Abstract
Williams syndrome (WS) is a complex developmental disorder resulting from the deletion of a large (∼1.5–2 Mb) segment of human chromosome 7q11.23. Physical mapping studies have revealed that this deleted region, which contains a number of known genes, is flanked by several large, nearly identical blocks of DNA. The presence of such highly related DNA segments in close physical proximity to one another has hampered efforts to elucidate the precise long-range organization of this segment of chromosome 7. To gain insight about the structure and evolutionary origins of this important and complex genomic region, we have constructed a fully contiguous bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) contig map encompassing the corresponding region on mouse chromosome 5. In contrast to the difficulties encountered in constructing a clone-based physical map of the human WS region, the BAC/PAC-based map of the mouse WS region was straightforward to construct, with no evidence of large duplicated segments, such as those encountered in the human WS region. To confirm this difference, representative human and mouse BACs were used as probes for performing fluorescence in situ hybridization (FISH) to metaphase and interphase chromosomes. Human BACs derived from the nonunique portion of the WS region hybridized to multiple, closely spaced regions on human chromosome 7q11.23. In contrast, corresponding mouse BACs hybridized to a single site on mouse chromosome 5. Furthermore, FISH analysis revealed the presence of duplicated segments within the WS region of various nonhuman primates (chimpanzee, gorilla, orangutan, and gibbon). Hybridization was also noted at the genomic locations corresponding to human chromosome 7p22 and 7q22 in human, chimpanzee, and gorilla, but not in the other animal species examined. Together, these results indicate that the WS region is associated with large, duplicated blocks of DNA on human chromosome 7q11.23 as well as the corresponding genomic regions of other nonhuman primates. However, such duplications are not present in the mouse.
Williams syndrome (WS; also known as Williams-Beuran syndrome) is a well-studied disorder with features involving multiple physiological systems [Online Mendelian Inheritance in Man (OMIM) 194050; http://www.ncbi.nlm.nih.gov/Omim]. The syndrome is characterized by congenital heart and vascular disease, dysmorphic facial features, infantile hypercalcemia, and a unique cognitive and personality profile (Burn 1986; Morris et al. 1988; Bellugi et al. 1990; Keating 1997). Major strides have been made in recent years to elucidate the genetic basis of WS. Following the landmark finding that WS is caused by hemizygous microdeletions within human chromosome 7q11.23 that include the elastin gene (ELN) (Ewart et al. 1993), numerous other genes have been found to reside within the commonly deleted interval. These include LIMK1 (encoding LIM kinase-1) (Frangiskakis et al. 1996; Tassabehji et al. 1996), RFC2 (encoding replication factor C subunit 2) (Peoples et al. 1996), FZD3 (encoding a homolog of the Drosophila frizzled wnt receptor) (Wang et al. 1997), CYLN2 (encoding cytoplasmic linker-2) (Hoogenraad et al. 1998), STX1A (encoding syntaxin 1A) (Osborne et al. 1997b; Nakayama et al. 1998), FKBP6 (Meng et al. 1998b), WSTF (Lu et al. 1998), GTF2I (encoding general transcription factor II-1) (Perez Jurado et al. 1998), BCL7B (Meng et al. 1998a; Jadayel et al. 1998), WS-βTRP (Meng et al. 1998a), WS-bHLH (Meng et al. 1998a), CPETR1 (Paperna et al. 1998), CPETR2 (Paperna et al. 1998), and several others (Osborne et al. 1996). The wide spectrum of phenotypic features observed in WS is likely a consequence of haploinsufficiency of some of the above genes and/or yet-to-be-identified genes in the deleted region.
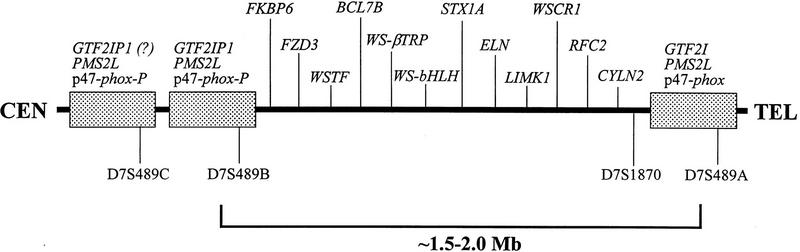
Various studies suggest that the microdeletions associated with WS span ∼1.5–2 Mb of DNA within 7q11.23 (Osborne et al. 1996; Perez Jurado et al. 1996; Robinson et al. 1996; Urban et al. 1996; Wang et al. 1997; Meng et al. 1998a; Wu et al. 1998). Although difficult to isolate in yeast artificial chromosome (YAC) clones, most of the deleted region (e.g., the segment immediately flanking ELN) has been relatively straightforward to map using bacterial-based clones [bacterial artificial chromosomes (BACs) (Shizuya et al. 1992) and P1-derived artificial chromosomes (PACs) (Ioannou et al. 1994)]. However, detailed physical mapping efforts of the broader genomic region encompassing the commonly deleted segment (i.e., the WS region) have been hindered greatly by the presence of large, duplicated segments of DNA. Our laboratory (Gorlach et al. 1997; E.D. Green and B.J. Trask, unpubl.) and others (Osborne et al. 1997a) have generated data suggesting the likely presence of three such duplicated blocks of DNA that reside close together in 7q11.23. These duplicated segments contain gene and pseudogene sequences of the p47–phox (NCF1) (Francke et al. 1990; Gorlach et al. 1997) and GTF2I (Perez Jurado et al. 1998) genes as well as members of the PMS2 mismatch repair gene family (Osborne et al. 1997a). Establishing the precise long-range organization of these duplicated regions relative to the WS-associated deletions has proven particularly difficult, although evidence emerging from our (E.D. Green and B.J. Trask, unpubl.) and several other (Osborne et al. 1996, 1997a,b; Perez Jurado et al. 1996, 1998; Robinson et al. 1996; Wang et al. 1997; Lu et al. 1998; Meng et al. 1998a) laboratories is converging towards a reasonably consistent model for the physical map of this region (see Fig. 1).
Figure 1.
Schematic representation of the human WS region. A working model of the long-range physical organization of the human WS region is depicted based on data generated in numerous studies (Osborne et al. 1996, 1997a,b; Perez Jurado et al. 1996, 1998; Robinson et al. 1996; Wang et al. 1997; Lu et al. 1998; Meng et al. 1998a; E.D. Green and B.J. Trask, unpubl.). The relative positions (not to scale) of key gene/pseudogene sequences and genetic markers are indicated. A middle, single-copy region contains numerous known genes [FKBP6 (Meng et al. 1998b), FZD3 (Wang et al. 1997), WSTF (Lu et al. 1998), BCL7B (Jadayel et al. 1998; Meng et al. 1998a), WS–βTRP (Meng et al. 1998a), WS-bHLH (Meng et al. 1998a), STX1A (Osborne et al. 1997b; Nakayama et al. 1998), ELN (Ewart et al. 1993), LIMK1 (Frangiskakis et al. 1996; Tassabehji et al. 1996), WSCR1 (Osborne et al. 1996), RFC2 (Peoples et al. 1996), CYLN2 (Hoogenraad et al. 1998)]. This region is flanked by several large (estimated at ∼200–300 kb) genomic segments of nearly identical composition (represented by stippled boxes), each of which contains the indicated gene/pseudogene sequences [GTF2I/GTF2IP1 (Perez Jurado et al. 1998), PMS2L (Osborne et al. 1997a), p47–phox/p47–phox-P (Gorlach et al. 1997)]. Note that the relative order of the latter within the duplicated segments has not been established nor has the presence of GTF2IP1 in all the duplicated segments (reflected by the ? after GTF2IP1 in the far left duplicated segment). The genomic segment commonly deleted in WS that spans ∼1.5–2.0 Mb is indicated along the bottom (Osborne et al. 1996; Perez Jurado et al . 1996; Robinson et al. 1996; Urban et al. 1996; Wang et al. 1997; Meng et al. 1998a; Wu et al. 1998).
Human chromosome 7q11.23 has conserved synteny to a segment on distal mouse chromosome 5 (DeBry and Seldin 1996). Several mouse homologs of the above-mentioned genes, including Eln (Wydner et al. 1994), Limk1 (Mao et al. 1996), Cyln2 (Hoogenraad et al. 1998), Cpetr1 (Paperna et al. 1998), Cpetr2 (Paperna et al. 1998), Gtf2i (Wang et al. 1998), and p47–phox (Jackson et al. 1994) (also see http://www.informatics.jax.org and http://www.ncbi.nlm.nih.gov/Homology), have been genetically or cytogenetically mapped to this region of the mouse genome. In light of the complex organization of the human WS region, we sought to gain insight into the structure of the WS region in a distantly related mammal. Here we report the results of comparative physical mapping of the human and mouse WS regions as well as fluorescence in situ hybridization (FISH) studies that demonstrate evolutionary differences in the presence of the duplicated segments residing within this complex genomic region.
RESULTS
Construction of a BAC/PAC Contig Map of the Mouse WS Region
The collective data generated by studies from several groups (Osborne et al. 1996, 1997a,b; Perez Jurado et al. 1996, 1998; Robinson et al. 1996; Wang et al. 1997; Lu et al. 1998; Meng et al. 1998a; E.D. Green and B.J. Trask, unpubl.) indicate that the WS region of human chromosome 7q11.23 consists of a unique segment harboring a number of known single-copy genes flanked by several large, closely related genomic segments (Fig. 1; see Introduction for specific references). The latter contain a number of known genes or related pseudogene sequences [e.g., p47–phox (Gorlach et al. 1997), GTF2I (Perez Jurado et al. 1998), PMS2 family members (Osborne et al. 1997a)]. Our interest in human–mouse comparative genome analysis prompted us to construct a BAC/PAC-based physical map of the region of mouse chromosome 5 corresponding to the human WS region.
Little pre-existing physical mapping information or DNA sequence was available for this region of the mouse genome. Using partial sequence data for the mouse Limk1 (GenBank accession no. X86569) and p47–phox (S.J. Chanock, pers. comm.) genes and information about an intronic polymorphism in the mouse Eln gene (Wydner et al. 1994; also see http://www.informatics.jax.org), sequence-tagged site (STS)-specific PCR assays were developed and used to screen mouse BAC and PAC libraries. Sequences were then derived from the insert ends of positive clones and used to develop new STS-specific PCR assays, which in turn were used to isolate additional clones. This process was repeated for several rounds, which in some cases also included hybridization-based screening of BAC/PAC libraries and the use of STSs derived from other sources. Eventually, this process and the resulting STS-content data allowed assembly of the BAC/PAC contig depicted in Figure 2.
Figure 2.
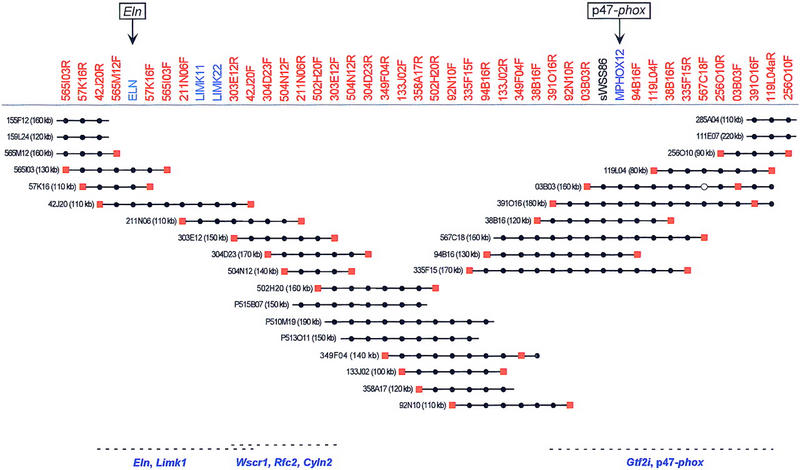
BAC/PAC-based STS-content map of the mouse WS region (oriented with centromere leftward and telomere rightward). The deduced positions of 44 STSs are depicted along the top, with the indicated clones shown as horizontal lines below. Relevant information about the STSs is available in GenBank (http://www.ncbi.nlm.nih.gov). The STSs were developed from clone insert ends (red), known genes (blue), or a conserved human DNA sequence (black). (●,█) The STS is confirmed to be present in that clone by PCR testing. When an STS corresponds to a clone insert end, a red square is present at the end of the clone from which it was derived. PAC clones are indicated with a P at the beginning of their names. Four of the BAC clones were isolated from the Genome Systems C57BL/6 mouse library (111E07, 285A04, 155F12, and 159L24), with the rest of the BACs coming from the Research Genetics CITB–CJ7-B mouse library. The size of each BAC/PAC, as assessed by pulsed-field gel analysis, is also provided. The indicated BAC/PAC overlaps were confirmed by restriction enzyme digest-based fingerprint analysis (Marra et al. 1997; data not shown). The depicted clones together span ∼680 kb of DNA from mouse chromosome 5. Preliminary genomic sequence analysis has revealed the presence of Eln, Limk1, and part of Wscr1 in BAC 42J20, Wscr1, Rfc2, and Cyln2 in BAC 303E12, and Gtf2i and p47–phox in BAC 391O16, as indicated along the bottom (U. DeSilva and E.D. Green, unpubl.).
The assembled contig of the mouse WS region contains 28 clones and provides ordering information for 44 STSs. Of the 44 STSs, 39 correspond to BAC insert ends, 4 to known genes, and 1 to a conserved human sequence. The average number of clones identified per STS is 4.9, whereas the average number of STSs within each clone is 7.7. Importantly, the clone overlaps depicted in the contig shown in Figure 2 were confirmed by restriction enzyme digest-based fingerprint analysis (Marra et al. 1997; data not shown). The latter analysis also revealed that the total length of the contig is ∼680 kb, as assessed by summing the size of all unique restriction fragments, and extends >100–200 kb on either side of the interval defined by Eln and p47–phox. Thus, with the mapping of 44 STSs across ∼680 kb, the average STS spacing within the contig map is ∼15 kb. Typical variability in the depth of clone coverage (ranging from two to seven positive clones per STS) was encountered across the mouse WS region, with no evidence of a particular region(s) with strikingly higher representation. This pattern is in sharp contrast to the mapping of the human WS region, where the ratio of p47–phox-containing to ELN-containing clones (e.g., YAC, BAC, P1) is typically ∼3:1 (Gorlach et al. 1997; E.D. Green and B.J. Trask, unpubl.) Preliminary sequence analysis of several mouse BACs has revealed the expected presence of known genes residing between Eln and p47–phox (e.g., Limk1, Wscr1, Rfc2, Cyln2, Gtf2i; compare the bottoms of Figs. 2 with 1), providing additional support for the colinearity of these regions in human and mouse. To date, we have not detected Pms2-related sequences within the mouse WS region.
FISH-Based Comparative Analysis of the WS Region
To gain additional insight into the structure of the human and mouse WS regions, comparative FISH analysis was performed using representative BAC clones. Markedly different results were obtained in each case. Hybridization of human metaphase and interphase chromosomes with a BAC containing ELN (RG030E19) yields results routinely encountered with single-copy genomic regions (Fig. 3). In contrast, FISH analysis with a BAC containing p47–phox (RG350L10) reveals the presence of multiple, closely spaced hybridizing segments in human 7q11.23 as well as additional weaker hybridizing segments in 7p22 and 7q22 (Fig. 3). The number of signals observed in interphase nuclei is consistent with the presence of three to four copies of the genomic region containing p47–phox sequences within 7q11.23 and confirms the single-copy nature of the ELN-containing region (Fig. 3; Table 1). In contrast, FISH analyses of mouse chromosomes using BACs containing Eln (42J20) or p47–phox (391O16) yield virtually indistinguishable results (Fig. 3). No evidence is seen with mouse metaphase or interphase chromosomes for the presence of duplicated p47–phox-containing segments (Fig. 3; Table 1).
Figure 3.
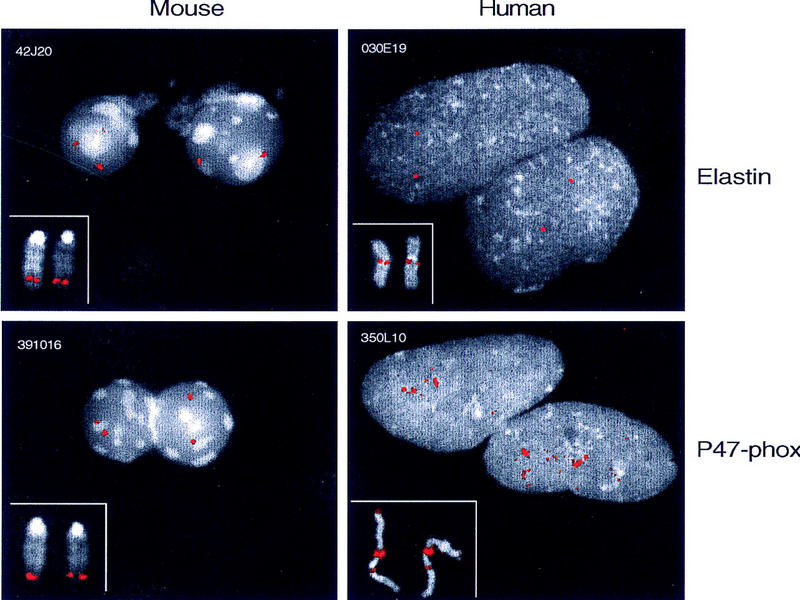
Human–mouse comparative FISH analysis of the WS region. Representative BAC clones from the ELN/Eln- and p47–phox-containing regions were selected from the physical maps of the human and mouse WS region. FISH analyses were performed on both metaphase chromosomes (insets) and interphase nuclei (main panels). Clones containing ELN/Eln (RG030E19 and 42J20) show a hybridization pattern on human chromosome 7q11.23 and mouse chromosome 5G2 typical of a single-copy segment. In mouse, the p47–phox-containing BAC (391O16) also produces a single-copy signal on metaphase and interphase chromosomes. In contrast, the human p47–phox-containing BAC (RG350L10) produces a very large and intense signal on metaphase chromosomes at 7q11.23 and multiple fluorescent dots on interphase chromosomes (also see Table 1). Sequences that cross-hybridize to the p47–phox BAC are encountered in 7p22 and 7q22 in human, in addition to the cluster in 7q11.23. We note that there is overlap between these locations and the sites detected with probes for PMS2-related sequences (Nicolaides et al. 1995).
Table 1.
Interphase FISH Analysis of BAC/PAC Clones from the Mouse and Human WS Regions
| Clone | Size (kb) | Gene within clone | No. of FISH “dots” per chromosome | |
|---|---|---|---|---|
| Average | s.d. | |||
| Human | ||||
| RG030E19 | 155 | ELN | 1.21 | 0.43 |
| RG350L10 | 180 | p47–phox | 4.85 | 1.23 |
| Mouse | ||||
| 42J20 | 107 | Eln | 1.48 | 0.69 |
| 303E12 | 163 | 1.44 | 0.55 | |
| P510M19 | 188 | 1.49 | 0.57 | |
| 92N10 | 111 | 1.37 | 0.56 | |
| 391O16 | 180 | p47–phox | 1.61 | 0.60 |
BAC/PAC DNA was labeled and hybridized to human G0/G1 interphase fibroblast cells or to mouse interphase cells prepared from spleen. The number of distinct fluorescent dots (i.e., a multilobed signal was scored as one dot) per cluster was scored in 100 interphase chromosomes for each probe. Note that an average of 1 is expected for a small, unreplicated single-copy sequence. Large BACs representing known single-copy loci often produce signals with some substructure, which produces a dot count of >1. The presence of some S and G2 cells in the preparations can also inflate the average dot-count above 1. These factors explain the observation that the average dot-count is 1.2–1.6 for the BACs that we conclude contain single-copy sequences. Of the clones listed, only the human p47–phox-containing BAC (RG350L10) contains sequences that are present in multiple copies. Note that the two human BACs were isolated from the Research Genetics human BAC library (E.D. Green; unpubl.). See Fig. 2 for additional information about the mouse clones.
Sequences separated by >100 kb can be resolved in interphase chromatin (Trask et al. 1989; van den Engh et al. 1992). Note that the similarity of FISH patterns obtained with the Eln- and p47–phox-containing BACs in interphase nuclei, both with respect to the signal intensity (Fig. 3) and number of resolvable dots (Table 1), argue against the possibility of even closely spaced duplicated segments in mouse. Three other BACs, which together with the Eln- and p47–phox-containing clones provide a tiling path across the ∼680-kb mouse WS region, gave similar results, consistent with the single-copy nature of the WS region in mouse (Table 1; data not shown).
To establish if duplications of the WS region are present in other primates, the chromosomes of chimpanzee, gorilla, orangutan, and gibbon were analyzed by FISH using human ELN- and p47–phox-containing BACs. The FISH patterns obtained with the ELN BAC are consistent with the presence of a single-copy segment in all these animal species (data not shown). The ELN sequence maps to the expected genomic locations corresponding to human chromosome 7q11.23, based on comparison of the banding patterns of chromosome-7 orthologs (Yunis and Prakash 1982; Jauch et al. 1992) (Fig. 4a,b). These chromosomes are referred to as PTR6, GGO6, PPY6, and HLA1, respectively (note that the locations are denoted henceforth using nomenclature for the human karyotype). The gorilla and orangutan chromosomes differ from the human and chimpanzee chromosomes by the presence of one and two inversions, respectively (Yunis and Prakash 1982). One breakpoint of the paracentric inversion that differentiates the gorilla and human chromosomes lies in or near 7q11.23. However, the position of the ELN-associated signal is unaffected, and thus it lies proximal to this evolutionary breakpoint. Relative to the human and chimpanzee 7, orangutan shows the same paracentric inversion seen in gorilla, plus a large pericentric inversion. The ELN-associated signal resides within the latter inverted unit, resulting in its location on the small short arm of the orangutan chromosome.
Figure 4.
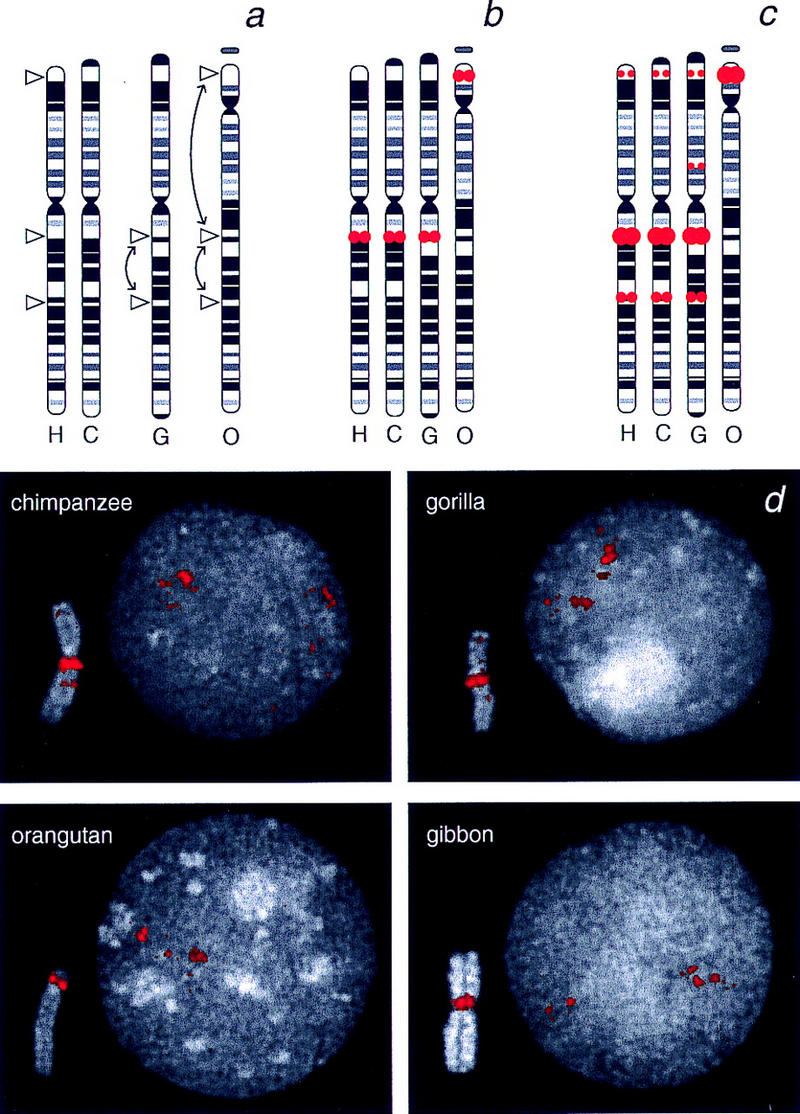
Comparative FISH analyses of WS-region segments on nonhuman primate chromosomes. (a) Idiograms of chromosome-7 orthologs in human (H), chimpanzee (C), gorilla (G), and orangutan (O) are taken from Yunis and Prakesh (1982) and used to indicate the positions of the breakpoints of inversions that have occurred during primate evolution. (b) Summary of the locations of FISH signals produced by a human ELN-containing BAC (RG030E19) on human, chimpanzee, gorilla, and orangutan chromosomes. Sequences in this ELN-containing segment appear single copy in all species and map to the expected locations, based on the known inversions. The ELN segment maps in gibbon near the centromere of a chromosome that has been shown by chromosome painting to contain sequences orthologous to human chromosome 7 (not shown; Jauch et al. 1992). (c) Summary of the locations of FISH signals produced by a human p47–phox-containing BAC on hominoid chromosomes (see Fig. 3 and d for corresponding FISH images). (d) Representative images of metaphase chromosomes (insets) and interphase nuclei (main panels) from chimpanzee, gorilla, orangutan, and gibbon after FISH with a human p47–phox-containing BAC (RG350L10). Local duplication of p47–phox sequences within the region corresponding to human chromosome 7q11.23 is evident in all species, both from the size of the signal in metaphase and the cluster of multiple dots in interphase. Additional cross-hybridizing sequences are detected in 7p22 and 7q22 in chimpanzee and in 7p22, 7p13, and 7q22 in gorilla. Bands are indicated using nomenclature from the human karyotype.
The FISH pattern obtained with the p47–phox-containing BAC is strikingly different than that with the ELN-containing BAC in the nonhuman primates. As in human, a very large FISH signal is seen in the region corresponding to 7q11.23 in metaphase chromosomes from the four nonhuman primates tested (Fig. 4). Multiple duplicated segments are also evident in interphase nuclei. In addition, signals are observed on chimpanzee and gorilla chromosomes at locations corresponding to 7p22 and 7q22. These additional copies of p47–phox-related sequence are not observed in either orangutan or gibbon (Fig. 4).
DISCUSSION
Elucidating the genetic bases of WS represents an important area of ongoing investigation. The general region of human chromosome 7q11.23 deleted in WS is complex in structure, which may in part account for its propensity to undergo interstitial deletions. As a complement to the intensive studies of the human WS region (Osborne et al. 1996, 1997a,b; Perez Jurado et al. 1996, 1989; Robinson et al. 1996; Wang et al. 1997; Lu et al. 1998; Meng et al. 1998a), we are pursuing the detailed characterization of the mouse WS region for several reasons. First, the resulting information should facilitate the identification of genes deleted in WS, thereby enhancing studies aiming to define the basis for each of the phenotypic features of this contiguous gene deletion syndrome. Whereas a number of deleted genes have been identified already [e.g., ELN (Ewart et al. 1993), LIMK1 (Frangiskakis et al. 1996; Tassabehji et al. 1996), RFC2 (Peoples et al. 1996), FZD3 (Wang et al. 1997), STX1A (Osborne et al. 1997b; Nakayama et al. 1998b), CYLN2 (Hoogenraad et al. 1998), FKBP6 (Meng et al. 1998a), WSTF (Lu et al. 1998), WSCR1 (Osborne et al. 1996), GTF2I (Perez Jurado et al. 1998), BCL7B (Meng et al. 1998a; Jadayel et al. 1998), WS-βTRP (Meng et al. 1998), WS-bHLH (Meng et al. 1998a), CPETR1 (Paperna et al. 1998a), CPETR2 (Paperna et al. 1998)], comparative mouse–human sequence analysis should provide a more robust approach for identifying all the encoded genes and their regulatory elements. Systematic sequencing of both the human and mouse WS regions is underway (U. DeSilva and E.D. Green unpubl.; see bottom of Fig. 2). Second, the resulting mapping and sequencing data generated from the mouse WS region should aid the generation of mouse models of WS, for example, the creation of mice lacking one or more of the genes located within the commonly deleted interval. Finally, comparative studies of the human and mouse WS regions should provide insight into the evolutionary origins of this structurally complex genomic region.
The findings reported here reveal that the human and mouse WS regions are strikingly different. Numerous genetic (Dutly and Schinzel 1996; Perez Jurado et al. 1996; Robinson et al. 1996; Urban et al. 1996; Baumer et al. 1998) and physical (Osborne et al. 1997a; Perez Jurado et al. 1998; E.D. Green and B.J. Trask, unpubl.) mapping studies have revealed the presence of large, duplicated blocks of DNA that flank the region deleted commonly in WS. Some studies also indicate that both inter- and intrachromosomal rearrangements are responsible for WS deletions (Dutly and Schinzel 1996; Perez Jurado et al. 1996, 1998; Robinson et al. 1996; Urban et al. 1996; Baumer et al. 1998). Presumably, the duplicated segments on chromosome 7 homologs or chromatids misalign and mediate meiotic or mitotic rearrangements. In contrast, our physical (Fig. 2) and cytogenetic (Fig. 3) mapping studies indicate the absence of such duplicated segments in the mouse WS region. These results are consistent with studies showing that Gtf2I (Wang et al. 1998) and p47–phox (S.J. Chanock, pers. comm.) sequences are present in a single-copy fashion in the mouse genome [whereas these gene sequences reside within the duplicated segments in the human WS region (see Fig. 1)]. The absence of such duplicated segments in the mouse genome indicates that the duplication event(s) that created the complex structure of the human WS region must be of recent evolutionary origin.
Our comparative FISH analyses of the WS region suggest that the duplication of the p47–phox sequences occurred sometime after the split of the ancestors of rodents and hominoids [∼65 million years ago (Mya)] and before the diversification of the hominoids (∼15–20 Mya) (see Kumar and Hedges 1998). Local duplication of this segment within the WS region on 7q11.23 (or its equivalent position) was observed in all animal species tested, except the mouse. Additional copies of p47–phox sequences appear to have spread to two locations elsewhere on chromosome 7 (7p22 and 7q22) in the ancestor common to human, chimpanzee, and gorilla.
The association of evolutionary breakpoints in the orthologs of human chromosome 7 and the locations of genomic segments containing p47–phox sequences is striking. The gorilla chromosome differs from the human and chimpanzee chromosomes by a paracentric inversion with breakpoints at 7q11.23-q21 and 7q22, and p47–phox sequences are present in both regions. The organization of chromosome 7 material in orangutan differs from that in the other hominoids by a pericentric inversion, with breakpoints in 7p22 and 7q11.23-q21. Again, p47–phox sequences are found in the vicinity of both breakpoints. This inversion event may explain the single cluster of p47–phox sequences on the short arm of the orangutan chromosome. It is, however, surprising that no p47–phox sequences are detected at the 7q22 site in orangutan as they are in gorilla, given that the banding pattern of the orangutan chromosome (Yunis and Prakash 1982) reveals the same paracentric inversion involving 7q11.23–7q22 as that seen in gorilla.
Our comparative analyses could be explained by a scenario in which a p47–phox-containing segment first duplicated locally and then some copies were distributed to three locations on chromosome 7 by inversion events. The presence of similar sequences at these various locations may lead to homologous recombination and subsequent intrachromosomal rearrangements. It is therefore possible that the inversions that occurred during primate evolution and the relatively frequent deletions that lead to WS in humans are mediated by the same duplicated sequences.
METHODS
Isolation of BAC/PAC Clones
BAC (Shizuya et al. 1992) and PAC (Ioannou et al. 1994) clones were isolated by PCR- and/or hybridization-based screening of the following libraries: (1) Research Genetics CITB-CJ7-B mouse BAC library (http://www.resgen.com); (2) Genome Systems C57BL/6 mouse BAC library (http://www.genomesystems.com); and (3) Roswell Park Cancer Institute RPCI-21 129SV mouse PAC library (http://bacpac.med.buffalo.edu). PCR screening was performed as recommended by the library supplier. For hybridization-based screening, the STS-specific PCR assay was used to generate an appropriate product, which was then isolated by gel electrophoresis, purified using a Geneclean II kit (BIO 101), and radiolabeled with [α-32P]dCTP using Ready-To-Go DNA labeling beads (Pharmacia). The resulting radiolabeled probe was then separated from unincorporated nucleotides by passage over a Sephadex-G50 column (Pharmacia). The purified probe was then hybridized with membrane filters containing the arrayed clones in 50% formamide buffer, essentially as described (Sambrook et al. 1989). Filters were then washed two times in 2× SSC at room temperature and two times in 0.1× SSC/0.2% SDS at 65°C prior to overnight exposure to Kodak X-Omat film. Candidate positive clones were then evaluated by additional hybridization and/or PCR analyses.
Sequencing of BAC Insert Ends
A freshly grown BAC colony was inoculated into 2 ml of 2× TY broth (Sambrook et al. 1989) containing 12.5 μg/ml of chloramphenicol and incubated shaking at 37°C for 4–5 hr. The culture was then added to 30 ml of 2× TY broth containing 12.5 μg/ml of chloramphenicol in a 50-ml tube and incubated shaking at 37°C for 16–22 hr. A total of 20 ml of the final culture was used to isolate BAC DNA with an Autogen 740 Plasmid Isolation System using the manufacturer’s protocol (v. 1.3). The resulting DNA from the 20 ml of culture was resuspended in 400 μl of water, and 5 μl of RNase cocktail (500 U/ml of RNase A and 20,000 U/ml of RNase T1; Ambion) was added. Following incubation at 37°C for 30 min, samples were transferred onto a Microcon-100 column (Amicon) and centrifuged at 800g for 15 min. The columns were then inverted into the collection tubes and centrifuged at top speed in a microcentrifuge for 5–10 sec. The resulting DNA was adjusted to a final concentration of ∼200–250 ng/μl.
DNA sequencing was performed using DYEnamic Energy Transfer Primers (Amersham) and an adaptation of the described protocol (Marra et al. 1996). Briefly, each sequencing reaction contained 2 μl of ThermoSequenase reagent mix (Amersham), 1 μl of DYEnamic Energy Transfer Primer (−40 universal or −28 reverse), and 5 μl of the above-purified BAC DNA. Thermal cycling was performed in either a Catalyst Molecular Biology Workstation (Perkin-Elmer) or an MJ Research PTC-200 DNA Engine using the following conditions: 95°C for 2 min followed by 20 cycles of 95°C for 10 sec, 50°C (−28 reverse primer) or 55°C (−40 universal primer) for 20 sec, and 72°C for 60 sec. Reactions corresponding to the four dideoxynucleotides were pooled, and the DNA precipitated by addition of 5 μl of glycogen and 132 μl of 100% ethanol. Samples were centrifuged at 3200 rpm for 30 min in a Jouan GR422 centrifuge. Following removal of the supernatant, the DNA pellets were dried and then resuspended in 5 μl (for analysis on an Applied Biosystems 373 sequencer) or 3 μl (for analysis on an Applied Biosystems 377 sequencer) of the supplied loading buffer (Amersham), heated to 60°C for 10 min, and analyzed on either an Applied Biosystems 373 or 377 sequencer.
Development of BAC Insert End-Specific STSs
Sequences derived from BAC insert ends were imported into MacVector DNA analysis program (Kodak), and BAC vector sequences were removed manually. The resulting sequence was analyzed for murine repeats using the Repeat Masker 2 Web server (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker), and PCR primers were designed from nonrepetitive sequences using the PCR primer design module of MacVector. Oligonucleotide primers were then synthesized commercially (Life Technologies). Information about the individual STS-specific PCR assays is available in GenBank (http://www.ncbi.nlm.nih.gov).
Determination of BAC/PAC Insert Sizes
BAC and PAC DNA was purified using the Autogen 740 Plasmid Isolation System as described above and digested to completion with NotI (New England Biolabs). The digested sample was then loaded onto a 1% SeaPlaque (FMC Bioproducts) agarose gel in 0.5× TBE and electrophoresed for 14 hr in a CHEF DRII pulsed-field gel apparatus (Bio-Rad) (5–12 sec ramped switching, 120 V, 12°C). Concatemerized λ DNA samples (New England Biolabs) were included on all gels to facilitate establishing the size(s) of the resulting BAC-derived band(s) following ethidium bromide staining (Riethman et al. 1997).
BAC/PAC Fingerprint Analysis
BACs and PACs were subjected to restriction enzyme digest-based fingerprint analysis, essentially as described (Marra et al. 1997). Specifically, purified BAC/PAC DNA was digested with HindIII, electrophoresed in a 1% SeaKem LE agarose (FMC Bioproducts) gel at 45 V for 16 hr, and stained with SYBR green (FMC Bioproducts). The gel was imaged using a Molecular Dynamics FluorImager. The resulting image was then transferred to a UNIX workstation for band calling and contig assembly. Restriction fragment bands were identified using the program Image 3.3, and contigs were constructed using FPC.
FISH Analysis
Purified BAC DNA was biotinylated by nick translation and hybridized (20 ng/slide in a 10-μl volume of hybridization solution) to immobilized mouse and human interphase and metaphase chromosomes. Mouse interphase cells were prepared from uncultured mouse splenocytes. Mouse metaphase spreads were prepared from mouse splenocytes after culture in lipopolysaccharide. Human G0/G1 interphase cells were prepared from growth-inhibited fibroblast cultures. Human metaphase spreads were prepared from PHA-stimulated peripheral blood lymphocyte cultures. Metaphase and interphase cells were harvested from actively growing cultures of chimpanzee (Pan troglodytes, CRL-1847), gorilla (Gorilla gorilla, CRL-1854), orangutan (Pongo pygmaeus, CRL-1850), and gibbon (Hylobates lar, TIB-201) cell lines obtained from the American Type Culture Collection (ATCC) (http://www.atcc.org). The culture and preparation of cells is described elsewhere (Trask 1999). All cells were incubated in hypotonic 75 mm KCl, fixed several times in 3:1 methanol:acetic acid, and dropped on clean slides. The same hybridization and wash stringencies were used in all experiments (hybridization solution: 50% formamide, 2× SSC, 10% dextran sulfate at 37°C, with washes in 50% formamide, 2× SSC at 42°C). Mouse or human Cot1 DNA was included in the hybridization solution to suppress labeling of highly repetitive sequences. Hybridization was detected by incubation of the slides in avidin-FITC, biotinylated goat anti-avidin, and avidin-FITC, with wash steps between each labeling step. In some experiments, two probes, one labeled with digoxigenin and one with biotin, were hybridized simultaneously and detected with different fluorochromes (Trask 1999). The nuclei were counterstained with DAPI. Images of the FITC and DAPI fluorescence were collected separately using Signal Analytics IPLab Spectrum software, a Princeton CCD camera, and a Zeiss Axiophot microscope equipped with separate excitation filters in a Ludl filter wheel mounted between the 100W lamp and the sample and a triple-band multichroic mirror and emission filter mounted between the sample and the detector (known as the Pinkel arrangement). The FITC image was pseudocolored in red and then overlaid on a gray-scale DAPI image. In some experiments, the number of hybridization dots was counted within the clusters of FISH signals observed in 100 interphase chromosomes.
Acknowledgments
We thank Johannah Doyle, Curt Jamison, Jackie Idol, Danny Wangsa, and Thomas Ried for various contributions to these studies. This work was supported in part by grants from the Department of Energy (DE-FG03-96ER62173) and the National Institutes of Health (GM57070).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL btrask@u.washington.edu; FAX (206) 685-7354.
E-MAIL egreen@nhgri.nih.gov; FAX (301) 402-4735.
REFERENCES
- Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA. High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum Mol Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S. Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome. Am J Med Genet (Suppl.) 1990;6:115–125. doi: 10.1002/ajmg.1320370621. [DOI] [PubMed] [Google Scholar]
- Burn J. Williams syndrome. J Med Genet. 1986;23:389–395. doi: 10.1136/jmg.23.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBry RW, Seldin MF. Human/mouse homology relationships. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- Dutly F, Schinzel A. Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams-Beuren syndrome. Hum Mol Genet. 1996;5:1893–1898. doi: 10.1093/hmg/5.12.1893. [DOI] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- Francke U, Hsieh C-L, Foellmer BE, Lomax KJ, Malech HL, Leto TL. Genes for two autosomal recessive forms of chronic granulomatous disease assigned to 1q25 (NCF2) and 7q11.23 (NCF1) Am J Hum Genet. 1990;47:483–492. [PMC free article] [PubMed] [Google Scholar]
- Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, et al. LIM–kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- Gorlach A, Lee PL, Roesler J, Hopkins PJ, Christensen B, Green ED, Chanock SJ, Curnutte JT. A p47–phox pseudogene carries the most common mutation causing p47–phox-deficient chronic granulomatous disease. J Clin Invest. 1997;100:1907–1918. doi: 10.1172/JCI119721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Eussen BHJ, Langeveld A, van Haperen R, Winterberg S, Wouters CH, Grosveld F, De Zeeuw CI, Galjart N. The murine CYLN2 gene: Genomic organization, chromosome localization, and comparison to the human gene that is located within the 7q11.23 Williams syndrome critical region. Genomics. 1998;53:348–358. doi: 10.1006/geno.1998.5529. [DOI] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- Jackson SH, Malech HL, Kozak CA, Lomax KJ, Gallin JI, Holland SM. Cloning and functional expression of the mouse homologue of p47phox. Immunogenetics. 1994;39:272–275. doi: 10.1007/BF00188790. [DOI] [PubMed] [Google Scholar]
- Jadayel DM, Osborne LR, Coignet LJA, Zani VJ, Tsui L-C, Scherer SW, Dyer MJS. The BCL7 gene family: Deletion of BCL7B in Williams syndrome. Gene. 1998;224:35–44. doi: 10.1016/s0378-1119(98)00514-9. [DOI] [PubMed] [Google Scholar]
- Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating MT. On the trail of genetic culprits in Williams syndrome. Cardiovasc Res. 1997;36:134–137. doi: 10.1016/s0008-6363(97)00027-8. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- Mao X, Jones TA, Williamson J, Gutowski NJ, Proschel C, Noble M, Sheer D. Assignment of the human and mouse LIM-kinase genes (LIMK1; Limk1) to chromosome bands 7q11.23 and 5G1, respectively, by in situ hybridization. Cytogenet Cell Genet. 1996;74:190–191. doi: 10.1159/000134411. [DOI] [PubMed] [Google Scholar]
- Marra M, Weinstock LA, Mardis ER. End sequence determination from large insert clones using energy transfer fluorescent primers. Genome Res. 1996;6:1118–1122. doi: 10.1101/gr.6.11.1118. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lu X, Li Z, Green ED, Massa H, Trask BJ, Morris CA, Keating MT. Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes. Hum Genet. 1998a;103:590–599. doi: 10.1007/s004390050874. [DOI] [PubMed] [Google Scholar]
- Meng X, Lu X, Morris CA, Keating MT. A novel human gene FKBP6 is deleted in Williams syndrome. Genomics. 1998b;52:130–137. doi: 10.1006/geno.1998.5412. [DOI] [PubMed] [Google Scholar]
- Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: Physical characteristics. J Pediatr. 1988;113:318–326. doi: 10.1016/s0022-3476(88)80272-5. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Matsuoka R, Kimura M, Hirota H, Mikoshiba K, Shimizu Y, Shimizu N, Akagawa K. Hemizygous deletion of the HPC-1/syntaxin 1A gene (STX1A) in patients with Williams syndrome. Cytogenet Cell Genet. 1998;82:49–51. doi: 10.1159/000015063. [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Carter KC, Shell BK, Papadopoulos N, Vogelstein B, Kinzler KW. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- Osborne LR, Martindale D, Scherer SW, Shi X-M, Huizenga J, Heng HHQ, Costa T, Pober B, Lew L, Brinkman J, et al. Identification of genes from a 500-kb region at 7q11.23 that is commonly deleted in Williams syndrome patients. Genomics. 1996;36:328–336. doi: 10.1006/geno.1996.0469. [DOI] [PubMed] [Google Scholar]
- Osborne LR, Herbrick J-A, Greavette T, Heng HHQ, Tsui L-C, Scherer SW. PMS2-related genes flank the rearrangement breakpoints associated with Williams syndrome and other diseases on human chromosome 7. Genomics. 1997a;45:402–406. doi: 10.1006/geno.1997.4923. [DOI] [PubMed] [Google Scholar]
- Osborne LR, Soder S, Shi X-M, Pober B, Costa T, Scherer SW, Tsui L-C. Hemizygous deletion of the syntaxin 1A gene in individuals with Williams syndrome. Am J Hum Genet. 1997b;61:449–452. doi: 10.1086/514850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paperna T, Peoples R, Wang Y-K, Kaplan P, Francke U. Genes for the CPE receptor (CPETR1) and the human homolog of RVP1 (CPETR2) are localized within the Williams-Beuren syndrome deletion. Genomics. 1998;54:453–459. doi: 10.1006/geno.1998.5619. [DOI] [PubMed] [Google Scholar]
- Peoples R, Perez-Jurado L, Wang Y-K, Kaplan P, Francke U. The gene for replication factor C subunit 2 (RFC2) is within the 7q11.23 Williams syndrome deletion. Am J Hum Genet. 1996;58:1370–1373. [PMC free article] [PubMed] [Google Scholar]
- Perez Jurado LA, Peoples R, Kaplan P, Hamel BCJ, Francke U. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet. 1996;59:781–792. [PMC free article] [PubMed] [Google Scholar]
- Perez Jurado LA, Wang Y-K, Peoples R, Coloma A, Cruces J, Francke U. A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-1 and BAP-135, a phosphorylation target of BTK. Hum Mol Genet. 1998;7:325–334. doi: 10.1093/hmg/7.3.325. [DOI] [PubMed] [Google Scholar]
- Riethman H, Birren B, Gnirke A. Preparation, manipulation, and mapping of HMW DNA. In: Birren B, et al., editors. Genome analysis: A laboratory manual. Vol. 1 Analyzing DNA. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 83–248. [Google Scholar]
- Robinson WP, Waslynka J, Bernasconi F, Wang M, Clark S, Kotzot D, Schinzel A. Delineation of 7q11.2 deletions associated with Williams-Beuren syndrome and mapping of a repetitive sequence to within and to either side of the common deletion. Genomics. 1996;34:17–23. doi: 10.1006/geno.1996.0237. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Metcalfe K, Fergusson WD, Carette MJA, Dore JK, Donnai D, Read AP. LIM-kinase deleted in Williams syndrome. Nat Genet. 1996;13:272–273. doi: 10.1038/ng0796-272. [DOI] [PubMed] [Google Scholar]
- Trask B. Fluorescence in situ hybridization. In: Birren B, et al., editors. Genome analysis: A laboratory manual. Vol. 4 Mapping genomes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 303–413. [Google Scholar]
- Trask B, Pinkel D, van den Engh G. The proximity of DNA sequences in interphase nuclei is correlated to genomic distance and permits ordering of cosmids spanning 250 kilobase pairs. Genomics. 1989;5:710–717. doi: 10.1016/0888-7543(89)90112-2. [DOI] [PubMed] [Google Scholar]
- Urban Z, Helms C, Fekete G, Csiszar K, Bonnet D, Munnich A, Donis-Keller H, Boyd CD. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am J Hum Genet. 1996;59:958–962. [PMC free article] [PubMed] [Google Scholar]
- van den Engh G, Sachs R, Trask BJ. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science. 1992;257:1410–1412. doi: 10.1126/science.1388286. [DOI] [PubMed] [Google Scholar]
- Wang Y-K, Harryman Samos C, Peoples R, Perez-Jurado LA, Nusse R, Francke U. A novel human homologue of the Drosophila frizzled wnt receptor gene binds wingless protein and is in the Williams syndrome deletion at 7q11.23. Hum Mol Genet. 1997;6:465–472. doi: 10.1093/hmg/6.3.465. [DOI] [PubMed] [Google Scholar]
- Wang Y-K, Perez-Jurado LA, Francke U. A mouse single-copy gene, Gtf2i, the homolog of human GTF2I, that is duplicated in the Williams-Beuren syndrome deletion region. Genomics. 1998;48:163–170. doi: 10.1006/geno.1997.5182. [DOI] [PubMed] [Google Scholar]
- Wu Y-Q, Sutton VR, Nickerson E, Lupski JR, Potocki L, Korenberg JR, Greenberg F, Tassabehji M, Shaffer LG. Delineation of the common critical region in Williams syndrome and clinical correlation of growth, heart defects, ethnicity, and parental origin. Am J Med Genet. 1998;78:82–89. doi: 10.1002/(sici)1096-8628(19980616)78:1<82::aid-ajmg17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Wydner KS, Sechler JL, Boyd CD, Passmore HC. Use of an intron length polymorphism to localize the tropoelastin gene to mouse chromosome 5 in a region of linkage conservation with human chromosome 7. Genomics. 1994;23:125–131. doi: 10.1006/geno.1994.1467. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]