC, Jr, Searls DB, Overton GC. Analysis of EST-driven gene annotation in human genomic sequence. Genome Res. 1998;8:362–376. doi: 10.1101/gr.8.4.362. [DOI] [PubMed] [Google Scholar]
- Birney E, Durbin R. Dynamite: a flexible code generating language for dynamic programming methods used in sequence comparison. Proc Fifth Int Conf Intelligent Systems Mol Biol. 1997;5:56–64. [PubMed] [Google Scholar]
- Boguski M, Lowe T, Tolstoshev C. dbEST—Database for “expressed sequence tags.”. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chao K-M, Zhang J, Ostell J, Miller W. A tool for aligning very similar DNA sequences. Comput Appl Biosci. 1997;13:75–80. doi: 10.1093/bioinformatics/13.1.75. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. The Caenorhabditis elegans Genome Project. In: Lamberti F, et al., editors. Advances in molecular plant nematology. New York, NY: Plenum Press; 1994. pp. 3–18. [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett N, King A, Higgs D. The relationship between chromosome structure and function at a human telomeric region. Nature Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Pevzner PA. Spliced alignment: A new approach to gene recognition. Proc Natl Acad Sci. 1996;93:9061–9066. doi: 10.1073/pnas.93.17.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory element: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Harris N. Genotator: A workbench for sequence annotation. Genome Res. 1997;7:754–762. doi: 10.1101/gr.7.7.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams M, Zhou H, Kerlavage A. A tool for analyzing and annotating genomic sequences. Genomics. 1997;6:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Zhang J, Boguski M. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 1996;6:846–857. doi: 10.1101/gr.6.9.846. [DOI] [PubMed] [Google Scholar]
- Miller W, Myers EW. A file comparison program. Software—Practice Experience. 1985;15:1025–1040. [Google Scholar]
- Mott R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Oeltjen J, Malley T, Muzny D, Miller W, Gibbs R, Belmont J. Large scale comparative sequence analysis of the human and murine Bruton’s tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Rubin GM. Around the genomes: The Drosophila genome project. Genome Res. 1996;6:71–79. doi: 10.1101/gr.6.2.71. [DOI] [PubMed] [Google Scholar]
- Ruddy D, Kronmal G, Lee V, Mintier G, Quintana L, Domingo R, Jr, Meyer N, Irrinki A, McClelland E, Fullan A, et al. A 1.1 Mb transcript map of the hereditary hemochromatosis locus. Genome Res. 1997;7:441–456. doi: 10.1101/gr.7.5.441. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Miller W, Yang C-M, Hardison RC. Software tools for analyzing pairwise alignments of long sequences. Nucleic Acids Res. 1991;19:4663–4667. doi: 10.1093/nar/19.17.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Lee M, Szabo C, Jerome N, McEuen M, Taylor M, Hood L, King M-C. Complete genomic sequence and analysis of 117 kb or human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- Uberbacher E, Xu Y, Mural R. Discovering and understanding genes in human DNA sequence using GRAIL. Methods Enzymol. 1996;266:259–281. doi: 10.1016/s0076-6879(96)66018-2. [DOI] [PubMed] [Google Scholar]
- Xu Y, Uberbacher EC. Automated gene identification in large-scale genomic sequences. J Comput Biol. 1997;4:325–338. doi: 10.1089/cmb.1997.4.325. [DOI] [PubMed] [Google Scholar]
- Zhang J, Madden T. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]