Abstract
Practice-changing evidence requires confirmation, preferably in multi-institutional clinical trials. The collection of tissue within such trials has enabled biomarker studies and evaluation of companion diagnostic tests. Tissue microarrays (TMAs) have become a standard approach in many cooperative oncology groups. A principal goal is to maximize the number of assays with this precious tissue. However, production strategies for these arrays have not been standardized, possibly decreasing the value of the study. In this article, members of the Cancer and Leukemia Group B Pathology Committee relay our experiences as array facility directors and propose guidelines regarding the production of high-quality TMAs for cooperative group studies. We also discuss statistical issues arising from having a proportion of patients available for TMAs and the possibility that patients with TMAs fail to represent the greater study population.
INTRODUCTION
Companion biomarker tests that qualify patients for specific therapies are the centerpieces of personalized medicine. Tissue samples and their analyses are critical in understanding which therapies are appropriate for individual patients and for delivering those therapies. It is becoming a standard for cooperative oncology groups and other multicenter consortia to collect tissue samples as a key part of the clinical trial, with the goal of using the material in correlative studies to help develop or validate companion diagnostic tests. Some trials are highly successful and collect specimens from nearly every patient enrolled onto the trial, and some even require tissue blocks for enrollment. However, the availability of a complete sample of patients from the population is not always possible. Sometimes pathology departments are reluctant to part with the tissue block. Reasons include paucity of tissue, state laws, hospital regulations, departmental policies, ambiguity regarding ownership of specimens, and lack of familiarity with cooperative groups and clinical trials more generally. A consequence is that the number of tissue blocks collected for a given trial may represent only a subset of the patients in the trial. This gives rise to problems that we will discuss.
Tissue microarrays (TMAs), first described in the late 1980s,1 were popularized after publication of a mechanized method of production.2 They are constructed by using a needle to core a tissue block and then placing that cored paraffin into a recipient, predrilled master block with as many as 800 other tissue cores. There are numerous reviews describing both the methods and advantages of this approach. Specific advantages of using TMAs include minimal damage to the source block, more cases in direct parallel analysis on the same slide, lower reagent costs, and faster results. Although it has been nearly 20 years since the first TMA was constructed, their use by clinical trials pathology coordinating centers is still in a relatively early stage, with each center using its own methods and format. A summary of the advantages and disadvantages of TMAs is provided in Table 1. There are no established standards for array construction and little consensus on best practices. Array facilities often use formats requested by pathologists who have little experience in array construction or scientific methods. Array maps and strategies for tissue placement have also varied depending on the anticipated method of analysis.
Table 1.
The Advantages and Disadvantages of TMAs
| Advantages | Disadvantages |
|---|---|
| Applies uniform testing methodology to all samples on the slide | Cannot control for variability in tissue collection and processing before analysis (preanalytic variables) |
| Uses small amounts of tissue for studies, minimizing consumption of material and increasing the number of assays per specimen | May not take into account tumor heterogeneity (minimized by core replication) |
| Reduces reagent costs, laboratory processing, and time | May introduce statistical bias if high level of missing data |
| Allows comparison of replicates for assessment of reproducibility | Variable tissue loss from individual patients through sectioning |
| Amenable to use of multiple techniques and markers | Techniques (like FISH) where different protease treatment is required for different tissue samples are challenging |
| Allows direct comparison of different tissue histotypes on a single slide | Requires validation of techniques specific to TMAs |
Abbreviations: TMA, tissue microarray; FISH, fluorescent in situ hybridization.
In this article, we provide a set of evidence-based guidelines (summarized in Table 2) for array construction. The following guidelines represent our cumulative experience as directors of TMA facilities and researchers who collaborate with those facilities. The goal is achieve uniform, productive, and valid analyses of valuable tissue resources. Our guidelines for TMAs are consistent with the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) more generally.3
Table 2.
Summary of Proposed Recommendations
| Recommendations |
|---|
| Collection of the blocks to form the cohort |
| Write a protocol for the proposed study, including prospective design of the collection methods, anticipated studies to be performed, and statistical analyses to be completed, if known at the time of array construction. |
| Determine whether tissue subset available for TMA is consistent with total cohort with respect to relative risk of effect of primary end point of study. If representative and powered, then proceed to block review. If not representative, consider conversion to case-control series or selected cases series, or consider repeating efforts to obtain more blocks. |
| HE-stained slides from each block must be reviewed by a qualified reviewer (usually a pathologist) to define the region of interest for coring. Establish the cohort of patients who have acceptable amounts of target tissue for the specified TMA design. |
| Array construction issues |
| For large cohorts, use the 0.6-mm core size unless tissue type or TMA need justifies larger core. |
| To prevent map orientation confusion, use asymmetric array construction. |
| Use at least two-fold nonadjacent redundancy. |
| Include appropriate controls on every TMA master block. |
| If greater than one core from each patient is to be used in a single master block, we recommend nonadjacent placement of the cores. |
| Postconstruction array validation |
| Slides should be stained as soon as feasible after cutting (within 5 days) of the blocks or appropriately preserved. To minimize loss, TMA master blocks should be cut in batches of at least 10 slides to minimize loss related to facing the block. |
| Quality control should include HE staining and pathologist review of at least every 20th cut. |
| Statistical review of the final cohort from whom biomarker data were obtained should include assessment of the subset of the cohort missing from the TMA to be sure missing patients as a result of technical issues do not introduce bias into the cohort analysis, and comparison of row means and column means on each TMA section and between TMA blocks stained for the same biomarker to identify any systemic bias in data collection. |
Abbreviations: TMA, tissue microarray; HE, hematoxylin and eosin.
COLLECTION OF BLOCKS
Whether one does a retrospective collection in a small institution or prospectively collects samples as part of an international intergroup study, the first step is to write a protocol. The protocol is prospective. It must give a detailed description of the various steps to be followed and the eventual statistical analyses to be used in reporting the results.
The protocol addresses the manner of collecting the tissue blocks. The most common approach is to obtain all blocks that can be retrieved for coring. Another possibility is to obtain only a random sample of blocks. A common reason for choosing this approach is when there are budgetary constraints prohibiting complete tissue collection. Another instance is when a first random sample will be used to generate a hypothesis, such as whether a particular biomarker predicts the effects of a therapy, with a planned second and possibly even a third sample that will be used to test that hypothesis.
Once block collection is completed, we strongly recommend that hematoxylin and eosin (HE) –stained slides from each block be reviewed by a qualified pathologist to define the region of interest for coring. This can be done by simply circling the relevant region to be cored using a felt-tip pen. The review process inevitably eliminates some samples from use in the array as a result of insufficient tumor or other issues. Regions of artifact or necrosis can be excluded, and subregions can be defined if necessary. This is also the point at which areas of interest can be highlighted. For example, arrays can be made that sample the leading edge versus the center of a tumor as defined by annotation directly on the slides made by a qualified pathologist.
The review process may be done directly on the slides or using a digital pathology platform, allowing scanned slides to be reviewed remotely. Remote review has the advantage of including more pathologists but the disadvantage of requiring that another researcher transfer the information to the slide or block being used for array construction. Concurrent with or after this HE review, the paraffin block should be evaluated to identify thin or scant material in a block and to determine whether the target tissue in the block will provide the correct number of cores designed for the array yet retain sufficient tumor in the original block if necessary. Once this review has occurred and each block is paired with an HE slide, the cohort is ready for TMA construction.
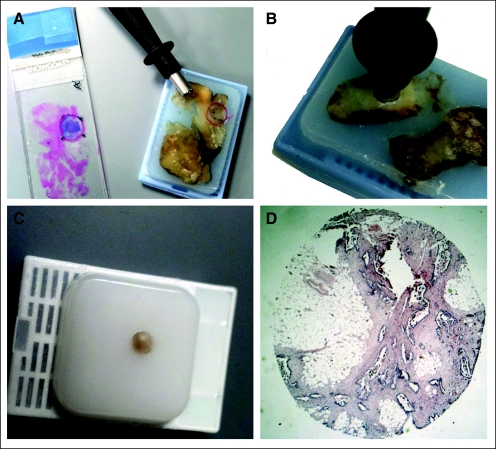
In some cases, the originating pathology department is not willing to release the block, even though the patient has given consent for the tissue to be released. One solution to this problem is for the originating institution to send the cooperative group a subsample of the tumor block. This can be done by using a skin punch biopsy (4-mm diameter) and then sending the resultant punch sample (still in the skin punch biopsy tool or transferred to an Eppendorf [Hamburg, Germany] tube or re-embedded in a new block) to the group bank. The resultant large core can then be re-embedded, and 0.6- to 1-mm cores can then be taken from the second block for inclusion in the study TMA (Fig 1). This approach should specifically be offered to reluctant pathology departments because they may not be aware of this option and it may significantly decrease missingness in the final collection. This approach is consistent with the College of American Pathologist's laboratory inspection guidelines requiring maintenance of formalin-fixed, paraffin-embedded diagnostic material for 10 years.
Fig 1.
A series of photos are shown that illustrate the process of coring a block and re-embedding the core in a new block to be sent to cooperative groups or others requesting formalin-fixed, paraffin-embedded tissue for research. The 4-mm core provides enough tissue to make a generous amount of DNA or RNA and can be either sectioned as is or cored up to four times with a 0.6-mm needle for tissue microarray construction.
STATISTICAL ISSUES REGARDING MISSING BLOCKS
Ideally, a representative block is available from every patient's tumor. This is achievable when a tumor block is mandatory for participating in the trial and probably only then. In studies involving retrospective tissue collection, the issue of missing blocks is inevitable. Any type of missing data in a clinical study is problematic. At best, the patient with missing data does not contribute to the study, and therefore, some statistical power is lost. More importantly, missingness introduces bias because the reason for missingness may be related to the end point in question. For example, small tumors may be associated with relatively good prognosis and may be disproportionately missing from the TMA precisely because they are small. When missingness is as low as 5%, for example, this bias may be negligible, and it might be reasonable to ignore it. When missingness is high, perhaps 30% or greater, the biases are potentially severe and may limit the ability to draw any conclusions. However, even when missingness is less than 30%, the possibility of bias looms large. Likewise, when missingness is greater than 30%, as it often is in existing cooperative group TMAs, valuable conclusions may still be drawn if the potential biases are addressed.
Reasons for missingness should be addressed. Sporadic missingness is more troublesome than whole subsets that are missing. For example, if 30% of the samples are missing and they are all from a cooperative group that refused participation in the correlative study, then power is decreased but there may be no concern about bias when analyzing questions of interactions between biomarkers and treatment effect. The best way to deal with missingness is to try to eliminate it. Although this is often impossible, second and third attempts to obtain tissue may be valuable, especially if alternative methods are offered.
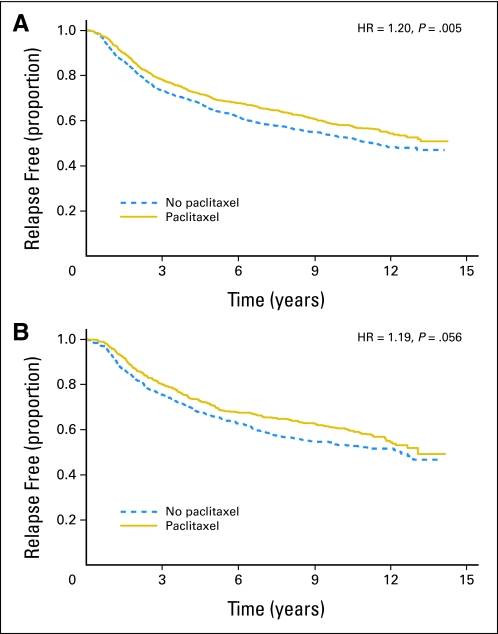
Figures 2, 3, and 4 show three examples of clinical trials with tissue retrospectively assessed using TMAs. Figures 2A (patients included in TMA) and 2B (patients not included in TMA) show relapse-free survival of patients with node-positive breast cancer who received adjuvant chemotherapy and did or did not receive paclitaxel in the Cancer and Leukemia Group (CALGB) 9344 trial.4 The missingness rate was 35%. Despite such a high rate, conclusions about whether a biomarker is predictive of paclitaxel's effect have greater credibility because the effect of paclitaxel is quite similar in the two subsets. One's comfort level can be further enhanced when the same markers (such as estrogen receptors and HER2/neu expression in this example) are assessed by other means for at least some of the patients in both subsets. Thus, concordance of the two methodologies can be assessed, and the missingness rate when assessing the possible role of these markers is substantially reduced.5
Fig 2.
Kaplan-Meier plots of relapse-free survival in patients with node-positive breast cancer who received adjuvant chemotherapy on the Cancer and Leukemia Group B 9344 trial split by receipt of paclitaxel. (A) Patients included in the tissue microarray (TMA). (B) Patients not included in the TMA. HR, hazard ratio.
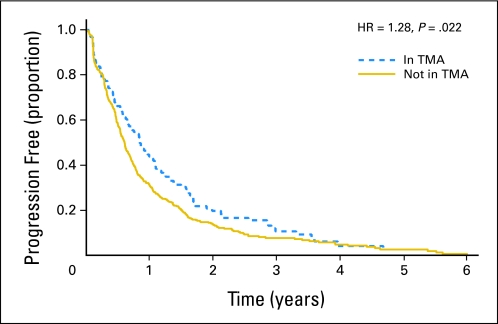
Fig 3.
Kaplan-Meier plot of patients from the Cancer and Leukemia Group B 9840 trial showing progression-free survival split by inclusion in the tissue microarray (TMA). Note that patients selected for inclusion in the TMA show statistically significant better outcome than patients not included in the TMA. HR, hazard ratio.
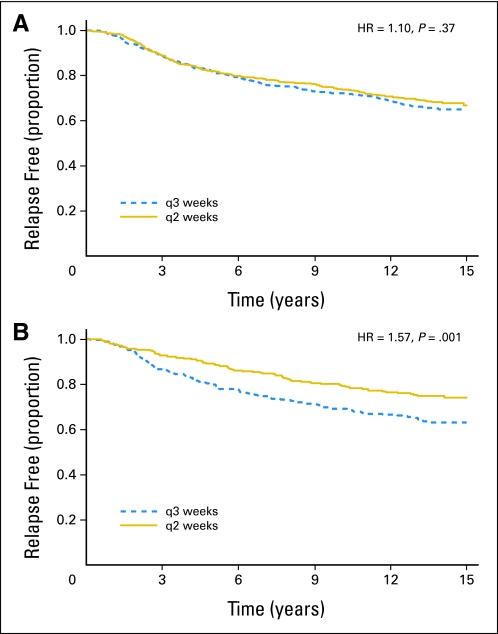
Fig 4.
Kaplan-Meier plots of patients from the Cancer and Leukemia Group B 9741 trial show that (A) patients for whom blocks were available for tissue microarray construction showed no improvement in relapse-free survival, even though (B) the whole study showed improved relapse-free survival with dose-dense doxorubicin (every 2 weeks [q2]) compared with dosing every 3 weeks (q3), the main finding of this clinical trial. HR, hazard ratio.
Figures 3 and 4 tell a different story. Figure 3 (progression-free survival) gives the results for CALGB 9840, a trial in metastatic breast cancer. The progression-free survival plot distinguishes between patients who were and were not included in the correlative sciences study. In this example, the missingness rate is 78%, which by itself challenges the credibility of any conclusions. To make matters worse, there is evidence of a bias in the selection of patients, with those patients included in the TMA having better prognoses than those not included in the TMA. Because of this concern, data obtained from this subset should only be considered exploratory.
Figures 4A (patients included in TMA) and 4B (patients not included in TMA) give the results of CALGB 9741.6,7 This trial showed that delivering chemotherapy every 2 weeks improves relapse-free survival for adjuvant, node-positive breast cancer compared with delivering chemotherapy every 3 weeks. We retrieved all available tissue blocks for TMAs, and we were interested in human epidermal growth factor receptor 2 status, among other markers. The missingness rate of 38% is similar to that for CALGB 9344 in Figure 2, but in Figure 4, the patients available for TMA show no benefit for chemotherapy delivered every 2 weeks, whereas the full study shows a significant benefit. This example is even more problematic than that of Figure 3. In that example, there was a bias in that the patients included in the TMA performed better overall than the patients not included in the TMA. However, any treatment benefit might well be the same in both groups. In Figure 4, there is evidence that the treatment effect was different in patients included in TMA than in patients not included in TMA. Any assessment of the role of human epidermal growth factor receptor 2, for example, in predicting the benefit of chemotherapy delivered every 2 weeks might be quite different for the patients included in TMA than for all patients in the study.
There is no ideal resolution to this conundrum. We retrieved all available blocks. Had we identified a sample of patients representative of the full trial and sought only those blocks, we would have gotten a subset of the patients we actually got, but we would not have gotten any of those who we were not able to get when we requested samples from all patients. Thus, we would have had the same bias in our sample of patients, but our sample would have been smaller.
One approach to consider before building the array with missing data is to determine whether the subset that is available is representative. Although representivity can be hard to define and results in subjective selection of variables that define representation, the most important variable in cooperative group trials is the issue addressed by the primary hypothesis or goal of the trial. For example, does drug X increase recurrence-free survival, or do patients receiving drug Y show a higher response rate than placebo? Thus, before making a TMA with a high degree of missingness, one can assess the relative risk, or protective value of the intervention (the drug) in the subset available for TMA construction. If the patients available for array construction show a relative risk for the primary end point that is significantly different than the entire cohort, then the subset cohort is not suitable for TMA production or the TMAs produced must only be used for exploratory studies.
Although there is no ideal resolution to the problem of missingness, there are strategies one can use in attempting to repair the problem. One is multiple imputation.8 A detailed description of the methodology is beyond our scope. Suffice it to say that the method produces a probability distribution of the full results based on the TMA results, outcome data, and covariate information, such as tumor size, for patients with TMA results and based on the outcome data and covariate information for patients with missing TMA results. The TMA results for the latter patients are imputed from a probability distribution that depends on the patient-specific information. This gives rise to a complete data set and enables finding measures such as statistical significance level of the interaction between biomarkers and treatment. Repeating this imputation process multiple times gives a probability distribution of statistical significance levels. This process delivers the strength of the data regarding questions to be addressed, and most importantly, it properly conveys the uncertainty in addressing those questions.
An advantage of multiple imputation is that it naturally incorporates the rate of missingness. If this rate is large, then the multiple imputation process results in a probability distribution of statistical significance that is wide, meaning that the study was not greatly informative. However, if the subset of missing can be proven to be biased by comparing relative risks of the missing and nonmissing data, then even multiple imputation will not resolve the missingness issue.
ARRAY CONSTRUCTION
Selection of Core Size
The diameter of the core for each spot on the array can range from 0.2 to 2 mm. The extremes are technically difficult, so most arrays use the 0.6-, 1.0-, or 1.5-mm cores. The appropriate core size may be driven by the target tissue. TMAs for some lesions, such as pancreatic carcinoma and carcinoma in situ of the breast, are more successful using larger cores.9,10 In addition, some pathologists feel comfortable with larger spots for conventional by-eye analysis. The disadvantage of larger spots is that fewer cores can be placed in each block, reducing some of the advantages of using a TMA. The larger the series to be analyzed, the more efficient it is to use smaller cores. Recent work has shown that when doing quantitative analysis, there is little difference in protein expression for core sizes of 0.6, 1.0, and 1.5 mm over a range of biomarkers.11 Therefore, for large cohorts, we recommend 0.6-mm cores.
Format
The format of TMAs has varied greatly in different facilities. Key issues related to formatting include array symmetry, placement of cores from the same patient (redundant cores), size of the array (row and column dimensions), and interspot distance and uniformity. We recommend asymmetric array construction to minimize errors in orientation. This can be achieved by design of the array, leaving an incomplete row at the bottom or inclusion of control tissue spots (often normal liver or kidney) outside of the row/column format to mark the top of the array. Level of redundancy is addressed later, but if more than one core from each patient is to be used in a single master block, we recommend nonadjacent placement of the cores. Optimally, the second core is placed some distance away from the first core so that artifacts that affect one core (staining, cutting, transfer, bubbles, and so on) do not affect the second core.12 This placement also decreases reading bias if stained spots are read by eye directly at the microscope. Although this technique requires more time constructing and scoring the TMA, quality of the test results should be the principal focus.
Inclusion of Controls
Nearly all TMAs include controls. Sometimes, they are normal tissue or normal tissue adjacent to tumors. Other times, the controls are from other species, other cell lines, other cell blocks, or even nontissue material. One group has used a series of eight to 12 immortalized cell lines, purchased from commercial sources such as the American Type Culture Collection (Manassas, VA) and validated by the American Type Culture Collection, for a standardization study.13 Some groups have used in-cohort controls in which certain patients are represented at higher frequencies or are present on every block of a series of TMAs to serve as standards between blocks.14 We recommend that controls be included on every TMA master block. However, control design should be based on the content, size, and goals of the study.
Level of Replication/Redundancy
With rare exceptions of extremely large cohorts, nearly all published work on TMAs is at least two-fold redundant (two or more spots for each patient). Replication gives a more robust estimate of biomarker expression, and it gives information about tissue and assay variability. Many studies have shown the representativeness of various numbers of spots. The most common replication in the literature seems to be two or three spots per sample.15–17 A handful of studies have used four or more spots.18 However, for most diagnostically useful biomarkers, the literature supports that two spots are sufficiently representative of the tissue sample as compared with whole-section analysis. Tumor biomarker density must also be considered in constructing the TMA. Just as highly heterogeneous tumors may require more cores, consideration of biomarker density should also be considered. For example, if a protein marker is only present in rare cells, a much larger number of cores may be required.
Definitive evidence on levels of redundancy and tumor and/or biomarker heterogeneity is not available. In the absence of direct evidence, we recommend at least duplicate nonadjacent sampling of tissue cores, with three-fold or greater replication when the scientific question includes issues related to tumor heterogeneity. Sister (replicate identical) blocks should be considered if sufficient tissue is available. Data reported must include all results, including replicates. Any replicates made based on observed results—such as when the original observation seemed to be unusual—must be reported with the original observations, however unusual they may be. For studies of heterogeneously expressed biomarkers that would require large numbers of cores for accurate assessment, TMAs may not be an appropriate methodology.
Array Maps
Accurate maps are necessary for each TMA. Commercially available algorithms exist to convert maps from serpentine to row-by-row format, but there is no consensus on the best method or orientation. Using automated digital image capture, the sector map may need to be tailored to the platform used.
ARRAY ANALYSIS AND VALIDATION
Array Cutting, Distribution, and Staining
Cutting a TMA master block always requires great care because each section is valuable. In particular, the process of facing the block should be done infrequently. The tumor loss as a result of facing the block must be weighed against the issue of cut TMA slide storage. For many antigens, cut slides lose antigenicity over time.19,20 Such loss may be a result of oxidation, exposure to water vapor, or some unknown reason. Several researchers recommend immediate staining21 or recommend storage conditions for both TMA cuts and routine formalin-fixed, paraffin-embedded slide cut sections20 to minimize artifactual loss of antigenicity generated by poorly defined variables associated with long intervals between cutting slides and staining them. To minimize this problem, we recommend staining slides within as short a time as possible after cutting the blocks. We also recommend that TMA master blocks be cut in batches of at least 10 slides to minimize loss from facing the block. This can be achieved by waiting until a number of assays are ready to be performed and then having a cutting day, on which all laboratories doing the assays proceed with their protocols. The protocols should be carefully developed on analogously handled but less valuable tissue resources. Some laboratories dip slides in paraffin to preserve them if they are not going to be stained within a few (5) days,20 whereas others wrap slides in paraffin and store at 4, −20, or −80°C (A. DeMarzo, personal communication, May 2006). Some new evidence suggests that paraffin dipping and preservation in a nitrogen box preserves slides for a year (Welsh et al, personal communication, August 2009), but the evidence on this topic is still insufficient to recommend a specific approach for preservation of cut sections.
Target Tissue Quality Control Review
After array construction, most facilities cut sections and stain with HE to review the array and assess its quality. We recommend HE staining and pathologist review at least every 20th cut. These reviews should be orthogonal in the sense that each row is represented the same number of times and each column is represented the same number of times. The estimated percentage of technically representative, usable cores should be recorded. High-quality TMAs should achieve 90% usability, although in our experience exceeding 95% is difficult to achieve. Percentages decrease as a block is sectioned. TMAs with less than 70% of patients may still be valuable, but missing data issues become paramount (see earlier discussion). Some TMAs with less than 70% of patients (and especially with < 50% of patients) may be worthless in drawing legitimate statistical conclusions. The problem is that the availability of cores may depend on important prognostic factors such as tumor size, and therefore, overall outcome measures may be biased. Statistical analyses may be able to partially adjust for such biases. However, in the worst case, an array may be strictly exploratory.
Analysis of the TMA
TMAs are most commonly used to assess protein expression by immunohistochemistry. Although there is no consensus in the committee regarding the best method to make this assessment, a range of methods can be acceptable if properly performed, standardized, and quality controlled. Perhaps most commonly, arrays may be read using traditional pathologist-based estimations of intensity and/or area of staining using chromogenic methods for visualization. Alternatively, quantitative measurements can be made on each spot. There are a number of commercially available tools for this task, including both chromogenic and fluorescence-based visualization techniques. There are advantages and disadvantages to each method. However, detailed exploration of the techniques and the strengths and weaknesses of each is beyond the scope of this study.
Regardless of the method of analysis, the TMA is especially susceptible to issues related to preanalytic variables. Specifically, warm and cold ischemic time, fixative time, processor, fixative and paraffin types, and myriad other variables can all affect the availability of protein for analysis on slides. TMAs made from tissue blocks from diverse sources can magnify these problems. We know of no well-documented, published method of standardization of preanalytic variables. Thus, we cannot make any global recommendations. However, it is important to be aware of preanalytic variables because they can represent a limitation of studies that use TMAs.
Analysis in the Presence of Missing Data
Spots are often lost in the process of cutting a TMA master block. The loss may be a result of technical reasons in the transfer or staining process but is often caused by tissue or tumor exhaustion in the core as a result of insufficient thickness of the original block in the region chosen for coring. This may give rise to a bias because the smallest tumors will be exhausted first. Such a bias is not necessarily limiting, but it requires careful comparison of included and excluded patients. This can be analyzed in a biostatistical process by looking for missingness-not-at-random. In the analysis of TMA data, we recommend biostatistical assessment of the subset of the cohort missing from the TMA to be sure missing samples as a result of technical issues do not introduce bias into the cohort analysis.
Analysis of Robustness of Each Assay (Row and Column Means and Block-to-Block Means)
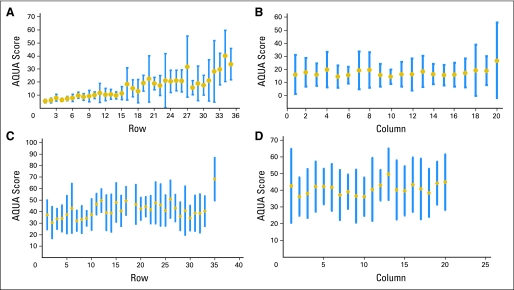
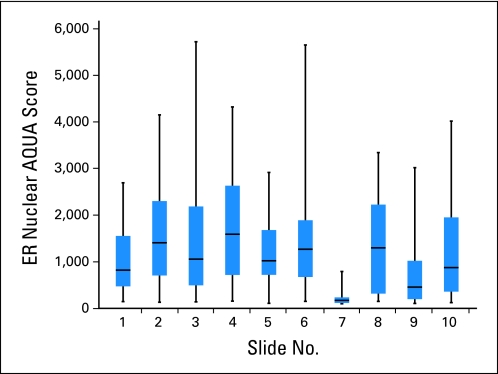
Analyses may fail to reveal a systematic technical flaw, leading to a spurious conclusion. Consistent with methods used to analyze nucleic acid arrays, one can calculate row and column means to look for regional differences or artifacts associated with the staining or other aspects of the assay. An example of calculated row and column means is shown in Figure 5. Large cohorts often must be split into multiple array blocks (eg, full assessment of the Southwest Oncology Group 9313 trial required data collection from 28 slides22). In cases where there are multiple blocks, the mean value for the assays can be compared to determine whether there is an outlier block (Fig 6). This group recommends a planned method for exclusion of systematic error. One option for quantitative data is the calculation and statistical comparison of row means and column means for each TMA and slide means comparisons for cohorts whose size requires representation on multiple master blocks. This information (or at least confirmation that the data have been assessed in this manner) should be included in Methods sections of reports of TMA data.
Fig 5.
Calculation of (A) row mean scores and (B) column mean scores show nonuniformity in the analysis of the tissue microarray (TMA) where rows 1 through 10 show dramatically lower average scores than rows 20 to 30, suggesting systematic error in the staining or analysis process. This observation suggests data from this array are not valid. Graphs showing (C) row means and (D) column means from another array analysis show the level of uniformity expected in analysis of TMAs. Row 35 in panel C shows a cell line/normal tissue control row that is an expected outlier compared with the tumor rows. The blue bars show the range of the data, and the gold dot shows the data mean for the row or column. AQUA, automated quantitative analysis.
Fig 6.
Assessment of average estrogen receptor (ER) quantitative scores on a per-slide basis suggests that slide 7 is an outlier. The difference may be caused by a systematic error in the process of generating the scores. The full data must be communicated, including those data points that seem unusual. Once that is done, such a slide might be excluded from one analysis of the data. A better approach is to prospectively define the use of a nonparametric method of data analysis. Similarly, if a fresh slide is used to repeat slide 7, then the original data must be communicated and suitably addressed in any analysis and then should be excluded from data analysis or, if possible, repeated on a fresh slide. AQUA, automated quantitative analysis.
Acknowledgment
We thank Allison Welsh and Bonnie Gould Rothberg for contributing data for the figures used in these guidelines. We also acknowledge Jason Bacher and Scott Hammond from the Cancer and Leukemia Group B Pathology Coordinating Office for their collective experience in managing, developing, and constructing tissue microarrays for the Cancer and Leukemia Group B.
Written on behalf of the Cancer and Leukemia Group B, Eastern Cooperative Oncology Group, Southwest Oncology Group, and North Central Cancer Treatment Group.
Research for Cancer and Leukemia Group B (CALGB) 9840, 9344, and 9741 trials was supported, in part, by National Cancer Institute (NCI) Grant No. CA31946 to the CALGB (Monica M. Bertagnolli, MD, Chair) and Grant No. CA33601 to the CALGB Statistical Center (Daniel Sargent, PhD). Also supported by NCI Grants No. CA16359, CA77658, CA33601, CA60138, CA04326, CA04457, and CA41287 and by the Breast Cancer Research Foundation.
Glossary Terms
- Formalin-fixed, paraffin-embedded:
Formalin-fixed, paraffin-embedded (FFPE) tissue is the standard for tissue preparation in anatomic pathology. The processing of tissue historically has included cutting into thin (5-mm) sections, then placing a cassette for fixation in formalin in a tissue processor, followed by infusion of paraffin and embedding on the block for subsequent sectioning for histologic evaluation or immunohistochemistry.
- Immunohistochemistry:
The application of antigen-antibody interactions to histochemical techniques. Typically, a tissue section is mounted on a slide and is incubated with antibodies (polyclonal or monoclonal) specific to the antigen (primary reaction). The antigen-antibody signal is then amplified using a second antibody conjugated to a complex of peroxidase-antiperoxidase (PAP), avidin-biotin-peroxidase (ABC) or avidin-biotin alkaline phosphatase. In the presence of substrate and chromogen, the enzyme forms a colored deposit at the sites of antibody-antigen binding. Immunofluorescence is an alternate approach to visualize antigens. In this technique, the primary antigen-antibody signal is amplified using a second antibody conjugated to a fluorochrome. On UV light absorption, the fluorochrome emits its own light at a longer wavelength (fluorescence), thus allowing localization of antibody-antigen complexes.
- Preanalytic variables:
Variables that occur before the time that the tissue is fixed in formalin. Most significantly, this includes biologic and artifactual changes to the tissue that occur during the time between the surgical ligation of the oxygen supply and tissue fixation. These are often divided into warm and cold ischemic time variables. However, preanalytic variables also include dozens of other variables beyond time, including room temperature, specimen thickness, surgical technique, transfer methods, and so on.
- Tissue microarray:
Used to analyze the expression of genes of interest simultaneously in multiple tissue samples, tissue microarrays consist of hundreds of individual tissue samples placed on slides ranging from 2 to 3 mm in diameter. Using conventional histochemical and molecular detection techniques, tissue microarrays are powerful tools to evaluate the expression of genes of interest in tissue samples. In cancer research, tissue microarrays are used to analyze the frequency of a molecular alteration in different tumor type, to evaluate prognostic markers, and to test potential diagnostic markers.
Appendix
The following institutions participated in this study: Christiana Care Health Services, Community Clinical Oncology Program (CCOP), Wilmington, DE–Stephen Grubbs, MD, supported by Grant No. CA45418; Dana-Farber Cancer Institute, Boston, MA–Harold J. Burstein, MD, supported by Grant No. CA32291; Dartmouth Medical School–Norris Cotton Cancer Center, Lebanon, NH–Konstantin Dragnev, MD, supported by Grant No. CA04326; Duke University Medical Center, Durham, NC–Jeffrey Crawford, MD, supported by Grant No. CA47577; Georgetown University Medical Center, Washington, DC–Edward P. Gelmann, MD, supported by Grant No. CA77597; Hematology-Oncology Associates of Central New York CCOP, Syracuse, NY–Jeffrey Kirshner, MD, supported by Grant No. CA45389; Long Island Jewish Medical Center, Lake Success, NY–Kanti R. Rai, MD, supported by Grant No. CA35279; Massachusetts General Hospital, Boston, MA–Jeffrey W. Clark, MD, supported by Grant No. CA32291; Medical University of South Carolina, Charleston, SC–Mark Green, MD, supported by Grant No. CA03927; Memorial Sloan-Kettering Cancer Center, New York, NY–Clifford A. Hudis, MD, supported by Grant No. CA77651; Mount Sinai Medical Center, Miami, FL–Rogerio C. Lilenbaum, MD, supported by Grant No. CA45564; Mount Sinai School of Medicine, New York, NY–Lewis R. Silverman, MD, supported by Grant No. CA04457; Nevada Cancer Research Foundation CCOP, Las Vegas, NV– John A. Ellerton, MD, supported by Grant No. CA35421; New Hampshire Oncology-Hematology PA, Concord, NH–Douglas J. Weckstein; North Shore-Long Island Jewish Health System, New Hyde Park, NY–Daniel Budman, MD, supported by Grant No. CA35279; Rhode Island Hospital, Providence, RI–William Sikov, MD, supported by Grant No. CA08025; Roswell Park Cancer Institute, Buffalo, NY–Ellis Levine, MD, supported by Grant No. CA59518; Southeast Cancer Control Consortium, CCOP, Goldsboro, NC–James N. Atkins, MD, supported by Grant No. CA45808; State University of New York Upstate Medical University, Syracuse, NY–Stephen L. Graziano, MD, supported by Grant No. CA21060; The Ohio State University Medical Center, Columbus, OH–Clara D. Bloomfield, MD, supported by Grant No. CA77658; University of Alabama Birmingham, Birmingham, AL–Robert Diasio, MD, supported by Grant No. CA47545; University of California at San Diego, San Diego, CA–Barbara A. Parker, MD, supported by Grant No. CA11789; University of California at San Francisco, San Francisco, CA–Charles J. Ryan, MD, supported by Grant No. CA60138; University of Chicago, Chicago, IL–Hedy L. Kindler, MD, supported by Grant No. CA41287; University of Illinois Minority-Based CCOP (MBCCOP), Chicago, IL–David J. Peace, MD, supported by Grant No. CA74811; University of Iowa, Iowa City, IA–Daniel A. Vaena, MD, supported by Grant No. CA47642; University of Maryland Greenebaum Cancer Center, Baltimore, MD–Martin Edelman, MD, supported by Grant No. CA31983; University of Massachusetts Medical School, Worcester, MA–William V. Walsh, MD, supported by Grant No. CA37135; University of Minnesota, Minneapolis, MN–Bruce A. Peterson, MD, supported by Grant No. CA16450; University of Missouri/Ellis Fischel Cancer Center, Columbia, MO–Michael C. Perry, MD, supported by Grant No. CA12046; University of Nebraska Medical Center, Omaha, NE–Anne Kessinger, MD, supported by Grant No. CA77298; University of North Carolina at Chapel Hill, Chapel Hill, NC–Thomas C. Shea, MD, supported by Grant No. CA47559; University of Tennessee Memphis, Memphis, TN–Harvey B. Niell, MD, supported by Grant No. CA47555; University of Vermont, Burlington, VT–Steven M. Grunberg, MD, supported by Grant No. CA77406; Wake Forest University School of Medicine, Winston-Salem, NC–David D. Hurd, MD, supported by Grant No. CA03927; Walter Reed Army Medical Center, Washington, DC–Brendan M. Weiss, MD, supported by Grant No. CA26806; Washington University School of Medicine, St. Louis, MO–Nancy Bartlett, MD, supported by Grant No. CA77440; Weill Medical College of Cornell University, New York, NY–John Leonard, MD, supported by Grant No. CA07968; Eastern Cooperative Oncology Group, Philadelphia, PA–Robert L. Comis, MD, Chairman; supported by Grant No. CA21115; North Central Cancer Treatment Group, Rochester, MN–Jan Buckner, MD, Chairman; supported by Grant No. CA25224; Southwest Oncology Group, San Antonio, TX–Laurence H. Baker, DO, Chairman; supported by Grant No. CA32102.
Footnotes
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: David L. Rimm, HistoRx (C), Metamark Genetics (C), Dako (C); Richard L. Schilsky, Foundation Medicine (C) Stock Ownership: David L. Rimm, HistoRx, Metamark Genetics; Torsten O. Nielsen, Bioclassifier Honoraria: None Research Funding: Torsten O. Nielsen, sanofi-aventis Canada; Anthony M. Magliocco, HistoRx Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: David L. Rimm, Torsten O. Nielsen, Scott D. Jewell, Baljit Singh, Gregory J. Tsongalis, Wendy L. Frankel, Jonathan F. Lara, Eric D. Hsi, Richard L. Schilsky, Ann Thor, Donald A. Berry
Collection and assembly of data: David L. Rimm, Torsten O. Nielsen, Scott D. Jewell, Daniel C. Rohrer, Gloria Broadwater, Frederic Waldman, Kisha A. Mitchell, Baljit Singh, Gregory J. Tsongalis, Anthony M. Magliocco, Jonathan F. Lara, Eric D. Hsi, Ira J. Bleiweiss, Sunil S. Badve, Beiyun Chen, Peter M. Ravdin, Ann Thor, Donald A. Berry
Data analysis and interpretation: David L. Rimm, Torsten O. Nielsen, Gloria Broadwater, Donald A. Berry
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Wan WH, Fortuna MB, Furmanski P. A rapid and efficient method for testing immunohistochemical reactivity of monoclonal antibodies against multiple tissue samples simultaneously. J Immunol Methods. 1987;103:121–129. doi: 10.1016/0022-1759(87)90249-3. [DOI] [PubMed] [Google Scholar]
- 2.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 3.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 4.Henderson IC, Berry DA, Demetri GD, et al. Improved outcomes from adding sequential paclitaxel but not from escalating doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol. 2003;21:976–983. doi: 10.1200/JCO.2003.02.063. [DOI] [PubMed] [Google Scholar]
- 5.Berry D, Thor A, Jewell S. Benefits of adding paclitaxel to adjuvant doxorubicin/cyclophosphamide depending on HER2 & ER status: Analysis of tumor tissue microarrays and immunohistochemistry in CALGB 9344 (Intergroup 0148) Cancer Res. 2009;69(suppl 24):606. abstr. [Google Scholar]
- 6.Citron ML, Berry DA, Cirrincione C, et al. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: First report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741. J Clin Oncol. 2003;21:1431–1439. doi: 10.1200/JCO.2003.09.081. [DOI] [PubMed] [Google Scholar]
- 7.Berry DA, Cirrincione C, Henderson IC, et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA. 2006;295:1658–1667. doi: 10.1001/jama.295.14.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York, NY: John Wiley & Sons; 1987. [Google Scholar]
- 9.Yang XR, Charette LA, Garcia-Closas M, et al. Construction and validation of tissue microarrays of ductal carcinoma in situ and terminal duct lobular units associated with invasive breast carcinoma. Diagn Mol Pathol. 2006;15:157–161. doi: 10.1097/01.pdm.0000213453.45398.e0. [DOI] [PubMed] [Google Scholar]
- 10.Hewitt SM. Tissue microarrays as a tool in the discovery and validation of tumor markers. Methods Mol Biol. 2009;520:151–161. doi: 10.1007/978-1-60327-811-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anagnostou VK, Lowery FJ, Syrigos KN, et al. Quantitative evaluation of protein expression as a function of tissue microarray core diameter: Is a large (1.5 mm) core better than a small (0.6 mm) core? Arch Pathol Lab Med. 2010;134:613–619. doi: 10.5858/134.4.613. [DOI] [PubMed] [Google Scholar]
- 12.Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: Progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26:5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- 13.McCabe A, Dolled-Filhart M, Camp RL, et al. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 14.Sherman ME, Rimm DL, Yang XR, et al. Variation in breast cancer hormone receptor and HER2 levels by etiologic factors: A population-based analysis. Int J Cancer. 2007;121:1079–1085. doi: 10.1002/ijc.22812. [DOI] [PubMed] [Google Scholar]
- 15.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest. 2000;80:1943–1949. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 16.Torhorst J, Bucher C, Kononen J, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin MA, Dunn R, Strawderman M, et al. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol. 2002;26:312–319. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Moeder CB, Giltnane JM, Harigopal M, et al. Quantitative justification of the change from 10% to 30% for human epidermal growth factor receptor 2 scoring in the American Society of Clinical Oncology/College of American Pathologists guidelines: Tumor heterogeneity in breast cancer and its implications for tissue microarray based assessment of outcome. J Clin Oncol. 2007;25:5418–5425. doi: 10.1200/JCO.2007.12.8033. [DOI] [PubMed] [Google Scholar]
- 19.Fergenbaum JH, Garcia-Closas M, Hewitt SM, et al. Loss of antigenicity in stored sections of breast cancer tissue microarrays. Cancer Epidemiol Biomarkers Prev. 2004;13:667–672. [PubMed] [Google Scholar]
- 20.DiVito KA, Charette LA, Rimm DL, et al. Long-term preservation of antigenicity on tissue microarrays. Lab Invest. 2004;84:1071–1078. doi: 10.1038/labinvest.3700131. [DOI] [PubMed] [Google Scholar]
- 21.Simon R, Mirlacher M, Sauter G. Immunohistochemical analysis of tissue microarrays. Methods Mol Biol. 2010;664:113–126. doi: 10.1007/978-1-60761-806-5_12. [DOI] [PubMed] [Google Scholar]
- 22.Porter PL, Barlow WE, Yeh IT, et al. P27(Kip1) and cyclin E expression and breast cancer survival after treatment with adjuvant chemotherapy. J Natl Cancer Inst. 2006;98:1723–1731. doi: 10.1093/jnci/djj467. [DOI] [PMC free article] [PubMed] [Google Scholar]