Abstract
Cdc2L1 and Cdc2L2 span ∼140 kb on human chromosome 1p36.3. The products of the Cdc2L genes encode almost identical protein kinases, the PITSLRE kinases, which have functions that may be relevant to the regulation of transcription/splicing and apoptotic signaling. These genes are deleted/translocated in neuroblastomas with MYCN gene amplification, a subset of malignant melanomas, and in a newly delineated deletion syndrome. Here we report that the p36.3 region of human chromosome 1 consists of two identical genomic regions, each of which contain a Cdc2L gene linked to a metalloprotease (MMP) gene in a tail-to-tail configuration. This duplicated genomic region is also linked tightly to D1Z2, a genetic marker containing a highly polymorphic VNTR (variable number tandem repeat) consisting of an unusual 40-bp reiterated sequence. Thus, these genes and the polymorphic marker D1Z2 are organized as follows: telomere–D1Z2–5′-MMP22-3′–3′-Cdc2L2-5′–5′-Cdc2L1-3′– 3′-MMP21-5′–centromere. Remarkably, the introns and exons of Cdc2L1 and Cdc2L2, as well as their flanking regions, are essentially identical. A total of 15 amino acid differences, 12 nonconservative and 3 conservative, can be found in the 773–786 amino acids specified by the various products of the Cdc2L genes. Two separate promoter/5′ untranslated (UT) regions, CpG1 and CpG2, are identical to a reported previously methylated genomic CpG sequence and are used to express >20 different Cdc2L transcripts from the two genes. The expression of CpG2 transcripts from Cdc2L1 and Cdc2L2 is tissue/cell-line specific. CpG1 transcripts are expressed ubiquitously from both genes, with perhaps some bias towards the expression of CpG1 Cdc2L1 mRNAs in certain hematopoietic cells.
[The sequence data described in this paper have been submitted to the GenBank data library under the following accession nos.: (cDNAs) AF067511–AF067529; (genomic) AF080678–AF080697.]
The human Cdc2L1 and Cdc2L2 genes were localized previously to chromosome band 1p36.3, and they are deleted/altered frequently in neuroblastomas with amplified MYCN genes, a subset of malignant melanoma, and a 1p36 deletion disorder (Lahti et al. 1994; Shapira et al. 1997; Nelson et al. 1998). In fact, these genes are in close proximity to p73, which like Cdc2L1/2 is located proximally to D1Z2 (Kaghad et al. 1997). The Cdc2L1/2 genes express >20 distinct PITSLRE protein kinase isoforms, many arising by alternative splicing (Xiang et al. 1994; this study). Indirect immunofluorescence has demonstrated that the larger p110 PITSLRE isoforms expressed by Cdc2L1 and Cdc2L2 are localized in the nucleus, with a high concentration in nuclear speckles (Loyer et al. 1998). The cell cycle-regulated RNA-binding protein RNPS1 was isolated with the PITSLRE p110 isoforms by two-hybrid cloning (Loyer et al. 1998). Overexpression of RNPS1 in mammalian cells results in the redistribution of this kinase, RNPS1, and certain splicing components into fewer, and much larger, nuclear speckles. Treatment of the same mammalian cells with transcriptional inhibitors (e.g., H8, DRB) results in an identical phenotype. In addition, overexpression of PITSLRE p110 does not disrupt the nuclear organization (i.e., nuclear speckle structure) of spliceosome components such as SC-35. These results suggest that PITSLRE p110/RNPS1 may be involved in some aspect of transcriptional regulation.
Following apoptotic stimuli a number of the PITSLRE isoforms, including selected p110 proteins, are processed by caspases to generate a p46–50 PITSLRE isoform (Lahti et al. 1995; Beyaert et al. 1997; Tang et al. 1998). Caspase processing of PITSLRE protein kinases following TNFα or Fas treatment can be blocked effectively by expression of CrmA (a broad spectrum viral caspase inhibitor) and bcl-2, or by use of the caspase peptide inhibitors (e.g., ZVAD–cmk, DEVD–fmk). Caspase processing of the PITSLRE p110 isoforms is accompanied by the rapid phosphorylation of p110, and it appears that phosphorylation of the amino-terminal domain may influence the ability of these proteases to cleave this region of the protein (Tang et al. 1998). The p46–50 PITSLRE isoform generated by caspase cleavage retains the catalytic protein kinase domain, and when immunoprecipated is capable of phosphorylating histone H1 (Lahti et al. 1995). This is of considerable interest, as the larger p110 PITSLRE isoforms cannot phosphorylate histone H1 and, conversely, the p46–50 PITSLRE isoform cannot phosphorylate p110 substrates. Thus, it is possible that caspase processing may alter the substrate specificity of the PITSLRE protein kinase.
Human chromosome 1 band p36.2–.3 also contains a number of additional genes involved in apoptotic processes, including DR3, DR3L, TNFR2, CD30, OX40, and the 4-1BB ligand, which all encode proteins related to the TNF/NGF death-receptor family and their pathways (Jensen et al. 1997; Grenet et al. 1998). More recently, a novel p53-related gene, p73, has been mapped to a region directly adjacent to the Cdc2L1/2 genes (Kaghad et al. 1997). Like the Cdc2L1/2 and DR3/DR3L genes, one allele of p73 is deleted frequently, but mutations have not been found in the remaining allele in neuroblastomas or lung tumors. Expression of p73 can also trigger apoptosis under certain circumstances (Jost et al. 1997). Comparative genomic hybridization and fluorescent in situ hybridization (FISH) analyses have shown that many neuroblastoma cell lines and patient samples (particularly those with amplified MYCN genes) have deleted varying amounts of the short arm of chromosome 1 (van Roy et al. 1997). Amplification of MYCN occurs rarely in neuroblastoma cell lines or patient samples in the absence of such 1p deletions, suggesting that this region must be lost to accommodate the high levels of N-Myc expression (Caron et al. 1993; Shapiro et al. 1993). One interpretation of these results is that multiple tumor-suppressor genes are involved in the genesis of neuroblastoma (Takeda et al. 1994; Caron et al. 1995). Loss of 1p36 also occurs in a number of other tumors, including ductal carcinoma of the breast, a subset of malignant melanomas, Merkel cell carcinoma, and colon carcinoma (Schwab et al. 1996).
Clearly, defining the structure and expression patterns of the genes localized to the 1p36 region will help to facilitate detailed molecular analysis of these deleted/translocated regions in tumors. Such information may also provide insight into the possible mechanisms responsible for the deletion of such a large region of 1p36. Here we report that the Cdc2L1/2, as well as metalloprotease (MMP21/22), gene loci localized to 1p36.3 consist of two identical, tandemly linked genomic regions. Each of these regions contains a Cdc2L gene linked to an MMP gene in a tail-to-tail configuration. The highly polymorphic genetic marker D1Z2, which contains an unusual reiterated 40-bp variable number tandem repeat (VNTR), is located at the distal end of this ∼140-kb region. The distal 1p36.3 deletion breakpoint observed in tumors and a newly delineated 1p36-deletion syndrome frequently occurs near D1Z2, resulting in the loss of one or more of the Cdc2L genes (Schwab et al. 1996; Shapira et al. 1997). Cdc2L1 and Cdc2L2 are almost identical, yet transcripts are expressed ubiquitously from these genes in most human cell lines and tissues examined. Finally, two distinct promoters, containing CpG-rich sequences corresponding to a previously isolated CpG genomic sequence characterized by Bird and colleagues (Cross et al. 1994), are responsible for the expression of transcripts from Cdc2L1 and Cdc2L2. These data indicate that the Cdc2L1/2 genes and the surrounding sequences have been duplicated in their entirety in the 1p36.3 region, and that each of these genes remains transcriptionally active. The functional significance of the reiteration of selected genes within this region of 1p36.3 is not known, but it might be relevant to the deletion of 1p36 during tumorigenesis.
RESULTS AND DISCUSSION
Structure of the Cdc2L1 and Cdc2L2 Genes
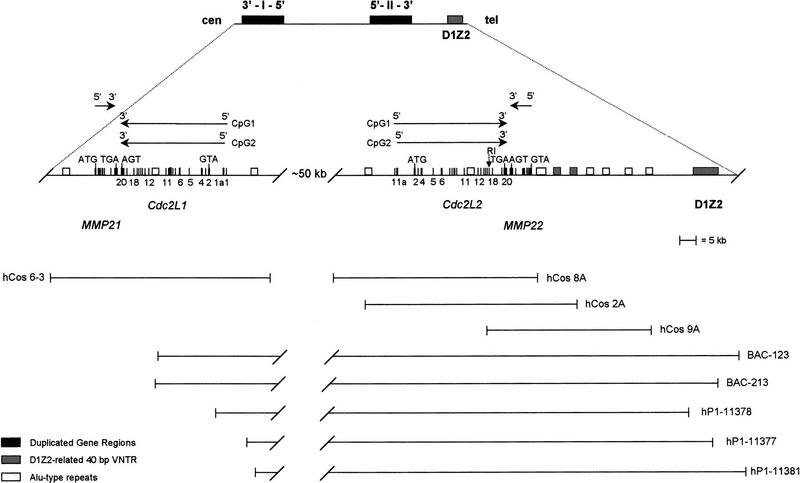
Four cosmids, hCos2A, hCos8A, hCos9A, and hCos6-3; three P1 phage clones, hP1-11377, hP1-11378, and hP1-11381; and two BAC clones, BAC-1 and BAC-2, containing the Cdc2L1 and Cdc2L2 genes and spanning ∼140 kb served as the starting point for determining the structure of these genes. Early on we discovered that the cosmids contained nearly identical DNA sequences corresponding to the 20 exons and 19 introns that constitute the most widely expressed products of the Cdc2L1 and Cdc2L2 genes (Fig. 1). Two distinct sets of alternatively spliced transcripts encoding ∼25 isoforms are generated from these genes. Whereas Cdc2L1 and Cdc2L2 express transcripts containing nearly identical 5′ untranslated regions (UTRs) and open reading frames (ORFs), a second set of transcripts containing a unique 5′ UTR identical to the consensus sequence of a previously isolated human CpG island is also expressed from these genes (Cross et al. 1994; see below). The very high level of identity between Cdc2L1 and Cdc2L2 created numerous technical problems (e.g., double priming that generated overlapping DNA sequences with occasional differences) when direct sequencing of cosmid/P1/BAC DNA was attempted. Therefore, each cosmid was divided into smaller fragments that were subcloned for the purpose of DNA sequence analysis. The Cdc2L1 and Cdc2L2 genes are separated by ∼50 kb. A highly polymorphic 40-bp VNTR found in D1Z2 is located on the distal end of this gene cluster.
Figure 1.
Schematic representation of the duplicated Cdc2L1/2 and MMP21/22 genes linked to D1Z2 on 1p36.3. Shown is the organization of two duplicated genomic segments on human chromosome 1p36.3; each segment contains a Cdc2L and MMP gene (Cdc2L1 and MMP2 or Cdc2L2 and MMP22). Also shown is their transcriptional orientation relative to one another, the mosaic structure of the Cdc2L genes (each having 21 exons and 20 introns), their linkage to D1Z2, and their relationship to telomeric and centromeric regions of the short arm of chromosome 1. The various human genomic cosmid, P1 phage, and BAC clones used to construct this detailed map are indicated below the map.
A single nucleotide difference in exon 17, resulting in a Lys residue at position 625 in Cdc2L1 and an Asn residue at the same position in Cdc2L2, accounts for an EcoRI restriction site that is present in the latter gene but not the former (Fig. 1). This restriction site provided a useful diagnostic tool that allowed us to begin the characterization of these genes. Cosmid hCos6-3, containing the Cdc2L1 gene, does not overlap the cosmids containing the Cdc2L2 gene (hCos2A, hCos8A, and hCos9A; Fig. 1). This, in part, made it clear that we were dealing with a locus containing reiterated and highly identical genes. The intron/exon boundary sequences of Cdc2L1 and Cdc2L2 agree with consensus sequences (see GenBank database entries for DNA sequence details). Each of these genes spans ∼20 kb (Fig. 1). The complete genomic sequence for these genes has been deposited in the GenBank database (accession nos. listed in Fig. 1).
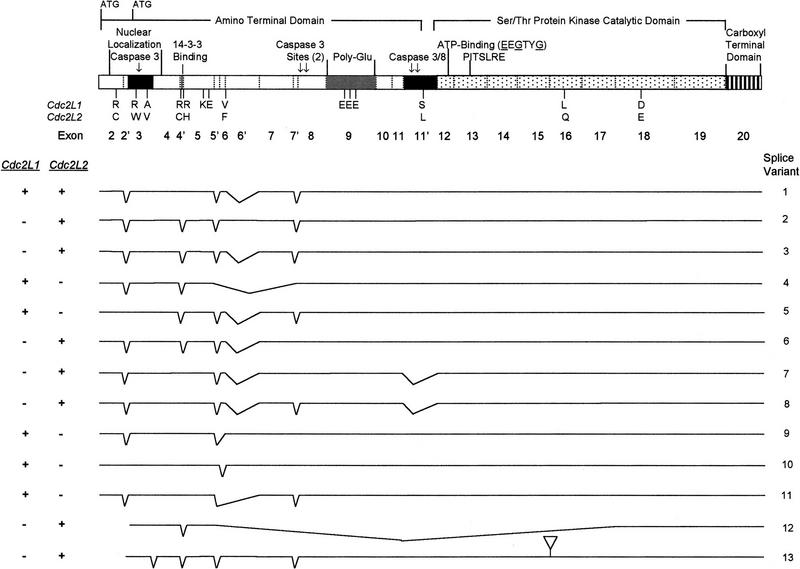
A schematic showing the nature and location of amino acid differences between the predicted ORFs of Cdc2L1 and Cdc2L2 is shown in Figure 2. A total of 15 amino acids of the 773–786 residues that encode the >20 PITSLRE protein kinase isoforms are unique to either Cdc2L1 or Cdc2L2. The majority of the 15 amino acid changes (11; all of which are nonconservative) are located in the amino-terminal domain, whereas only four differences (one nonconservative and three conservative) are located in the protein kinase catalytic and carboxy-terminal domains (Fig. 2). Transcripts corresponding to both genes have been detected in all human cell lines and tissues examined thus far (see Figs. 4 and 5, below). The significance of redundant Cdc2L/PITSLRE expression is unknown at this time, but the differences in these gene products may be relevant to their cellular function(s).
Figure 2.
Detailed schematic comparing the relationship between the structure of the PITSLRE protein kinase and Cdc2L1/Cdc2L2. The structure of the PITSLRE protein kinase and the Cdc2L exons that encode the protein is shown in detail at top. Amino acid differences between the proteins encoded by Cdc2L1 and Cdc2L2 are indicated below the protein schematic. Directly below this schematic are representations of the various PITSLRE mRNAs (denoted as splice variants) generated by alternative splicing from Cdc2L1, Cdc2L2, or both.
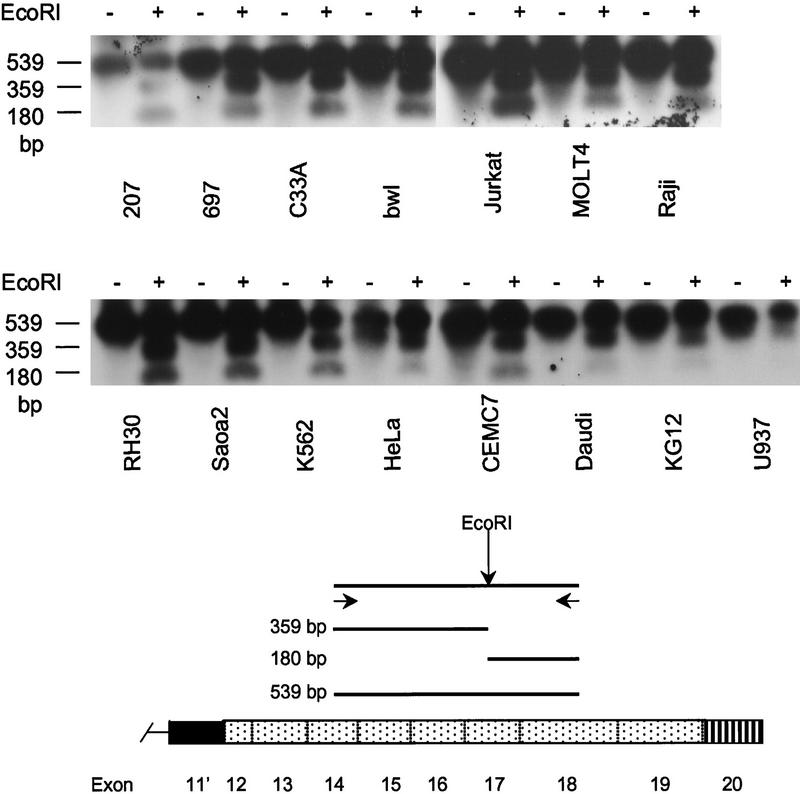
Figure 4.
PCR–LR analysis of Cdc2L1 and Cdc2L2 mRNAs expressed by various cell lines. An analysis of the expression of Cdc2L1 and Cdc2L2 using a unique EcoRI restriction site found in exon 17 of Cdc2L2 (bottom) was performed by RT–PCR. The undigested (−) and EcoRI-digested (+) RT–PCR products were separated by agarose gel electrophoresis, transferred to a membrane, and hybridized with the PITSLRE cDNA. Whereas most cell lines express both Cdc2L1 and Cdc2L2 transcripts, Daudi and U937 cells appear to express primarily Cdc2L1 mRNA, which lacks the diagnostic EcoRI site. (Bottom) A schematic of Cdc2L structure relative to the location of the PCR primers used in this analysis. The expected sizes of the undigested (539 bp) and digested (359 and 180 bp) products is also indicated.
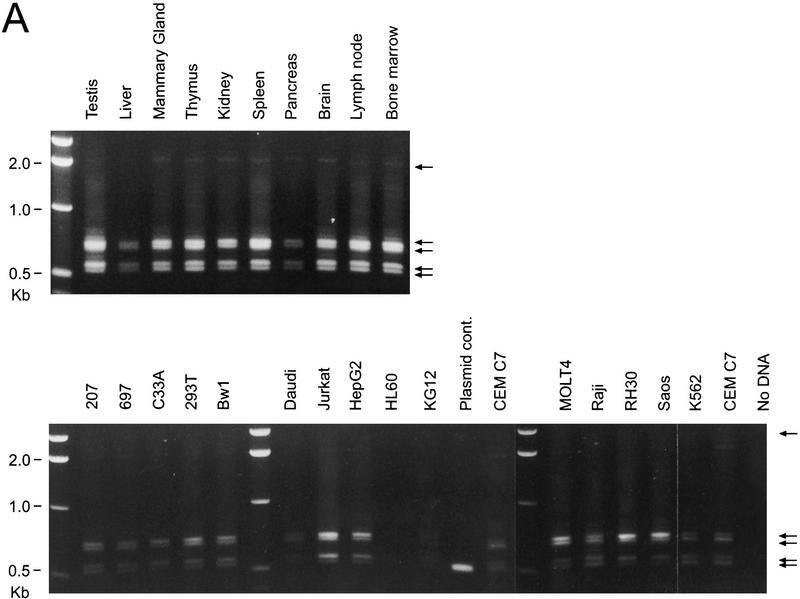
Figure 5.
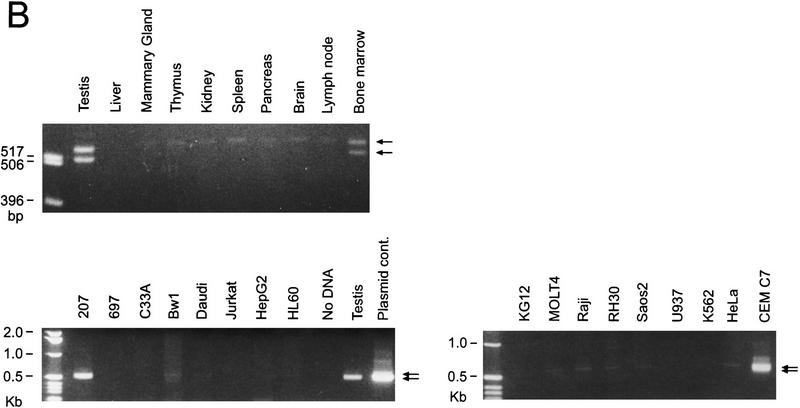
RT–PCR analysis of the expression of Cdc2L transcripts originating from the two different CpG promoter regions in various human tissues and cell lines. (A) RT–PCR analysis of CpG1 and CpG2 (see Fig. 1) transcripts expressed by Cdc2L1 and Cdc2L2. The various tissues and cell lines are indicated above each lane, whereas five different sized products (see text for explanation) are indicated on the right of each panel. (B) RT–PCR analysis of the expression of CpG1 transcripts corresponding to the Cdc2L1 gene, using the same set of tissue and cell line RNAs. The arrows at right indicate that these CpG1 mRNAs are expressed in a few, but not all, of the sources examined. Decreased resolution of the two RT–PCR products (bottom) is caused by the short run of the agarose gel used to analyze the RNAs from the various cell lines.
Several of the 11 amino acid differences found in the amino-terminal domain of the PITSLRE protein kinases are notable. These include Arg-94(Cdc2L1) → Cys-94(Cdc2L2), Arg-109(Cdc2L1) → Cys-109(Cdc2L2), Arg-112(Cdc2L1) → His-112(Cdc2L2), Arg-130(Cdc2L1) → Trp-130(Cdc2L2), the insertion of Lys-152–Glu-153 and Glu-347–Glu-348–Glu-349 in Cdc2L1, and Ser-402(Cdc2L1) → Leu-397(Cdc2L2). The Arg residues at positions 109 and 112 in Cdc2L1 are found within a putative 14-3-3 consensus-binding site [Arg–Arg–His–Arg–Ser–His–Ser–Ala (Aitken 1998)] that is eliminated by the changes in these amino acids in 151–349 occur in Cdc2L2, as does the Ser → Leu change found near a previously mapped caspase cleavage site (Asp-397 in Cdc2L1; Beyaert et al. 1997). The latter change is of possible interest because it resides within a region of the p110 PITSLRE protein containing sites known to be phosphorylated by casein kinase II (Malek and Desiderio 1994). Finally, a polyglutamic acid region spanning ∼30 amino acids is encoded by exon 9 of Cdc2L1 and Cdc2L1 (Fig. 2). Trinucleotide repeats (i.e., GAAn and GAGn) that span 39–48 nucleotides are associated with this region of these genes. A number of genes associated with neurological disorders, as well as certain tumors, undergo expansion of similar trinucleotide repeats as part of the disease process (Sanpei et al. 1996). Intriguingly, the number of CAG trinucleotide repeats, and thus the number of Gln residues, present in the Huntingtin protein appears to influence its ability to be cleaved by caspases during apoptosis (Goldberg et al. 1996; Wellington et al. 1998). This may be of interest as we have shown previously that a number of the p110 PITSLRE isoforms are cleaved by multiple caspases during TNF- and Fas-mediated cell death (Beyaert et al. 1997; Tang et al. 1998). Exon 9 of CdcL1 contains three more Glu residues than the same region of Cdc2L2 because of a difference in the number of GAA repeats (Fig. 2). At this time we do not know whether this difference influences the ability of the respective Cdc2L proteins to be cleaved by caspases.
Regions adjacent to Cdc2L1 and Cdc2L2, but outside the transcribed portion of the genes, contain several interesting sequences (Fig. 1). Most prominently, we noticed a single-copy DNA sequence immediately adjacent to the 3′ ends of both Cdc2L1 and Cdc2L2. Comparison of this sequence to the databases revealed that it was homologous to several metalloproteinases (Fig. 1; Gururajan et al. 1998). There are two MMP genes in this region of chromosome 1p36.3, each of which is linked tightly (∼500–800 bp) to the 3′ end of a Cdc2L gene. This is consistent with the duplication of the genomic region encompassing both the Cdc2L and MMP genes in the same area of the short arm (p36.3) of human chromosome 1. An unusual, reiterated 40-bp sequence (repeated >40 times) was found distal to Cdc2L2 (Fig. 1). A comparison of this sequence to those in available databases indicates that this sequence is ∼90% identical to a similar sequence contained in the anonymous DNA fragment D1Z2 (Buroker et al. 1987; Nakamura et al. 1987). Additionally, a number of Alu-type repeated sequences are found concentrated near the 5′ ends of these genes as well as several of the larger introns (Fig. 1).
Fiber FISH Analysis of Cdc2L1, Cdc2L2, and D1Z2
To establish a more precise genomic map of the Cdc2L1/2–MMP21/22 loci, and to confirm our preliminary linkage/organization of these genes, a technique employing the stretching of genomic DNA coupled with fluorescent in situ hybridization (fiber FISH) was used. Others have shown that this procedure allows high-resolution mapping of large DNA fragments, particularly with regard to establishing the physical distance from a fixed reference point (Parra and Windle 1993). Using this strategy we determined that the four cosmids, hCos6-3, hCos8A, hCos2A, and hCos9A, were ∼70, 22, 17, and 10 kb, respectively, from D1Z2 (Fig. 3). These results are consistent with what was determined independently by restriction mapping and DNA sequence analysis, and confirm the linkage arrangement of these genes (Fig. 1).
Figure 3.
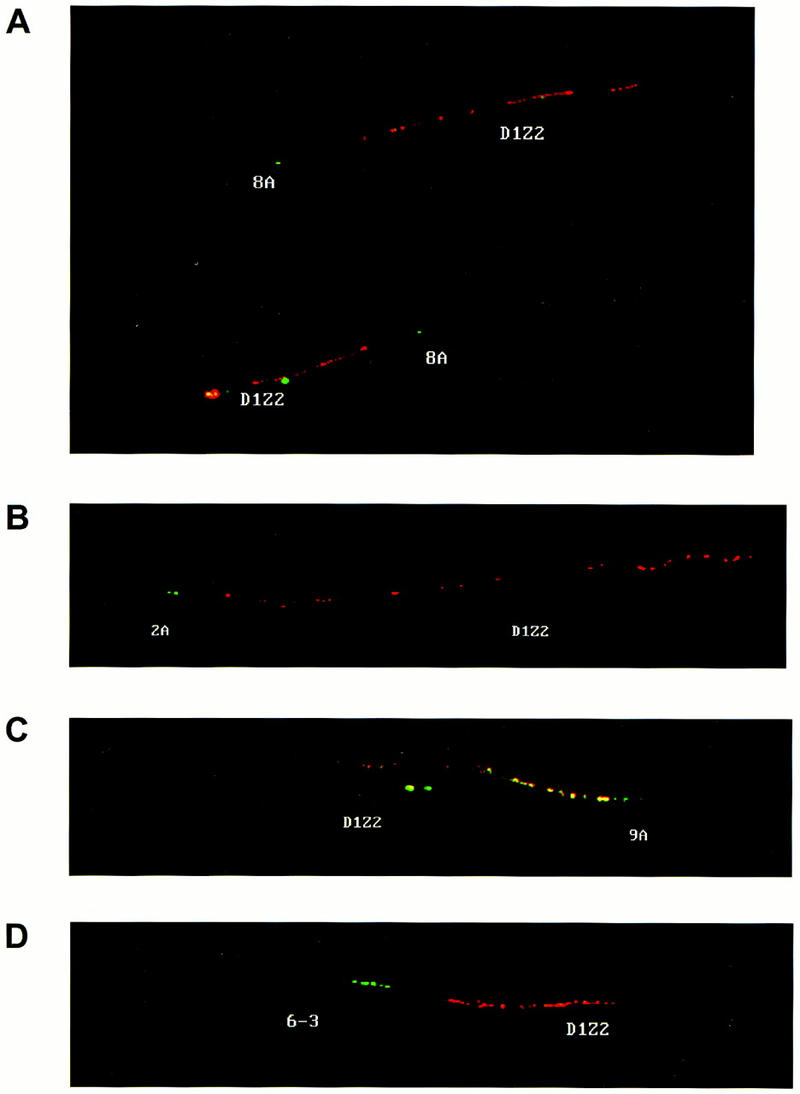
Fiber FISH analysis of the Cdc2L cosmids and the chromosome 1p36.3 marker D1Z2. Fiber FISH analysis using (A) hCos8A and D1Z2, (B) hCos2A and D1Z2, (C) hCos9A and D1Z2, and (D) hCos6–3 and D1Z2. Cosmids 2A, 8A, and 9A contain Cdc2L2; cosmid 6–3 contains Cdc2L1. The D1Z2 probe is red and the cosmid DNA probes are green. Significant overlap is indicated by yellow, as in C.
Expression of Cdc2L1 and Cdc2L2 in Human Cell Lines and Tissues
What are the products of the Cdc2L1 and Cdc2L2 genes, and are they expressed concomitantly in all cells? To answer this question we analyzed cDNAs by direct DNA sequencing from human HeLa and K562 cell lines and testis, as well as by PCR analysis of RNA from a number of additional human cell lines and tissues (Figs. 2, 4, and 5). Detailed sequence analysis of >30 cDNAs and RT–PCR products revealed that Cdc2L1 and Cdc2L2 transcripts are expressed in HeLa and K562 cells as well as testis. In addition, this analysis revealed that a number of Cdc2L1 and Cdc2L2 transcripts are generated by rather complex alternative splicing involving exons located in the amino-terminal domain, but not in the protein kinase catalytic domain (Fig. 2). Two rather intriguing transcripts were found in human testis. One contains a unique 5′ UTR, which is encoded by an alternative promoter, that corresponds to a previously reported CpG island isolated by Bird and colleagues (Fig. 1; Cross et al. 1994; see below). A second unusual transcript is generated apparently by alternative splicing and contains exons 2–5 and 18–20 (Fig. 2). This alternatively spliced transcript encodes the amino-terminal region of the p110 PITSLRE kinase extending from amino acids 58 (Met) to 165, but the protein kinase catalytic domain is removed in its entirety. Three novel amino acids that result from a frameshift between exons 5 and 18 are also introduced, followed by a premature stop codon. Thus far, this transcript has only been found in testis. Its functional implications are not known at this time, but it is possible that the product it encodes might function in a dominant negative manner during certain stages of development. In support of this notion, at least eight different PITSLRE protein kinase transcripts are expressed at varying levels in Drosophila melanogaster during development (Sauer et al. 1996).
To determine whether transcripts corresponding to Cdc2L1 and Cdc2L2 are expressed concomitantly in a variety of different human cell lines and tissues a PCR–RL experiment was performed (Fig. 4). This experiment was designed to take advantage of the EcoRI restriction site present in exon 17 of Cdc2L2, but not Cdc2L1 (Fig. 1). PCR primers flanking either side of the EcoRI site, placing the restriction site in an asymmetric position to allow easy detection of the digested 359 and 180 bp products, were used. Examination of the PCR–RL products reveals that almost equivalent amounts of the uncut and digested products are present in most of the cell lines and tissues analyzed (Fig. 4). For comparison, undigested RT–PCR products from the same cell lines are also shown. To demonstrate that the conditions we used for EcoRI digestion of the PCR products were appropriate for the samples, PCR products generated from the cosmid templates containing Cdc2L1 and Cdc2L2 were treated independently, or together, with the enzyme and the products analyzed (data not shown). At this time, we do not know whether the few differences in the ratio of Cdc2L1/Cdc2L2 transcripts in several cell types is relevant to the proliferative or death status of these cells. However, the cell lines that appear to express primarily Cdc2L1 transcripts, Daudi and U937 (Fig. 4), represent a particularly late stage of hematopoiesis. Whether this stage of hematopoietic development is relevant to the observed differences in Cdc2L expression is not known at this time.
It is not yet clear whether any of the alternatively spliced products are restricted to specific cell types, as it appears that most cell lines and tissues express many of these isoforms (Fig. 4). Discrimination of specific alternatively spliced transcripts has proven to be very difficult. A few, rather unusual, alternatively spliced transcripts (e.g., the transcript found in testis that removes the majority of the protein kinase catalytic domain) appear to be restricted to certain cell types. The most common alternatively spliced transcripts generate proteins with/without the putative 14-3-3 consensus-binding site mentioned earlier, as well as proteins with/without the previously mapped caspase cleavage sites at Asp-387 and Asp-391, which are used during TNF- and Fas-mediated apoptosis (Beyaert et al. 1997; Tang et al. 1998). The latter change would render the resulting protein insensitive to caspase cleavage at this site during apoptosis. Another result of some of the changes in the predicted proteins introduced by alternative splicing is the disruption of coiled–coiled structural motifs. These motifs are most likely involved in protein–protein interactions, suggesting that such changes might help to determine what proteins interact with the various isoforms of the PITSLRE kinases. The elucidation of the differences between the various alternatively spliced transcripts generated by Cdc2L1 and Cdc2L2 should now facilitate comprehensive analysis of this gene locus in human tumors as well as functional studies of these proteins in mammalian cells.
A Separate Set of Cdc2L Transcripts with an Alternatively Spliced 5′ UTR
The identification of an alternatively spliced Cdc2L transcript from testis containing a CpG island in its 5′ UTR (which matches a previously reported CpG sequence: GenBank accession no. Z61267) suggested that a distinct group of Cdc2L transcripts might be expressed in some cells. To examine this, we used the unique nature of this 5′ UTR to design PCR primers to amplify the CpG1 Cdc2L mRNAs (containing exon 1, but not exon 1a), or the CpG2 Cdc2L1 transcript (containing exon 1a, but not exon 1) specifically. Two sets of CpG2 doublets were generated from many of the cell lines examined (Fig. 5A). The distance between the bands in the doublet corresponds precisely to the difference generated by the presence/absence of exon 4′ (Fig. 2), which was confirmed by DNA sequence analysis of these products (data not shown). The second set of doublets was larger than expected, but the distance between the bands of this doublet once again corresponds to what is expected when alternative spicing of exon 4′ occurs. These products were also cloned to determine why they were larger in size. DNA sequence analysis of these CpG2 transcripts revealed that the difference in size was caused by the amount of CpG sequence retained in the 5′ UTR (∼250 bp or 1 kb), once again as a result of alternative splicing. None of this sequence was found to encode an extension of the p110 ORFs, as judged by several in-frame termination codons located immediately upstream of the previously identified translational start site.
The use of this particular CpG sequence, isolated in a search for methylated genomic DNA (Cross et al. 1994), as a promoter for the Cdc2L transcripts suggested that their expression might be subject to regulation by DNA methylation. If this were true, tissue-specific expression of Cdc2L1 and Cdc2L2 transcripts might occur. To determine whether this was the case, we designed PCR primers that took advantage of the significant differences between the 5′ UTRs in exon 1 of these genes. In fact, the CpG1 transcripts derived from Cdc2L1 are expressed only in human testis, bone marrow, and the hematopoietic cell lines CEM C7 and 207 (Fig. 5B). Both of these cell lines are characteristic of early hematopoietic progenitors; CEM C7 is of early T-cell lineage, whereas 207 is of early B-cell lineage. This may be of tremendous functional significance, as Cdc2L1 contains the putative 14-3-3 binding site, whereas Cdc2L2 does not.
Concluding Remarks
Differences between Cdc2L1 and Cdc2L2 may dramatically influence protein localization and/or function during proliferation and/or apoptosis. These differences, including a putative 14-3-3 binding site and changes in coiled–coiled domains, may also help to explain why two nearly identical genes express functional transcripts. It also appears that these genes are expressed in a tissue-specific manner, possibly because of the methylation of CpG sequences in their promoter/5′ UTR regions. Alternatively spliced transcripts that remove two essential caspase cleavage sites from the protein have also been identified, suggesting that a caspase-resistant form of the PITSLRE protein kinase may also be required in cells. It should be noted that the Cdc2L gene locus in several vertebrates, including chicken, mouse, and human, consists of at least two duplicated genes (Li et al. 1995; H. Li, J.M. Lahtia, and V.J. Kidd, unpubl.; this study). In addition, at least nine distinct PITSLRE mRNAs have been observed in D. melanogaster (Sauer et al. 1996). Gene duplications similar to what is described herein (i.e., involving more than a single gene), although infrequent, have also been described others. Examples include the genes encoding glucocerebrosidase (GBA) and metaxin (MTX) on human chromosome 1 band q21 (Winfield et al. 1997) and the X and Y genes in the iduronate-2-sulfate sulfatase (IDS) locus on human chromosome X band q28 (Timms et al. 1995). The duplication of the GBA and MTX genes resulted in a nonfunctional pseudogene adjacent to a functional copy of each of these genes (i.e., 5′ GBA–psMTX ... psGBA–MTX 3′). Conversely, the ∼27.5 kb region of the IDS locus containing the X and Y genes was duplicated in an inverted fashion (i.e., 5′ X–Y 3′ ... 3′ Y′–X′ 5′) with apparently functional genes in each repeated region. This latter duplication is very similar to what we have observed in the Cdc2L locus.
Finally, a number of the genes localized to 1p36.3, including Cdc2L1/2, MMP21/22, and DR3/DR3L, are tandemly duplicated (Grenet et al. 1998; Gururajan et al. 1998). As this region of chromosome 1p is subject to frequent deletion and/or translocation, this might suggest an essential function for these genes. Conversely, it is possible that these gene-duplication events reflect the relative instability of this chromosome region and that they are relevant to the deletions and translocations that are frequently mapped to 1p36.3. Karsten and colleagues have suggested that a double deletion involving the IDS and W genes in a Hunter syndrome patient may be the result of homology-associated nonhomologous recombinations caused by the presence of similarly large duplicated regions on human chromosome X band q28 (Karsten et al. 1997). Either way, it will be of interest to determine whether additional tandemly duplicated genes are localized to this region of chromosome 1 and if such duplications are a common feature of other human chromosomes.
METHODS
Cosmids, P1 Phage, and BAC Clone Isolation
The isolation of cosmids spanning Cdc2L1 and Cdc2L2 has been described earlier (Lahti et al. 1994). P1 phages and BAC clones were obtained from Genome Systems, Inc. PCR primers corresponding to the 5′ UTRs of Cdc2L1 and Cdc2L2 were used to generate 185- (Cdc2L1) and 390- (Cdc2L2) bp PCR products from human genomic DNA. The primer sequences are as follows: Cdc2L1, 197F, 5′-CAGATGAATGTGGCTCAGG-3′; 199R, 5′-CGAGAGTCTCTCCAGGTC-3′; Cdc2L2, T308, 5′-GGTAGACAGAAGGGACTC-3′; 198R, 5′-GGCTTGACCCACAGCCTC-3′. For the first primer set (197F/199R) the PCR conditions were as follows: initial denaturation at 95°C for 5 min, denaturation at 95°C for 15 sec, annealing at 56°C for 15 sec, and extension at 72°C for 30 sec for a total of 30 cycles. For the second and third primer sets (T308/198R and 196F/198R) the conditions were hot start, initial denaturation at 95°C for 5 min, denaturation at 95°C for 15 sec, annealing at 56°C for 15 sec, and extension at 72°C for 30 sec for a total of 30 cycles. The PAC clone addresses are PAC-166-15I, PAC-254-8O, and PAC-23-22D.
Isolation of PITSLRE cDNAs
A human testis Lambda ZAP express cDNA library (Stratagene) was screened independently with random primed 32P-labeled cDNA probes derived from a region corresponding to nucleotides 1124–2471 (carboxy-terminal half; all numbering is based on GenBank sequence U04816) and nucleotides 106–768 (amino-terminal region). The filters were hybridized at 42°C in standard hybridization buffer containing 50% formamide overnight and washed with 2× SSC/0.5%SDS at 68°C. Phagemids from positive-phage clones were rescued according to manufacturer’s instructions (Stratagene). Genomic DNA subclones were constructed by digesting cosmid DNA with the desired restriction enzyme and subcloning fragments into pBS KS(+) (Stratagene).
To obtain full-length clones corresponding to the 5′ region of PITSLRE p110 cDNA, RT–PCR was performed using total RNA from HeLa cells. The RNA was reverse transcribed using random primers with Superscript II (Stratagene). PITSLRE cDNA was obtained from the cDNA pool by PCR using primers GSPRT1, 5′-GTGTTGATATCACTCAAATGGGTGATGAAAAGGACTC-3′ (spans nucleotides 106–131 of GenBank sequence U04816) and GSPRT2, 5′-AGGATGGTGTTGATCTCCCTCAG-3′ (spans nucleotides 1525–1505). The PCR conditions used for this reaction were as follows: initial denaturation at 94°C for 4 min, denaturation at 94°C for 1 min, annealing at 65°C for 45 sec, and extension at 72°C for 4 min for a total of 35 cycles. The resulting ∼1.4-kb PCR product was cloned into PCR II vector (Invitrogen).
To further identify cDNA clones derived from Cdc2L1 or Cdc2L2, the cDNA clones were sequenced rapidly using two nucleotides with primer 6822, 5′-GTGCCCTCCTCGATCCTG-3′ (corresponding to nucleotides 1400–1383). Clones derived from Cdc2L1 have a G at nucleotide position 1298, whereas those from Cdc2L2 have an A at the same position. Truncated, or alternatively spliced, cDNA clones were assigned to either gene by comparison to the genomic sequence of Cdc2L1 and Cdc2L2.
Sequence Analysis
The cDNA and genomic clones were sequenced using T3 and T7 primers and various internal primers. The resulting genomic sequence were compared to PITSLRE cDNA sequences by GAP and BESTFIT analyses (GCG) to ascertain the intron/exon boundaries, as well as the extent of alternative splicing. The genomic DNA sequences were further analyzed using FASTA (GCG) and BLAST to identify repeat sequences.
Fiber FISH Analysis
Genomic DNA was stretched as described by others (Parra and Windle 1993). Chromosome preparations were performed according to standard procedures. DNA probes (the human Cdc2L cosmids 6–3, 2A, 8A, and 9A, and D1Z2) were prepared by nick translation with 30 μm of each of four deoxynucleotide triphosphates and 100 μm of either biotin–dUTP (Boehringer Mannheim) or digoxigenin-11–dUTP (Boehringer Mannheim). Slides were hybridized using a modified fluorescent in situ hybridization protocol (Lestou et al. 1996). Briefly, slides were dehydrated by ethanol passage and then air-dried. Hybridization mixture (10–20 μl) containing 50% formamide, 10% dextran sulfate (Pharmacia AB) in 2× SSC, 0.3 m sodium citrate (pH 7), 0.3 μg sonicated salmon sperm DNA (Sigma), and 2–4 ng/μl of each labeled probe. Standard fluorescent detection procedures were used. Fluorescence microscopy utilized a Leitz Ortholux II microscope equipped with filter blocks A2, N3, and I3 for viewing the DAPI, TRITC, and FITC signals, respectively, and with double or triple band-pass filters (Omega Optical and AF-Analysentechnik), allowing the simultaneous visualization of fluorescence by DAPI, fluorescein, and Texas red or rhodamine.
Estimations of length and distance on microscopically observed DNA images were based on the predicted span of 0.34 nm per base pair for relaxed duplex DNA. These measurements were performed with a 0.01-mm Nikon Objective Micrometer. Fluorescence images were analyzed and stored using the ISIS software (MetaSystems, Altlussheim, Germany).
RT–PCR and PCR–Restriction Length Analysis
Total RNA (5 μg) from different tissues was reverse transcribed with random primers using Superscript II (Stratagene). PITSLRE cDNA-specific primers E5FR, 5′-GACCTCAAGAGCCTGATGGAGACC-3′, corresponding to nucleotides 1621–1644 and E9RP, 5′-GTTCATGAGGTCGAAGCCCTGGTC-3′, corresponding to nucleotides 2160–2137, were then used for the PCR–LR reaction. The PCR conditions were as follows: initial denaturation at 94°C for 4 min, denaturation at 94°C for 45 sec, annealing at 62°C for 45 sec, and extension at 72°C for 80 sec for a total of 25 cycles. The resulting 537-bp PCR product was purified away from free nucleotides by centrifugation through a microcon 100 (Amicon), and an aliquot of this material digested with EcoRI. Undigested PCR product was electrophoresed alongside the EcoRI-digested product on a 2% agarose gel for comparison. Cdc2L2-derived PCR product can be digested by EcoRI and visualized as 359- and 180-bp products. The corresponding Cdc2L1-derived PCR product is not digested by EcoRI. PCR analysis of CpG1 transcripts expressed from Cdc2L1 and Cdc2L2 in human tissues and cell lines was performed with the following primers: A–EA (Cdc2L1-specific), 5′-TTCTCTGGCTCCGGGACCGGCG-3′ corresponding to nucleotides 45–65 of GenBank accession no. U04816; EERP (Cdc2L1 and Cdc2L2), 5′-TCCTGGAATGCTCCCTTGCCATTTC-3′ corresponding to nucleotides 566–541 of GenBank accession no. U04816; and CpGA (Cdc2L1 and Cdc2L2), 5′-GACTGAACCCGAGGAAATAGCCCA-3′. PCR analysis of PITSLRE CpG2 transcript expression was performed with the CpGA and EERP primers as follows: initial denaturation at 94°C for 3 min, denaturation at 94°C for 45 sec, annealing at 74°C for 30 sec, and extension at 72°C for 2.1 min for a total of 35 cycles. PCR analysis of PITSLRE Cdc2L1 transcript expression was performed with the A–EA and EERP primers as follows: initial denaturation at 94°C for 3 min, denaturation at 94°C for 45 sec, annealing at 67°C for 45 sec, and extension at 72°C for 2.1 min for a total of 35 cycles.
Acknowledgments
We thank G. Richmond for help with manual DNA sequencing and the Molecular Resource Facility (SJCRH) for assistance with oligonucleotide synthesis and automated DNA sequence analysis. This research was supported by grants from the National Institutes of Health (NIH) (GM44088 and CA67938) to V.J.K., an NIH Cancer Center Core grant (CA21765) to SJCRH, and by the American Lebanese Syrian Associated Charities (ALSAC) to V.J.K. and J.M.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL vincent.kidd@stjude.org; FAX (901) 495-2381.
REFERENCES
- Aitken A. 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 1998;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- Beyaert R, Kidd VJ, Cornelis S, Van de Craen M, Denecker G, Lahti JM, Gururajan R, Vandenabeele P, Fiers W. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. J Biol Chem. 1997;272:11694–11697. doi: 10.1074/jbc.272.18.11694. [DOI] [PubMed] [Google Scholar]
- Buroker N, Bestwick R, Haight G, Magenis RE, Litt M. A hypervariable repeated sequence on human chromosome 1p36. Hum Genet. 1987;77:175–181. doi: 10.1007/BF00272388. [DOI] [PubMed] [Google Scholar]
- Caron H, Peter M, van Sluis P, Speleman F, de Kraker J, Laureys G, Michon J, Brugieres L, Voute PA, Westerveld A, Slater R, Delattre O, Versteeg R. Evidence for two tumor suppressor loci on chromosomal bands 1p35-36 involved in neuroblastoma: One probably imprinted, another associated with N-myc amplification. Hum Mol Genet. 1995;4:535–539. doi: 10.1093/hmg/4.4.535. [DOI] [PubMed] [Google Scholar]
- Caron H, van Sluis P, van Hoeve M, de Kraker J, Bras J, Slater R, Mannens M, Voute PA, Westerveld A, Versteeg R. Allelic loss of chromosome 1p36 in neuroblastoma is of preferential maternal origin and correlates with N-myc amplication. Nat Genet. 1993;4:187–190. doi: 10.1038/ng0693-187. [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird A. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, Graham RK, Bromm M, Kazemi-Esfarjani P, Thornberry NA, Vaillancourt JP, Hayden MR. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- Grenet J, Valentine V, Kitson J, Li H, Farrow SN, Kidd VJ. Duplication of the DR3 gene on human chromosome 1p36 and its deletion in human neuroblastoma. Genomics. 1998;49:385–393. doi: 10.1006/geno.1998.5300. [DOI] [PubMed] [Google Scholar]
- Gururajan, R., J. Grenet, J.M. Lahti, and V.J. Kidd. 1998. Isolation and characterization of two novel metalloproteinase genes linked to the Cdc2L locus on human chromosome 1p36.3. Genomics (in press). [DOI] [PubMed]
- Jensen SJ, Sulman EP, Maris JM, Matise TC, Vojta PJ, Barrett JC, Brodeur GM, White PS. An integrated transcript map of human chromosome 1p35–p36. Genomics. 1997;42:126–136. doi: 10.1006/geno.1997.4714. [DOI] [PubMed] [Google Scholar]
- Jost CA, Marin MC, Kaelin WG., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Karsten SL, Lagerstedt K, Carlberg B-M, Kleijet WJ, Zaremba J, Van Diggelen OP, Czartoryska B, Petterson U, Bondeson M-L. Two distinct deletions in the IDS gene and the gene W: A novel type of mutation associated with the Hunter syndrome. Genomics. 1997;43:123–129. doi: 10.1006/geno.1997.4811. [DOI] [PubMed] [Google Scholar]
- Lahti JM, Valentine M, Xiang J, Joens B, Amann J, Grenet J, Richmond G, Look AT, Kidd VJ. Alterations in the PITSLRE protein kinase gene complex on chromosome 1p36 in childhood neuroblastoma. Nat Genet. 1994;7:370–375. doi: 10.1038/ng0794-370. [DOI] [PubMed] [Google Scholar]
- Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ. PITSLRE protein kinase activity is associated with apoptosis. Mol Cell Biol. 1995;15:1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestou VS, Strehl S, Lion T, Gadner H, Ambros PF. High-resolution FISH of the entire integrated Epstein-Barr virus genome on extended human DNA. Cytogen Cell Genet. 1996;74:211–217. doi: 10.1159/000134416. [DOI] [PubMed] [Google Scholar]
- Li H, Grenet J, Valentine M, Lahti JM, Kidd VJ. Structure and expression of the chicken PITSLRE protein kinase gene and mRNAs. Gene. 1995;153:237–242. doi: 10.1016/0378-1119(94)00801-x. [DOI] [PubMed] [Google Scholar]
- Loyer P, Trembley J, Lahti JM, Kidd VJ. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J Cell Sci. 1998;111:1495–1506. doi: 10.1242/jcs.111.11.1495. [DOI] [PubMed] [Google Scholar]
- Malek SN, Desiderio S. A cyclin-dependent kinase homologue, p130PITSLRE, is a phosphotyrosine-independent SH2 ligand. J Biol Chem. 1994;269:33009–33020. [PubMed] [Google Scholar]
- Nakamura T, Julier C, Wolff R, Holm T, O’Connell P, Leppert M, White R. Characterization of a human “midisatellite” sequence. Nucleic Acid Res. 1987;15:2537–2547. doi: 10.1093/nar/15.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, M.A., M.E. Ariza, J.M. Yang, F.H. Thompson, R. Taetle, J.M. Trent, J. Wymer, K. Massey-Brown, M. Broome-Powell, J. Easton, J.M. Lahti, and V.J. Kidd. 1998. Abnormalities in the p34cdc2-related PITSLRE protein kinase gene complex on chromosome band 1p36 in melanoma. Cancer Genet. Cytogenet. (in press). [DOI] [PubMed]
- Parra I, Windle B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nat Genet. 1993;5:17–21. doi: 10.1038/ng0993-17. [DOI] [PubMed] [Google Scholar]
- Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- Sauer K, Weigmann K, Sigrist S, Lehner CF. Novel members of the cdc2-related kinase family in Drosophila: cdk4/6, cdk5, PFTAIRE, and PITSLRE kinase. Mol Biol Cell. 1996;7:1759–1769. doi: 10.1091/mbc.7.11.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Praml C, Amler L. Genomic instability in 1p and human malignancies. Genes Chromosomes Cancer. 1996;16:211–229. doi: 10.1002/(SICI)1098-2264(199608)16:4<211::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Shapira SK, McCaskill C, Northrup H, Spikes AS, Elder FFB, Sutton VR, Korenberg JR, Greenberg F, Shaffer LG. Chromosome 1p36 deletions; the clinical phenotype and molecular characterization of a common newly delineated syndrome. Am J Hum Genet. 1997;61:642–652. doi: 10.1086/515520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro DN, Valentine MB, Rowe ST, Sinclair AE, Sublett JE, Roberts WM, Look AT. Detection of N-myc gene amplification by fluorescence in situ hybridization. Am J Path. 1993;142:1339–1346. [PMC free article] [PubMed] [Google Scholar]
- Takeda O, Homma C, Maseki N, Sakurai M, Kanda N, Scwab M, Nakamura Y, Kaneko Y. There may be two tumor suppressor genes on chromosome arm 1p closely associated with biologically distinct subtypes of neuroblastoma. Genes Chromosomes Cancer. 1994;10:30–39. doi: 10.1002/gcc.2870100106. [DOI] [PubMed] [Google Scholar]
- Tang D, Gururajan R, Kidd VJ. Phosphorylation of PITSLRE p110 isoforms accompanies their processing by caspases during Fas-mediated cell death. J Biol Chem. 1998;273:16601–16607. doi: 10.1074/jbc.273.26.16601. [DOI] [PubMed] [Google Scholar]
- Timms KM, Lu F, Shen Y, Pierson CA, Muzny DM, Gu Y, Nelson DL, Gibbs RA. 130 kb of DNA sequence reveals two new genes and a regional duplication distal to the human iduronate-2-sulfate sulfatase locus. Genome Res. 1995;5:71–78. doi: 10.1101/gr.5.1.71. [DOI] [PubMed] [Google Scholar]
- van Roy N, Jauch A, van Gele M, Laureys G, Versteeg R, de Paepe A, Cremer T, Speleman F. Comparative genomic hybridization analysis of human neuroblastomas: Detection of distal 1p deletions and further molecular genetic characterization of neuroblastoma cell lines. Cancer Genet Cytogenet. 1997;97:135–142. doi: 10.1016/s0165-4608(96)00362-7. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- Winfield SL, Tayebi N, Martin BM, Ginns EI, Sidransky E. Identification of three additional genes contiguous to the glucocerebrisidase locus on chromosome 1q21: Implications for Gaucher disease. Genome Res. 1997;7:1020–1026. doi: 10.1101/gr.7.10.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Lahti JM, Grenet J, Easton J, Kidd VJ. Molecular cloning and expression of alternatively spliced PITSLRE protein kinase isoforms. J Biol Chem. 1994;269:15786–15794. [PubMed] [Google Scholar]