Abstract
Purpose
BRAF mutations occur in non–small-cell lung cancer. Therapies targeting BRAF mutant tumors have recently been identified. We undertook this study to determine the clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations.
Patients and Methods
We reviewed data from consecutive patients with lung adenocarcinoma whose tumors underwent BRAF, EGFR, and KRAS mutation testing as well as fluorescence in situ hybridization for ALK rearrangements. Patient characteristics including age, sex, race, performance status, smoking history, stage, treatment history, and overall survival were collected.
Results
Among 697 patients with lung adenocarcinoma, BRAF mutations were present in 18 patients (3%; 95% CI, 2% to 4%). The BRAF mutations identified were V600E (50%), G469A (39%), and D594G (11%). Mutations in EGFR were present in 24%, KRAS in 25%, and ALK translocations in 6%. In contrast to patients with EGFR mutations and ALK rearrangements who were mostly never smokers, all patients with BRAF mutations were current or former smokers (P < .001). The median overall survival of advanced-stage patients with BRAF mutations was not reached. In comparison, the median overall survival of patients with EGFR mutations was 37 months (P = .73), with KRAS mutations was 18 months (P = .12), and with ALK rearrangements was not reached (P = .64).
Conclusion
BRAF mutations occur in 3% of patients with lung adenocarcinoma and occur more commonly in current and former smokers. The incidence of BRAF mutations other than V600E is significantly higher in lung cancer than in melanoma.
INTRODUCTION
The identification of activating mutations in the epidermal growth factor receptor (EGFR)1–3 that predict for response to EGFR tyrosine kinase inhibitors (TKIs) has changed how lung adenocarcinoma is managed. Subsequent work has identified other driver mutations that can be targeted with drugs, confirming the validity of this approach. The most recent example of this is with the oncogenic translocation between the anaplastic lymphoma kinase (ALK) and echinoderm microtubule like-4 (EML-4) genes.4,5 The immediate availability of an oral inhibitor of the ALK tyrosine kinase, crizotinib (PF-02341066), led to the rapid accrual of an early-phase clinical trial of this drug in patients with lung adenocarcinomas harboring an ALK rearrangement. The overall response rate from this study was 64%.6 Recent data suggest that EGFR mutations are present in 15% to 20% of lung adenocarcinomas, translocations between EML4 and ALK in 3% to 7%,4 and mutations in KRAS in 25%.7 Mutations in HER2, BRAF, FGFR2, and PIK3CA have been identified at lower frequencies. With a known driver mutation now identifiable in the majority of lung adenocarcinoma cases, individualized treatment is, in part, limited by the availability of proven targeted therapies.
Coincident with the discovery of EGFR mutations and EML4–ALK was the identification of subgroups that had higher frequencies of these mutations, which provided strategies for clinically enriching populations for mutation-positive patients. Hence EGFR mutations, although found in just 10% of unselected patients with non–small-cell lung cancer (NSCLC), are present in 50% of patients with lung adenocarcinoma who are never smokers. EML4–ALK translocations, found in approximately 5% of patients with NSCLC, are present in 20% of patients with lung adenocarcinoma who are wild-type (WT) for EGFR and KRAS, with an even higher frequency in patients who are never smokers. Observations such as these have been important, allowing us to select enriched subpopulations for genotyping and in whom early trials of targeted therapies might be more efficiently performed.
Although the identification of BRAF mutations in lung cancer predated the discovery of EML4–ALK translocations, few clinical studies of BRAF mutant lung cancer have been completed. As a member of the Ras/mitogen-activated protein kinase signaling pathway, BRAF lies downstream of KRAS, and directly phosphorylates MEK, which in turns phosphorylates ERK. The pathway culminates in the transcription of genes favoring proliferation and survival. A number of BRAF mutations have been identified. The most common is a valine to glutamate substitution at codon 600 (V600E), which accounts for more than 90% of the BRAF mutations in melanoma.8 Substitution of this negatively charged amino acid for valine is thought to eliminate a protein–protein interaction between the activation segment and glycine P-loop that normally maintains BRAF in an inactive conformation.9 Preclinical work by two groups has confirmed a role for mutant BRAF in lung adenocarcinoma initiation and maintenance. An inducible transgenic mouse model of BRAF V600E developed by Ji et al10 demonstrated that mutant BRAF was sufficient for the development of lung adenocarcinomas. The growth of these tumors was dependent on persistent oncogene expression, suggesting that mutant BRAF may also be necessary for maintenance. A mouse model containing a conditional knock-in of BRAF V600E generated by Dankort et al11 similarly led to the development of adenomatous tumors.
Because the incidence of BRAF mutations is highest in melanomas (50% to 70%), the bulk of the clinical trials to date have focused on this disease, targeting either BRAF itself or MEK 1/2, the latter of which is associated with growth-dependency in BRAF mutant cell lines.12,13 Specific agents have included PLX4032, XL281, selumetanib, and GSK2118436.14–17 The most promising of these has been PLX4032, which was associated with an 80% response rate in the extension phase of a recent multicenter phase I study that included 32 patients with advanced-stage melanoma with BRAF V600E mutations.18 On the basis of these results, a randomized phase III study of PLX4032 versus dacarbazine in untreated patients with metastatic melanoma harboring BRAF V600E mutations has opened.
These data, which suggest that mutant BRAF is a driver mutation in lung adenocarcinoma, coupled with the encouraging clinical trial work of RAF inhibitors in patients with metastatic melanoma, provide an impetus for the further study of BRAF targeted therapy in NSCLC. Since 2009, testing for BRAF mutations in lung adenocarcinomas has been performed at Memorial Sloan-Kettering Cancer Center through an ongoing institutional lung cancer mutation analysis program.19 To understand the natural history of this disease, we sought to summarize the clinical features of patients with lung adenocarcinomas who harbor BRAF mutations, comparing them with those from patients with mutations in EGFR and KRAS and rearrangements in ALK.
PATIENTS AND METHODS
Study Design and Patients
Patients with lung adenocarcinoma who underwent molecular testing for EGFR, KRAS, and BRAF mutations and ALK rearrangements between May 2009 and May 2010 were identified for review. Clinical characteristics including age, sex, race (reported by the patient), stage, treatment history, and Karnofsky performance status were recorded. Smoking history was obtained through review of a prospectively administered questionnaire given at the time of initial encounter. Computed tomography scans of advanced-stage (IIIB/IV) patients with BRAF mutations were reviewed before and during treatment with first-line therapy and until the time of radiographic disease progression to determine best overall response by Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1.
All chart review/tissue collection was carried out under institutional review board/privacy board–approved protocols or waivers.
Genotype Analysis
BRAF mutation analysis was performed using the MassARRAY system (Sequenom, San Diego, CA) based on matrix-assisted laser desorption/ionization time of flight mass spectrometry.20 Amplification and extension primers were designed using the Sequenom Assay Designer v3.1 software to target mutations involving codons V600, D594, and G469 of the BRAF gene. Amplification primers were designed with a 10-mer tag sequence to increase their mass so that they fall outside the range of detection of the matrix-assisted laser desorption/ionization time of flight mass spectrometry. The primer sequences are listed in Appendix Table A1 (online only), and a detailed description of the protocol is published elsewhere.21 EGFR exon 19 deletion and exon 21 L858R mutations were detected through a polymerase chain reaction–based assay, as previously described.22 KRAS exon 2 mutations were identified through direct sequencing.23 Rearrangements of ALK were detected through fluorescence in situ hybridization (FISH) using a dual-color break-apart FISH assay (Ang et al, manuscript submitted for publication). Positive cases were defined as the presence of a split signal indicating rearrangement of the ALK locus at 2p23 or the presence of a single red signal indicating loss of the 5′ DNA sequence in ≥ 5% of cells. Where tissue was available, including all cases with only 5% to 15% of FISH-positive cells, polymerase chain reaction for specific EML4–ALK transcripts or immunohistochemistry with an ALK-specific antibody (clone D5F3, gift of Cell Signaling Technology, Beverly, MA) was performed to confirm the presence of an EML4–ALK translocation. Technical details of EML4–ALK testing are presented elsewhere (Ang et al, manuscript submitted for publication). Because of the nonoverlapping nature of mutations in EGFR and KRAS and translocations in EML4–ALK, only patients who were WT for EGFR and KRAS underwent reflex testing for ALK rearrangement.
Statistical Methods
Groups determined by mutation status (EGFR, KRAS, EML4-ALK, BRAF) were compared with respect to clinical characteristics using Fisher's exact test and Wilcoxon-Mann-Whitney test. Overall survival (OS) was calculated using the Kaplan-Meier method. Patients were followed from the date of diagnosis of stage IIIB/IV or recurrent disease until death or last available follow-up. Survival data were obtained through existing medical records or Social Security death index and updated as of June 2010. Group comparison was performed with log-rank tests. Statistical analyses were performed using SAS statistical software (SAS Institute, Cary, NC).
RESULTS
Patient Characteristics
Patient characteristics are summarized in Table 1. The individual clinical characteristics of patients with BRAF mutations are listed in Tables 2 and 3. No unique histologic phenotype was associated with a BRAF mutant genotype. Mutation testing identified five genotype categories (BRAF mutant, EGFR mutant, KRAS mutant, ALK rearrangement, and unknown genotype). There were no significant differences in stage, sex, age, or Karnofsky performance status between patients with BRAF mutations and those with either EGFR or KRAS mutations.
Table 1.
Patient Clinical Characteristics (N = 687)
| Characteristic |
BRAF Mutation |
EGFRMutation |
KRASMutation |
ALK Rearrangement |
Unknown Genotype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | |
| Total patients | 18 | 3 | 165 | 24 | 169 | 25 | 44 | 6 | 291 | 42 |
| Stage | ||||||||||
| Early (I-IIIA) | 8 | 44 | 62 | 38 | 54 | 32 | 6 | 14 | 91 | 31 |
| Advanced (IIIB/IV) | 10 | 56 | 103 | 62 | 115 | 68 | 38 | 86 | 200 | 69 |
| Sex, female | 11 | 61 | 116 | 70 | 121 | 72 | 22 | 50 | 182 | 63 |
| Age, years | ||||||||||
| Median | 64 | 65 | 66 | 60 | 67 | |||||
| Range | 44-81 | 39-88 | 31-90 | 32-84 | 33-89 | |||||
| ≥ 70 | 1 | 6 | 61 | 37 | 56 | 33 | 14 | 32 | 94 | 32 |
| KPS (%)* | ||||||||||
| Median | 80 | 80 | 80 | 90 | 80 | |||||
| Range | 70-90 | 60-90 | 50-90 | 60-90 | 30-90 | |||||
| ≤ 70 | 2 | 20 | 17 | 17 | 29 | 28 | 5 | 13 | 48 | 26 |
| Race, white | 18 | 100 | 134 | 81 | 153 | 91 | 34 | 77 | 252 | 87 |
| Smoking history | ||||||||||
| Never | 0 | 0 | 111 | 67 | 12 | 7 | 35 | 80 | 143 | 49 |
| Current and former | 18 | 100 | 54 | 33 | 157 | 93 | 9 | 20 | 148 | 51 |
| Pack years† | ||||||||||
| Median | 38 | 18 | 32 | 15 | 30 | |||||
| Range | 14-75 | 1-90 | 1-150 | 1-60 | 0-150 | |||||
Abbreviation: KPS, Karnofsky performance status.
Among patients with advanced-stage cancer only.
Among ever smokers.
Table 2.
Individual Patient Characteristics, Early Stage
| Patient | BRAF Mutation | Age(years) | Sex | Smoking Status | Pack Years | Stage | Neoadjuvant Treatment | Response* | Adjuvant Treatment | Survival (months) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D594G | 44 | F | Current | 25 | IA | Carboplatin + pemetrexed | SD | No | 16+ |
| 2 | G469A | 64 | M | Current | 56 | IIIA | Cisplatin + docetaxel | PR | No | 74+† |
| 3 | G469A | 54 | F | Current | 60 | IIIA | No | Cisplatin + vinorelbine | 5+ | |
| 4 | G469A | 66 | M | Current | 39 | IIIA | Carboplatin + paclitaxel | SD | No | 9+ |
| 5 | G469A | 68 | F | Current | 45 | IIIA | No | Carboplatin + vinorelbine | 10+ | |
| 6 | V600E | 68 | M | Former | 19 | IA | No | No | 12+ | |
| 7 | V600E | 70 | F | Former | 26 | IA | No | No | 9+ | |
| 8 | V600E | 66 | F | Former | 30 | IA | No | No | 6+ |
Abbreviations: F, female; SD, stable disease; M, male; PR, partial response.
Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1.
Recurred.
Table 3.
Individual Patient Characteristics, Advanced Stage
| Patient | BRAF Mutation | Age | Sex | KPS (%) | Smoking Status | Pack Years | Brain Metastasis | Lines of Therapy | First-Line Treatment | Response* | Survival (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D594G | 64 | F | 80 | Former | 68 | Yes | Unknown | Unknown | NA | 5+ |
| 2 | G469A | 64 | M | 80 | Current | 55 | No | 1 | Carboplatin + pemetrexed + bevacizumab | SD | 18+ |
| 3 | G469A | 61 | M | 80 | Current | 42 | Yes | 1 | Carboplatin + pemetrexed | PR | 7 |
| 4 | G469A | 68 | M | 90 | Former | 15 | Yes | 1 | Cisplatin + pemetrexed | PR | 6+ |
| 5 | V600E | 62 | F | 90 | Former | 13.5 | No | 3 | Erlotinib + pemetrexed + bevacizumab | PR | 19 |
| 6 | V600E | 67 | M | 90 | Former | 72.5 | No | 3 | Erlotinib | SD | 75+ |
| 7 | V600E | 58 | F | 90 | Former | 16.5 | No | 4 | Erlotinib | PD | 51+ |
| 8 | V600E | 81 | F | 70 | Former | 37 | No | 2 | Erlotinib | PD | 15+ |
| 9 | V600E | 55 | F | 80 | Current | 30 | No | 1 | Paclitaxel + pemetrexed + bevacizumab | SD | 8+ |
| 10 | V600E | 63 | F | 70 | Current | 75 | No | 2 | Carboplatin + pemetrexed | PR | 7+ |
Abbreviations: KPS, Karnofsky performance status; F, female; NA, not applicable; M, male; SD, stable disease; PR, partial response; PD, progressive disease.
Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1.
All patients with BRAF mutations were current or former smokers, with a median smoking history of 38 pack years. Using a cutoff of 15 or fewer pack years,24 only two of 19 patients were “light smokers.” This was significantly different from patients with EGFR mutations, the majority of whom were never smokers (0% v 67% never smokers, P < .001), and patients with ALK rearrangements (0% v 80%, P < .001). Patients with KRAS mutations were also predominantly smokers, with no significant difference in smoking history between them and those with BRAF mutations (93% v 100%, P = .61).
Although 86% of the overall population was white, all patients who tested positive for a BRAF mutation were white. The proportion of patients with BRAF mutations who were white was higher than the proportion of patients with EGFR mutations or ALK rearrangements who were white (EGFR v BRAF: 81% v 100%, P = .047; ALK versus BRAF: 77% v 100%, P = .025). The majority of nonwhite patients were Asian.
BRAF Mutation Genotypes
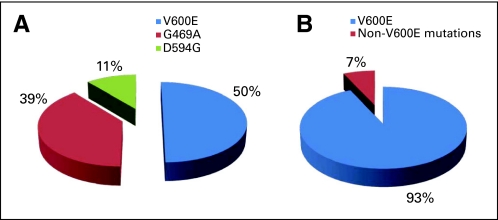
Three BRAF mutation genotypes were identified: V600E mutations (exon 15), G469A mutations, (exon 11), and D594G mutations (exon 15). The majority of mutations were V600E mutations (50%, n = 9), followed by G469A mutations (39%, n = 7) and D594G mutations (11%, n = 2; Fig 1). No patient with a BRAF mutation had a concomitant mutation in EGFR or KRAS or a translocation in ALK.
Fig 1.
Relative frequency of BRAF mutations in (A) lung adenocarcinoma versus (B) melanoma.
Clinical Outcomes
Of the 10 patients with advanced-stage disease who had BRAF mutations, two died during study follow-up. There were no significant differences in OS for advanced-stage patients with BRAF mutations compared with patients with other driver mutations (Fig 2). Median OS was not reached in patients with advanced disease who had BRAF mutations. In comparison, the median OS was 37 months for patients with EGFR mutations (P = .73), 18 months for patients with KRAS mutations (P = .12), and was not reached for patients with ALK rearrangements (P = .64). The 2-year OS for patients with BRAF mutations was 57% (95% CI, 24% to 100%). In comparison, patients with EGFR mutations, KRAS mutations, and ALK rearrangements had 2-year OS rates of 69% (95% CI, 53% to 81%), 40% (95% CI, 25% to 54%), and 91% (95% CI, 60% to 98%), respectively. Patients with EGFR mutations did have a longer OS as compared with patients with KRAS mutations (P = .001). Patients with ALK rearrangements also had a longer OS as compared with patients with KRAS mutations (P < .0001). Multivariate analysis was not feasible given the relatively small number of deaths in the BRAF mutation group. Four of 10 patients with advanced disease had a radiographic response to first-line chemotherapy (Table 3). Survival data for early-stage patients with BRAF mutations were not sufficiently mature for analysis, with 75% of patients censored at 12 months of follow-up and no deaths.
Fig 2.
Kaplan-Meier curve for overall survival in patients with advanced stage (IIIB/IV) disease.
DISCUSSION
Somatic activating BRAF mutations were first described by Davies et al8 in 2002. Their series showed an incidence of 8% across all cancers and 3% in lung cancer. Worldwide, this equates to some 35,000 patients who might benefit from a RAF inhibitor, which is similar in scope to the 45,000 patients who are projected to benefit from treatment with ALK inhibitors. Testing of lung adenocarcinoma tumors at Memorial Sloan-Kettering Cancer Center for BRAF mutations as well as EGFR and KRAS mutations and rearrangements in ALK has provided us with what is, to our knowledge, the largest clinical analysis of BRAF mutant lung adenocarcinoma to date.
The incidence of BRAF mutations in our series was 3% (95% CI, 2% to 4%), which is similar to other data.25 Remarkably, all patients with a BRAF mutation were current or former smokers. This absence of never smokers is striking when compared with patients with EGFR mutations and ALK rearrangements, in whom never smokers comprise 67% and 80%, respectively, of patients with these mutations (P < .001 v BRAF mutations for both). The relative paucity of BRAF mutations in nonwhite populations has been previously suggested, with one series showing just one of 97 Japanese patients with lung adenocarcinoma (1%) harboring a BRAF mutation (V600E).26
We also found a considerably smaller proportion of V600E mutations (due to a T→A transversion) than has been reported for melanomas (50% v > 90%; Fig 2). Notably, 39% of BRAF mutations in our series involved a G→C transversion (G469A), which is found, in contrast, in only 0.4% of melanomas.27,28 The higher relative frequency of G469A G→C transversions in lung cancers compared with melanomas may reflect a tobacco-related carcinogenic effect, although G→T transversions in KRAS and P53 have the strongest relationship to smoking7 (Dogan et al, manuscript submitted for publication). This lower incidence of V600E mutations is important, as current second-generation RAF inhibitors, in light of the near ubiquity of the V600E mutation in melanoma, have been tailored to have specific activity against the V600E mutant kinase. The clinical activity of these drugs against the G469A and D594G mutant kinases is unknown. Indeed, in vitro data have shown that cell lines with non-V600E mutations, including H1755 lung cancer cells harboring G469A mutations, are resistant to the growth-suppressive effects of PLX4032.29 These non-V600E mutations may, however, be targets for other existing inhibitors of RAF and MEK1/2. Data from Wan et al9 have shown that cells expressing low- or intermediate-activity non-V600E mutant kinases have increased C-RAF activity, and are, as a result, sensitive to sorafenib through inhibition of C-RAF dependent ERK activation. This is an important observation, as the first clinical trials of RAF inhibitors in melanoma used sorafenib, which was found to be ineffective against the V600E mutant isoform.30 There were too few patients with BRAF mutations to perform a comparison of the clinical characteristics and outcomes among BRAF mutation subtypes. Preclinical data demonstrate that both the V600E and G469A mutation are associated with increased BRAF kinase activity and downstream ERK1/2 phosphorylation.9 D594G mutants may have, in contrast, lower kinase activity.31 It will be interesting to see whether a comparable difference in clinical behavior is seen among these mutations, either within or outside the context of a specific treatment, as has been demonstrated with the two predominant EGFR mutation subtypes after treatment with erlotinib (exon 19 deletion, exon 21 L858R substitution).32
We note that other BRAF mutations in lung adenocarcinoma have been identified, including mutations in amino acids 421, 436, 459, 466, 471, and 597.28 These individual mutations represent 1% to 3% of all BRAF mutations reported, however. As such, it is unlikely that our reported BRAF mutation rate significantly under-represents the true mutation rate.
With a median follow-up of 10 months for the entire cohort and 16 months for patients with BRAF mutations, we found no significant differences in the OS of advanced-stage patients with BRAF mutations versus those with EGFR or KRAS mutations or ALK rearrangements. A comparison of the Kaplan-Meier curves suggests that the natural history of patients with BRAF mutations may be relatively favorable, even in the absence of treatment with a RAF inhibitor. These data are preliminary, however, and require longer follow-up for confirmation. The retrospective nature of this study and the recent availability of BRAF mutation testing raise the possibility of a bias in which the longest living patients preferentially underwent mutation testing, thereby enriching for patients with better outcomes. Because the inclusion criterion for the date of mutation testing was constant, however, all genotypes were at risk for this. Although outliers existed, it is unlikely that this bias disproportionately affected the BRAF group, as the median times from the diagnosis of disease to mutation testing for patients with EGFR, KRAS, and BRAF mutations were similar at 1.1 months, 1.2 months, and 1.5 months, respectively.
In conclusion, our data show that BRAF mutations occur in approximately 3% of patients with lung adenocarcinoma (Fig 3). All BRAF mutations we identified were mutually exclusive of EGFR, KRAS, and EML4–ALK. Our data have additionally defined subgroups with relatively higher proportions of these mutations, particularly smokers, in whom the frequency approaches 5% and doubles to 10% in those smokers who are WT for EGFR and KRAS and who do not harbor an ALK rearrangement. Many agents targeting the BRAF pathway are in clinical development, such as PLX4032, XL281, selumetanib, and GSK2118436.14–17 Comprehensive prospective genotyping rather than clinical enrichment for future studies of drugs targeting this pathway is essential in light of recent data noting a paradoxical RAF inhibitor-mediated activation of the RAS signaling pathway in BRAF WT cell lines.33
Fig 3.
Relative frequency of driver mutations in patients with lung adenocarcinoma. Driver mutations can now been identified in the majority of patients with lung adenocarcinoma.
Appendix
Table A1.
PCR Primers and Extension Primers
| Mutation | Primer |
|---|---|
| PCR* | |
| BRAF_1799_PCR_F | 5′-acgttggatgTCTTCATGAAGACCTCACAG-3′ |
| BRAF_1799_PCR_F | 5′-acgttggatgATGGATCCAGACAACTGTTC-3′ |
| BRAF_1406G | 5′-acgttggatgTGGGCAGATTACAGTGGGAC-3′ |
| BRAF_1798G | 5′-acgttggatgATGGGACCCACTCCATCGAG-3′ |
| Extension | |
| BRAF_1799_F | 5′-GGTGATTTTGGTCTAGCTACAG-3′ |
| BRAF_1799_R | 5′-CCCACTCCATCGAGATTTC-3′ |
| BRAF_1406G | 5′-CACTTTCCCTTGTAGACTGTT-3′ |
| BRAF_1798G | 5′-GTGATTTTGGTCTAGCTACA-3′ |
Abbreviation: PCR, polymerase chain reaction.
With 10-bp 5′ overhang lowercase (working solution, 0.5 μmol/L).
Footnotes
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Mark G. Kris, GlaxoSmithKline (C), AstraZeneca (C) Stock Ownership: None Honoraria: Vincent A. Miller, GlaxoSmithKline, Roche Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Paul K. Paik, Mark G. Kris, Gregory J. Riely
Administrative support: Mark G. Kris
Provision of study materials or patients: Mark G. Kris, Vincent A. Miller, Gregory J. Riely
Collection and assembly of data: Paul K. Paik, Maria E. Arcila,Michael Fara
Data analysis and interpretation: Paul K. Paik, Maria E. Arcila, Michael Fara, Camelia S. Sima, Vincent A. Miller, Mark G. Kris, Marc Ladanyi, Gregory J. Riely
Manuscript writing: Paul K. Paik, Maria E. Arcila, Michael Fara, Camelia S. Sima, Vincent A. Miller, Mark G. Kris, Marc Ladanyi, Gregory J. Riely
Final approval of manuscript: Paul K. Paik, Maria E. Arcila, Michael Fara, Camelia S. Sima, Vincent A. Miller, Mark G. Kris, Marc Ladanyi, Gregory J. Riely
REFERENCES
- 1.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bang Y, Kwak EL, Shaw AT, et al. Clinical activity of the oral ALK inhibitor PF-02341066 in ALK-positive patients with non-small cell lung cancer (NSCLC) J Clin Oncol. 2010; 28(suppl):6s. abstr 3. [Google Scholar]
- 7.Riely GJ, Kris M, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 9.Wan P, Garnett M, Roe M, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 10.Ji H, Wang Z, Perera SA, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 11.Dankort D, Filenova E, Collado M, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellbrock C, Hurlstone A. BRAF as therapeutic target in melanoma. Biochem Pharmacol. 2010;80:561–567. doi: 10.1016/j.bcp.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaherty K, Puzanov I, Sosman J, et al. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009; 27(suppl):461s. abstr 9000. [Google Scholar]
- 15.Schwartz G, Robertson S, Shen A, et al. A phase I study of XL281, a selective oral RAF kinase inhibitor, in patients (Pts) with advanced solid tumors. J Clin Oncol. 2009; 27(suppl):149s. abstr 3513. [Google Scholar]
- 16.Patel S, Lazar A, Mahoney S, et al. Clinical responses to AZD6244 (ARRY-142886)-based combination therapy stratified by gene mutations in patients with metastatic melanoma. J Clin Oncol. 2010;28(suppl):611s. doi: 10.1002/cncr.27790. abstr 8501. [DOI] [PubMed] [Google Scholar]
- 17.Kefford R, Arkenau H, Brown M, et al. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol. 2010; 28(suppl):611s. abstr 8503. [Google Scholar]
- 18.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kris M, Lau C, Ang D, et al. Initial results of LC-MAP: An institutional program to routinely profile tumor specimens for the presence of mutations in targetable pathways in all patients with lung adenocarcinoma. J Clin Oncol. 2010; 28(suppl):516s. abstr 7009. [Google Scholar]
- 20.Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry. Mol Biotechnol. 2004;26:147–164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 21.Arcila M, Lau C, Nafa K, et al. Detection of KRAS and BRAF mutations in colorectal carcinoma: Roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham D, Kris MG, Riely GJ, et al. Use of cigarette-smoking history to estimate the likelihood of mutations in epidermal growth factor receptor gene exons 19 and 21 in lung adenocarcinomas. J Clin Oncol. 2006;24:1700–1704. doi: 10.1200/JCO.2005.04.3224. [DOI] [PubMed] [Google Scholar]
- 25.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS Mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 26.Sasaki H, Kawano O, Endo K, et al. Uncommon V599E BRAF mutations in Japanese patients with lung cancer. J Surg Res. 2006;133:203–206. doi: 10.1016/j.jss.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Flaherty KT, McArthur G. BRAF, a target in melanoma: Implications for solid tumor drug development. Cancer. 2010;116:4902–4913. doi: 10.1002/cncr.25261. [DOI] [PubMed] [Google Scholar]
- 28.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): A resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, Higgins B, Kolinsky K, et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010;70:5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- 30.Eisen T, Ahmad T, Flaherty KT, et al. Sorafenib in advanced melanoma: A phase II randomised discontinuation trial analysis. Br J Cancer. 2006;95:581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalley KSM, Xiao M, Villanueva J, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28:85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 33.Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]