Abstract
The cytoprotective response to thermal injury is characterized by transcriptional activation of “heat shock proteins” (hsp) and proinflammatory proteins. Expression of these proteins may predict cellular survival. Microarray analyses were performed to identify spatially distinct gene expression patterns responding to thermal injury. Laser injury zones were identified by expression of a transgene reporter comprised of the 70 kD hsp gene and the firefly luciferase coding sequence. Zones included the laser spot, the surrounding region where hsp70-luc expression was increased, and a region adjacent to the surrounding region. A total of 145 genes were up-regulated in the laser irradiated region, while 69 were up-regulated in the adjacent region. At 7 hours the chemokine Cxcl3 was the highest expressed gene in the laser spot (24 fold) and adjacent region (32 fold). Chemokines were the most common up-regulated genes identified. Microarray gene expression was successfully validated using qRT- polymerase chain reaction for selected genes of interest. The early response genes are likely involved in cytoprotection and initiation of the healing response. Their regulatory elements will benefit creating the next generation reporter mice and controlling expression of therapeutic proteins. The identified genes serve as drug development targets that may prevent acute tissue damage and accelerate healing.
Keywords: heat shock protein, Hsp70, thermal stress, laser-tissue interactions, gene expression analysis, microarray, RT-polymerase chain reaction, skin
Introduction
Thermal injury is associated with a pronounced catabolic response in tissue reflecting both reduced protein synthesis and stimulated protein breakdown.1 The effect of acute heat shock in cells ranges from survival and adaptation through alterations in gene expression to cellular death by apoptosis and necrosis after severe thermal stress.2 The effects of thermal stress on tissue can be due to a direct effect on cellular constituents (denaturation of proteins) or as a secondary effect mediated by adaptive mechanisms like the expression of heat shock proteins (hsp) and inflammation. The effects of moderate heat on cellular structure and function have been well established, and hsp expression has been linked to numerous physiological changes in the cell that can affect cell cycle, cellular differentiation, and apoptosis.3, 4, 5, 6
Thermal effects have been largely studied in cultured cells, and there is a need for a better understanding of tissue responses to hyperthermia. In tissues, the cellular stress response does not occur in isolation but rather occurs in the context of the tissue matrix, multiple cell types, and integrated physiology leading to an inflammatory response. The epidermis is a physiological barrier that protects the organism against constant irradiation of the skin, which would result in DNA damage, apoptosis, and transcriptional changes. Cutaneous defensive mechanisms are highly complex and involve DNA repair, release of pro-inflammatory intercellular signaling molecules, and activation of specific signal transduction cascades that result in activation of transcription factors and regulation of gene expression.7, 8, 9, 10, 11, 12, 13 The increased usage of lasers in medicine, research, and military applications has caused an increase in laser injury, particularly to the eye.14 Laser damage to tissue can be categorized into three basic types: photomechanical, photochemical, and photothermal. Of these basic categories, photothermal damage seems to be the most important.
Gene expression profiling of cultured cells has shown that the pathophysiology following laser damage closely parallels the response following burn injuries.14, 15, 16 In comparison to scalpel incisions, those made by lasers in surgery generally have the disadvantage of causing a delay in wound healing due to collateral thermal damage to surrounding tissues. This was demonstrated in a study where differences in gene expression between free electron laser (FEL) and scalpel incisions in mouse skin were analyzed. It was shown that there was significant differential expression between scalpel and FEL wounds of 89 genes using a 15000-gene microarray.17
DNA microarray analysis has proven to be an informative tool for multiplexed gene expression studies and was applied here to assess acute tissue responses to laser-induced thermal stress. Enk et al.18 have investigated differential expression of ultraviolet-regulated genes in intact human epidermis following in vivo exposure with microarray profiling of 12500 genes. This allowed for differentiation between irradiated and nonirradiated epidermis allowing for the identification of 800 ultraviolet-regulated genes.
It has been widely shown that changes in gene expression are integral for the cellular response to thermal stress. Although genes encoding hsps have been well studied, thermal stress leads to increased expression of a substantial number of genes not normally considered to be hsps.19, 20 These genes can be affected by a variety of different stressors and therefore represent a nonspecific cellular response to stress. Analysis of the inactivation of keratinocytes from short exposure (on the order of seconds) to high temperatures (50 to 60°C) indicated that the absorbing species and/or the injury and response mechanisms may be different from injuries caused by prolonged exposure to moderately elevated temperatures (40 to 50°C for 10 to 20 min).21
To study the tissue response to thermal stress, we developed a transgenic reporter mouse where the transgene is comprised of the promoter from hsp70A1 and the coding sequence for luciferase fused to green fluorescent protein (GFP).22 Tissues in this reporter mouse, called hsp70-L2G, respond with bioluminescence and fluorescence to exposure to elevated temperatures. Using this mouse we were able to precisely identify the times and temperatures that induced an hsp response after irradiation with a CO2 laser. Hsp70-L2G expression demarcated the region of extreme thermal injury, the response of surrounding tissue, and the unaffected regions. We used this expression as a guide to study thermal stress in the mouse epidermis. Gene expression patterns in the two stress regions were compared with that of normal epidermis using DNA microarrays. We identified twenty-three genes with a ten-fold change of expression in the laser-treated region and thirteen genes with a ten-fold change in the adjacent region. Expression of the chemokine Cxcl3 was increased to the greatest extent among all of the genes examined. Altered expression levels for eleven specific genes, including Cxcl3, were validated using quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Materials and Methods
Transgenic Reporter Mouse
The transgenic mouse used for bioluminescent analysis of laser stress was first described by O’Connell-Rodwell et al.22 Briefly, the transgenic mouse employed the dual reporter gene L2G (Luc-2A-eGFP) consisting of a modified firefly luciferase gene (pGL3, Promega Inc., Madison, Wisconsin) joined to 54 base pairs (bp) of the foot-and-mouth disease virus 2A sequence and, via 24 bp of polylinker sequence, to the eGFP gene at the 3′ end. The reporter genes are driven by the murine HSP70A1 promoter sequence. The reporter gene construct is shown in Fig. 1a. Potential founder mice were screened by PCR and tested for thermal response by exposure to mild thermal stress and bioluminescence imaging using an in vivo imaging system (IVIS) imaging system (a Xenogen product form Caliper Life Sciences, Alameda, California) (see below). The selected transgenic mouse line [friend virus B (FVB).Hsp70-luc_2A-eGFP; short form: Hsp70-L2G] was bred to homozygosity, and four to eight week old female mice were used for the in vivo experiments. All research performed in this study was approved by the Stanford University Administrative Panel for Laboratory Animal Care and were conducted under strict adherence to institutional guidelines for animal care.
Figure 1.
(a) Hsp70 reporter construct. (b) Bioluminescent imaging of the 6-mm laser spot at the 7-h time point. (c) This figure is color coded to reveal genes largely involved in angiogenesis, apoptosis, inflammation, and cellular stress. The numbers in the inner circle indicate the number of up-regulated genes (>4-fold change) related to each functional region in the 6-mm laser treated spot. The numbers in the “donut” region surrounding the inner circle are the number of up-regulated genes (>4-fold change) related to each colored functional region in the location adjacent to the laser treated spot.
Laser Thermal Stress/Damage
Tissue thermal stress/damage was produced with a 100-Watt CO2 laser (PLX-100, Parallax Technology Incorporated, Waltham, Massachusetts) and was evaluated with bioluminescent imaging (BLI) with the Hsp70-L2G transgenic mouse. The dorsal skin of the transgenic mice was irradiated with a wavelength of 10.6 μm. Mice were shaved and depilated 24 h prior to irradiation with the laser. It was determined that 24 h was sufficient to prevent residual expression of Hsp70 caused by shaving. Previous research has shown that the peak increase in Hsp70 transcription occurs at 7-h post-irradiation, therefore, the collection of bioluminescence data and tissue sampling were performed at 7-h post-irradiation.
Previous research was performed using a 1-s pulse duration, and it was identified that a 4.4 joules per square centimeter (J/cm2) laser pulse was sufficient to cause thermal damage to the dorsal epidermis of the transgenic mice.22 In this study, a 6-mm diameter flat-top spot was created by irradiating the mouse with the central part of a 1-inch Gaussian beam with a 6-mm hole drilled through an aluminum plate. The mouse back was placed in contact to the bottom of the aluminum plate prior to the laser pulse following. The mice were anesthetized prior to all experiments with an intraperitoneal (i.p.) injection of ketamine/xylazine. It was determined through the use of a forward looking infrared (FLIR, FLIR Thermography) camera that this laser pulse would raise the temperature of the skin to 68.7°C by the end of the 1-s pulse. Other research identifying the thermal effects of laser stress on mouse skin has been performed by Mackanos et al.23 using 1, 10, 30, 100, and 500 ms laser pulse durations.
In Vivo BLI of Laser Stress/Damage
The laser pulse delivered through the 6-mm diameter hole in the aluminum plate was used to generate an affected area large enough to provide adequate resolution to determine the expression profile using an IVIS 200 imaging system (Caliper Life Sciences) with high resolution binning4 and a 3.9-cm field of view to obtain a 60-μm resolution across each treated location. BLI was performed 10 min after an i.p. injection of 5 μl/1 g body weight of 30 mg/ml stock solution of D-luciferin (Biosynth L-8220, Staad, Switzerland) with a 1-min integration time. Imaging was performed at 7-h post-irradiation on a minimum of four mice per group.
Microarray Genomic Analysis
Once the region of interest was identified through BLI of luciferase patterns in the transgenic reporter mouse, the laser treatment was performed on nontransgenic mice (strain FVB) in the six to eight week age range. One 1-s 6-mm 4.4 J/cm2 laser spot was applied to the back of each mouse 24 h after shaving and depilation. At seven hours after laser irradiation, the mice were euthanized with CO2 and the dorsal tissue of interest was immediately harvested using forceps and surgical scissors. Initially, the 6-mm diameter laser spot (thermal damage) was removed followed by an adjacent 3-mm donut shaped region (thermal stress). A third 6-mm diameter region at a minimum of 1-cm distance from the laser spot was removed as normal tissue for comparison.
Once removed, the tissue samples were stored in 1-ml of RNAlater, ribonucleic acid (RNA) stabilization reagent (Qiagen, Valencia, California) in an Eppendorf tube at −20°C. A total of five of each type of tissue sample were collected and analyzed by the Protein and Nucleic Acid (PAN) facility at Stanford University for microarray analysis. Microarray analyses were performed as recommended by the manufacturer (Affymetrix, Santa Clara, California). RNA was first isolated from the dorsal skin tissue using the RNeasy Fibrous Tissue kit (Qiagen). The RNA was then analyzed for quality using a Bioanalyzer 2100 (Agilent Technologies, Foster City, California). A total of four samples for the laser and normal regions were used, while a total of five samples were used for the adjacent region.
The GeneChip Mouse Genome 430 2.0 array (Affymetrix) was used to perform the microarray experiments to analyze expression of the entire mouse genome with 39000 transcripts. The fluorescence of the transcripts is measured with a GeneChip Scanner 3000 7G using GeneChip Operating Software (GCOS) (Affymetrix).
Target preparation is performed by synthesizing double-stranded cDNA from total RNA isolated from the mouse dorsal tissue. An in vitro transcription reaction is used to produce biotin-labeled cRNA from the cDNA before cRNA is then fragmented prior to hybridization. A hybridization cocktail is prepared, which includes the fragmented target, probe array controls, bovine serum albumin and herring sperm DNA. This is then hybridized to the probe array with a 16-h incubation in the GeneChip Hybridization Oven 640 (Affymetrix). A fluidics station (GeneChip Fluidics Station 450, Affymetrix) was primed with appropriate buffers and the probe array was prepared using an automated washing and staining procedure following hybridization. The arrays were scanned using GCOS software, which defines the probe sites and computes the intensity for each cell on the gene chip. The data is stored on DVDs from the PAN facility and was then analyzed using GeneSpring GX 7.3.1 software (Agilent Technologies, Santa Clara, California). For analysis with the software, the measurements less than 0.01 were set to 0.01, each chip was normalized to the 50th percentile and each gene was normalized to the median. A default interpretation was used for all samples and the cross-gene error model using on-color data with a deviation from one was used. The final analysis was performed using a log of the ratio for each transcript. The genes that showed a 2-fold change (up or down) were filtered by student's t-test for significance at a threshold of p < 0.05. Fold inductions were represented as the average of heated samples over the median of the unheated control values for each gene. A Benjamini-Hochberg multiple testing correction (MTC) was used prior to the t-test analysis. A false discovery rate of 0.05 was used. Our analysis focused on the most highly up-regulated genes, which have inherently less probability of being false positives than lower expressing genes.
Relative Expression Quantification by Real Time qRT-PCR
Expression of putatively differentially expressed genes was quantified by SYBR Green real time qRT-PCR on laser treated and control tissue. RNA was isolated using the RNeasy fibrous tissue kit (Qiagen, Valencia, California). DNase treated total RNA was reverse transcribed using Multiscribe Reverse Transcriptase and random hexamers (AppliedBiosystems, Alameda, California) according to the manufacturer's instructions. Quantitative qRT-PCR analyses were performed with an iCycler iQ(Bio-Rad, Hercules, California) using Quantitect PCR Master Mix (Qiagen). Individual expression values were normalized to 18S rRNA expression (Pre-Developed TaqMan Assay Reagents Control Kit; AppliedBiosystems). Primer design was done with help of the “Primer Express 2.0” software (AppliedBiosystems), according to the manufacturer's guidelines. To prevent amplification of genomic sequences at least one primer of each pair was designed spanning two exons where possible. The primer sequences that were used are shown in Table 1. Specificity of amplification was furthermore checked by melting curve analysis with help of the iCycler iQ software (Bio-Rad). Primer oligonucleotides were purchased from Operon (Huntsville, Alabama). PCR conditions were: initial denaturation for 10 min at 95°C followed by 45 cycles consisting of 15 s at 95°C and 1 min at 60°C. Expression ratios between two samples were calculated from differences in threshold cycles at which an exponential increase in reporter fluorescence could first be detected (CT-values). Results of triplicates were averaged for tissue collected at 1-, 3-, and 7-h post-irradiation.
Table 1.
Forward and reverse primer sequences used for SYBR Green qRT-PCR analysis of gene expression.
| Entrez Gene | Entrez Gene | |||
|---|---|---|---|---|
| Gene ID | Symbolb | Gene Name | Forward | Reverse |
| 330122 | Cxcl3 | chemokine (C-X-C motif) ligand 3 | AGTCATAGCCACTCTCAAGGATGG | TGGACTTGCCGCTCTTCAGTA |
| 16193 | Il6 | interleukin 6 | CGCTATGAAGTTCCTCTCTGCAA | TGGTATCCTCTGTGAAGTCTCCTCTC |
| 11910 | Atf3 | activating transcription factor 3 | AGTCACCAAGTCTGAGGCGG | CTCCAGTTTCTCTGACTCTTTCTGC |
| 18787 | Serpine1 | serine peptidase inhibitor, clade 3, member 1 | TGGGATTCAAAGTCAATGAGAAGG | GCATCCGCAGTACTGATCTCATTC |
| 15116 | Has1 | hyaluronan synthase 1 | GTGGACTACGTGCAGGTCTGTG | CTCAAGAAGCTGACCCAGGAGT |
| 16878 | Lif | leukemia inhibitory factor 3 | TAATGAAGGTCTTGGCCGCAG | TGATCTGGTTCATGAGGTTGCC |
| 193740 | Hspa1a | heat shock protein 1a (Hsp70) | ATCGAGGAGGTGGATTAGAGGC | ACCTTGACAGTAATCGGTGCCC |
| 15525 | Hspa4 | heat shock protein 4 | CTGTGTCATCTCAGTCCCATCCTT | GCAACAGCCGTCATGTCATTC |
| 11838 | Arc | activity regulated cytoskeletal-associated protein | GCTGAAGGTGAAGACAAGCCAG | CTGCTCAAGCTGCAGAGGCTAA |
| 17384 | Mmp10 | matrix metallopeptidase 10 | CCCACATCACCACAGGATTGTG | CCTTCAGAGATCCTGGAGAAAGTG |
| 17082 | Il1rl1 | interleukin 1 receptor-like 1 | CTTGTGTTATCAGAAGCCCCAACT | CCAAGCTGCAATATCCCTGATTA |
| – | pGL3a | firefly luciferase | GGATTACCAGGGATTTCAGTCGAT | GGTAGATGAGATGTGACGAACGTGT |
Promega luciferase reporter vector
Entrez Gene: gene-centered information at NCBI version v.33, official Gene ID, symbol, and name
Results
The first step in identifying differentially regulated genes following laser thermal stress with a CO2 laser was to image luciferase expression patterns in irradiated hsp70-L2G transgenic mice after stress induction. BLI identified the zones of differential hsp expression and guided tissue selection for microarray analysis. Previous studies that used a 1-s laser pulse duration had shown large differences in the stress response of tissue among the 23 different energies used. It was determined that 4.4 J/cm2 of laser energy with a 1-s pulse duration resulted in two very different regions of interest for analysis. By using a 6-mm diameter laser spot, we were able to obtain a thermally damaged region of sufficient size to provide enough RNA for analysis. The images reveal that there is limited expression of hsp70-L2G at the laser spot, but expression levels in the surrounding region were high [Fig. 1b]. The signal intensity in the region surrounding the laser spot showed a significant increase in the bioluminescent signal and clearly demarcated the normal and laser treated regions; the region surrounding the laser spot can be seen as a ring of increased luciferase activity. These two regions and an untreated nearby control region were collected for comparison.
BLI revealed three zones of differential hsp expression and served as a guide for tissue selection. The laser treatment was repeated with wildtype mice and the tissues were collected for each of the zones and analyzed for gene expression patterns. Mice were treated with one 6-mm flat-top laser spot at the center of the shaved dorsum, and allowed to recover for seven hours. BLI indicated that luciferase expression from the hsp70 promoter peaked at seven hours. Data analysis showed distinctively different results between the two regions, the laser spot and the adjacent laser region, in comparison to untreated tissue.
Analysis of raw expression data from 14000 mouse genes identified a 4-fold induction of 596 genes for the laser versus untreated analysis compared with 69 genes for the adjacent laser region versus untreated. When comparing these regions for genes with a 10-fold change, 23 genes were found for the laser versus normal analysis compared to 13 genes for the adjacent laser region versus normal tissue.
The list of 4-fold, or greater, induced genes was then analyzed to classify genes according to their role in angiogenesis, apoptosis, inflammation, and stress for the two regions of interest compared to normal tissue. The number of genes for each of the four biological processes is shown in the pie chart in Fig. 1c where the central region represents the laser region and the surrounding donut represents the adjacent laser region. Figure 2 shows a cluster analysis (heat map) of both the 4- and 10-fold changes in genes for both regions of interest when compared to untreated tissue. The genes that displayed a 4-fold or greater increase are shown in yellow, orange, and red coloring at the top of the map. Down-regulated genes are indicated in blue. The adjacent region did not show any genes that were decreased in expression. The gene symbols were added for those genes that are increased 10-fold or greater for reference. In addition, labels were added, where appropriate, to show the genes that are characterized as those involved in apoptosis, inflammation, and stress.
Figure 2.
The highest expressed genes from the laser spot and the adjacent tissue are shown in a gene cluster at the 4- and 10-fold ranges. The green gene tree shows a hierarchical relationship and gene clustering. The gene symbols of the 10-fold up- or down-regulated genes are labeled at their location in the cluster. Each gene is labeled related to apoptosis, inflammation, and stress. No angiogenesis was seen at or above the 10-fold change.
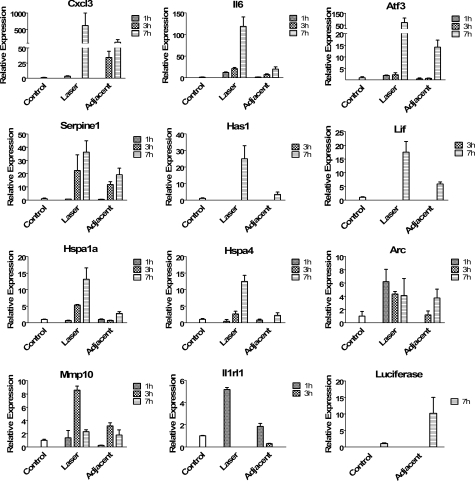
Microarray analyses are useful for assessing global changes and identifying genes of interest, and expression patterns of selected genes can then be verified using qRT-PCR. We selected eleven genes for further analysis based on either their level of up-regulation or their function. The genes that were selected for their high levels of transcriptional induction were Cxcl3, Il6, Atf3, Serpine1, Has1, Lif, Hspa1a (Hsp70), Hspa4, Arc, Mmp10, and Il1rl1. Since we were interested in the early tissue response prior to inflammatory infiltrates appearing, we performed qRT-PCR at 1- and 3-h after laser treatment, in addition to the 7-h time point (peak Hsp70-luc expression). The results of the qRT-PCR analysis are shown in Fig. 3 starting with the highest levels of increased expression in the upper left and continuing down to the lower right of the figure. In addition to the eleven genes of interest, qRT-PCR was performed on luciferase for comparison with tissues from the transgenic hsp70-L2G mouse. A minimum of three mice were used for each analysis at each of the three time points. Missing bars indicate RNA levels below detection threshold.
Figure 3.
qRT-PCR analysis was performed at 1-, 3-, and 7-h post-laser treatment of the laser spot and the adjacent tissue region compared to control for the eleven up-regulated genes of interest. In addition to the genes of interest, qRT-PCR on luciferase for the transgenic mouse was performed at seven hours as a control. Luciferase up-regulation is the highest at the 7-h time-point, so its analysis was only performed at that time for comparison. The results have been plotted in order from highest to lowest up-regulation of the genes of interest and are shown with relative expression at the three time points where available. qRT-PCR was performed for each time-point shown for each specific gene. In instances where data are not shown for a specific gene, there were no measureable expression changes for that gene (minimum of n = 3).
Discussion
The overall goal of this study was to identify genes that show differential gene expression immediately following laser stress/damage. We used the peak of hsp70-luc transcription (7-h) as a guide for early gene activation in vivo, and since induction of transcription takes at least one hour, we focused on the 1- to 7-h time period. Genes that are activated during this period have utility in assessing early tissue responses to stress, and their promoters could be used to drive reporter gene expression as indicators of tissue response to stress or to drive therapeutic gene expression in a directed fashion for use in genetic therapies. Directed therapies using thermal control of transcription could be used in a variety of applications where expression could be controlled by lasers or focused ultrasound. The promoters that most rapidly respond to thermal stress have the greatest utility and were therefore the focus of this study. Such genetic constructs could also be used to improve wound healing and survival following thermal injury. Alternatively, the products of the early response gene could be used as targets to develop small molecules that can modulate the healing response.
The thermal stress/damage of the laser spot was at a level that limited the immediate transcriptional activation of nearly all genes studied; however, by comparing the expression patterns in this region with that of surrounding regions we were able to identify genes that mark the acute response. The qRT-PCR at different time-points revealed increased expression of selected genes in the most thermally damaged laser region. The high levels of transcriptional activation for the chemokine Cxcl3 was confirmed by qRT-PCR. The microarray data showed a 24-fold increase in the laser spot and a 32-fold increase in the adjacent laser region. qRT-PCR indicated a more than 600-fold increase for the laser spot and a more than 100-fold increase for the adjacent laser region. These extremely high levels of transcriptional induction indicated that the promoter from Cxcl3 may have tremendous utility as a reporter for driving the expression of therapeutic genes. In reporter constructs, the significant dynamic range of this pattern would enable easy visualization of thermal stress. In therapeutic constructs, this promoter could be used to drive immediate and extreme levels of expression of a targeted gene. This would constitute another method of gene control in vivo and advance the field of optogenetics.24, 25
The gene with the next highest levels of activation as studied by qRT-PCR was that encoding the cytokine IL6 which showed a 119-fold increase at the laser spot and a 20-fold increase in the adjacent laser region. While Cxcl3 was the only chemokine analyzed with qRT-PCR, the microarrays indicated that there was a large number of members of this group that were up-regulated by 4-fold or more due to laser thermal stress. These included Cxcl2, Cxcl5, Cxcl13, Ccl3, Cxcl1, Ccl7, Ccl12, Ccl4, Ccl9, Ccl2, and Ccr1, and indicate an extreme and broad pro-inflammatory response comprised of a number of immune cell types. Of the 145 up-regulated genes in the laser spot, 11 were chemokines or chemokine related genes. This is not an unexpected finding since chemokines play an important role in regulating processes involved in wound healing including attracting immune cells and keratinocytes to the injury site and regulating epithelialization and angiogenesis.26, 27, 28, 29
While Hspa1a (Hsp70), the source of the promoter for our reporter construct, was up-regulated by 13.5 fold in the laser spot and 2.8 fold in the adjacent laser region, as shown by qRT-PCR, Hspa1a was only the seventh highest up-regulated gene of the ones that we had tested with qRT-PCR. Of the genes found to be elevated in our study, there are a number that could be considered for their role in prolonging cellular function after severe thermal stress. To identify these we performed a meta data analysis to identify genes associated with biological processes that are regarded as relevant for wound healing. These included angiogenesis, apoptosis, inflammation, and general stress response. Although these categories are not hard and fast and there is significant overlap and redundancy within the categories, they serve as a guide for understanding the stress response. A complete functional analysis could be conducted for selected genes in future studies.
In the category of angiogenesis, we found four genes with increased expression in stressed regions, and three genes that were down-regulated in this category, which were seen in the laser spot zone only. These results are shown in Table 2. Serpine1 is the principal inhibitor of tissue plasminogen activator (tPA) and urokinase (uPA), and is therefore an inhibitor of fibrinolysis, which is known to play multiple roles in angiogenesis.30 Serpine 1 was reported to be up-regulated by Cxcl12 stimulation.31
Table 2.
Genes determined by microarray analysis showing a four-fold or greater up-regulation or downregulation related to angiogenesis.
| Up-regulated Genes | |||||
|---|---|---|---|---|---|
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| Control | vs. Control | Gene ID | Symbold | Gene Name | |
| High | – | 5.238c | 16007 | Cyr61 | cysteine rich protein 61b |
| 4.7 | – | 13809 | Enpep | glutamyl aminopeptidase | |
| 4.4 | 4.8 | 18787 | Serpine1 | serine (or cysteine) peptidase | |
| inhibitor, clade E, member 1 | |||||
| Low | 4.4 | – | 12922 | Crhr2 | corticotropin releasing |
| hormone receptor 2 | |||||
| Down-regulated Genes | |||||
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| |
Control |
vs. Control |
Gene ID |
Symbol |
Gene Name |
| High | 0.12 | – | 20356 | Sema5a | sema domain, seven thrombospondin |
| repeats (type 1 and type 1-like), | |||||
| transmembrane domain (TM) | |||||
| and short cytoplasmic | |||||
| domain, (semaphorin) 5A | |||||
| 0.24 | – | 14183 | Fgfr2 | fibroblast growth factor receptor 2b | |
| Low | 0.25 | – | 19211 | Pten | phosphatase and tensin homolog |
duplicate Gene ID, different gene chip probe
less than a 4-fold difference from a different probe for the same gene at another location on the gene chip
bold type indicates the fold change that is used to designate the rank on the table
Entrez Gene: gene-centered information at NCBI version v.33, official Gene ID, symbol, and name
We also analyzed the category of apoptosis-associated genes, and determined that a total of six genes were up-regulated and nine genes were down-regulated in the laser spot. These results are shown in Table 3. In the apoptosis category, Il6 was analyzed by qRT-PCR and shown to be the second highest up-regulated gene in the set affected by thermal stress. The cytokine Il6 is known to be primarily produced at sites of acute and chronic inflammation, where it is secreted into the serum and induces a transcriptional inflammatory response through interleukin 6 receptor, alpha. Specifically, Il6 induces differentiation of Th17 effector T cells. Il6 activity has also been implicated in keloid formation.32
Table 3.
Genes determined by microarray analysis showing a four-fold or greater up-regulation or downregulation related to apoptosis.
| Up-regulated Genes | |||||
|---|---|---|---|---|---|
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| Control | vs. Control | Gene ID | Symbold | Gene Name | |
| High | 6.5 | 13.7c | 16193 | Il6 | interleukin 6 |
| 6.3 | – | 11801 | Cd5l | CD5 antigen-like | |
| 5.1 | 5.8 | 20750 | Spp1 | secreted phosphoprotein 1 | |
| 4.6 | – | 15951 | Ifi204 | interferon activated gene 204 | |
| 4.4 | 5.8 | 20296 | Ccl2 | chemokine (C-C motif) ligand 2 | |
| Low | 4.1 | – | 110876 | Scn2a1 | sodium channel, voltage-gated, type II, alpha 1 |
| Up-regulated Genes | |||||
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| |
Control |
vs. Control |
Gene ID |
Symbol |
Gene Name |
| High | 0.07 | – | 11891 | Rab27a | RAB27A, member RAS oncogene family |
| 0.11 | – | 56637 | Gsk3b | glycogen synthase kinase 3 beta | |
| 0.12 | – | 19116 | Prlr | prolactin receptor | |
| 0.15 | – | 12421 | Rb1cc1 | RB1-inducible coiled-coil 1 | |
| 0.18 | – | 19116 | Prlr | prolactin receptora | |
| 0.18 | – | 12683 | Cidea | cell death-inducing DNA fragmentation factor, | |
| alpha subunit-like effector A | |||||
| 0.19 | – | 19291 | Purb | purine rich element binding protein B | |
| 0.19 | – | 19877 | Rock1 | Rho-associated coiled-coil containing protein kinase 1 | |
| 0.21 | – | 11920 | Atm | ataxia telangiectasia mutated homolog (human) | |
| 0.21 | – | 12236 | Bub1b | budding uninhibited by benzimidazoles | |
| 1 homolog, beta (S. cerevisiae)b | |||||
| 0.22 | – | 217169 | Tns4 | tensin 4 | |
| 0.22 | – | 674070 | LOC674070 | similar to Ig heavy chain V region 1B43 precursor | |
| 0.22 | – | 20682 | Sox9 | SRY-box containing gene 9 | |
| 0.22 | – | 12189 | Brca1 | breast cancer 1 | |
| 0.22 | – | 12365 | Casp14 | caspase 14 | |
| 0.24 | – | 11799 | Birc5 | baculoviral IAP repeat-containing 5 | |
| 0.24 | – | 19411 | Rarg | retinoic acid receptor, gamma | |
| 0.24 | – | 227541 | Camk1d | calcium∕calmodulin-dependent protein kinase | |
| Low | 0.25 | – | 19211 | Pten | phosphatase and tensin homolog |
duplicate Gene ID, different gene chip probe
less than a 4-fold difference from a different probe for the same gene at another location on the gene chip
bold type indicates the fold change that is used to designate the rank on the table
Entrez Gene: gene-centered information at NCBI version v.33, official Gene ID, symbol, and name
In the inflammation category, we identified 25 genes with a 4-fold increase in the thermal stress areas and eight genes that showed a 4-fold, or greater, decrease in expression. These results are shown in Table 4.
Table 4.
Genes determined by microarray analysis showing a four-fold or greater up-regulation or downregulation related to inflammation.
| Up-regulated Genes | |||||
|---|---|---|---|---|---|
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| Control | vs. Control | Gene ID | Symbold | Gene Name | |
| High | 30.6 | 49.86c | 20310 | Cxcl2 | chemokine (C-X-C motif) ligand 2 |
| 28.7 | 43.2 | 20311 | Cxcl5 | chemokine (C-X-C motif) ligand 5 | |
| 6.8 | 15.5 | 14825 | Cxcl1 | chemokine (C-X-C motif) ligand 1b | |
| – | 10.5 | 16176 | Il1b | interleukin 1 beta | |
| 4.2 | 9.9 | 56644 | Clec7a | C-type lectin domain family 7, member a | |
| – | 9.9 | 20202 | S100a9 | S100 calcium binding protein A9 (calgranulin B) | |
| 8.2 | – | 55985 | Cxcl13 | chemokine (C-X-C motif) ligand 13b | |
| 8.0 | – | 20302 | Ccl3 | chemokine (C-C motif) ligand 3 | |
| 4.3 | 7.4 | 12475 | Cd14 | CD14 antigen | |
| – | 7.2 | 20344 | Selp | selectin, plateleta b, b | |
| – | 6.7 | 12655 | Chi3l3 | chitinase 3-like 3 | |
| – | 6.1 | 12986 | Csf3r | colony stimulating factor 3 receptor (granulocyte) | |
| 4.2 | 5.9 | 14825 | Cxcl1 | chemokine (C-X-C motif) ligand 1a b, b | |
| 5.1 | 5.8 | 20750 | Spp1 | secreted phosphoprotein 1 | |
| 4.4 | 5.8 | 20296 | Ccl2 | chemokine (C-C motif) ligand 2 | |
| 4.3 | 5.7 | 12768 | Ccr1 | chemokine (C-C motif) receptor 1b | |
| 5.7 | – | 12167 | Bmpr1b | bone morphogenetic protein receptor, type 1B | |
| 4.3 | – | 16644 | Kng1 | kininogen 1 | |
| 4.9 | – | 20306 | Ccl7 | chemokine (C-C motif) ligand 7 | |
| 4.8 | – | 20293 | Ccl12 | chemokine (C-C motif) ligand 12 | |
| 4.6 | – | 21897 | Tlr1 | toll-like receptor 1 | |
| 4.6 | – | 20303 | Ccl4 | chemokine (C-C motif) ligand 4 | |
| 4.6 | 15945 | Cxcl10 | chemokine (C-X-C motif) ligand 10 | ||
| 4.3 | – | 12266 | C3 | complement component 3 | |
| Low | 4.2 | – | 79221 | Hdac9 | histone deacetylase 9 |
| Down-regulated Genes | |||||
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| |
Control |
vs. Control |
Gene ID |
Symbol |
Gene Name |
| High | 0.13 | – | 17840 | Mup1 | major urinary protein 1b |
| 0.17 | – | 18829 | Ccl21a | chemokine (C-C motif) ligand 21A | |
| 0.17 | – | 20298 | Ccl21b | chemokine (C-C motif) ligand 21B | |
| 0.17 | – | 65956 | Ccl21c | chemokine (C-C motif) ligand 21C (leucine) | |
| 0.21 | – | 19883 | Rora | RAR-related orphan receptor alphab | |
| 0.22 | – | 674070 | LOC674070 | similar to Ig heavy chain V region 1B43 precursor | |
| 0.24 | – | 54450 | Il1f5 | interleukin 1 family, member 5 (delta) | |
| Low | 0.24 | – | 20299 | Ccl22 | chemokine (C-C motif) ligand 22 |
duplicate Gene ID, different gene chip probe
less than a 4-fold difference from a different probe for the same gene at another location on the gene chip
bold type indicates the fold change that is used to designate the rank on the table
Entrez Gene: gene-centered information at NCBI version v.33, official Gene ID, symbol, and name
Last, we examined the class of genes associated with stress responses. This process, by far showed the highest number of genes that were increased or decreased. A total of 39 stress response related genes showed increased expression between the two regions of interest, and 28 genes appeared to be decreased in expression within the laser spot. These results are shown in Table 5. Within this process, Il6, Hspa1a, and Hspa4 were up-regulated and qRT-PCR confirmed these observations (Fig. 3).
Table 5.
Genes determined by microarray analysis showing a four-fold or greater up-regulation or down regulation related to stress response.
| Up-regulated Genes | |||||
|---|---|---|---|---|---|
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| Control | vs. Control | Gene ID | Symbold | Gene Name | |
| High | 30.6 | 49.86c | 20310 | Cxcl2 | chemokine (C-X-C motif) ligand 2 |
| 28.7 | 43.2 | 20311 | Cxcl5 | chemokine (C-X-C motif) ligand 5 | |
| 6.8 | 15.5 | 14825 | Cxcl1 | chemokine (C-X-C motif) ligand 1b | |
| 6.5 | 13.7 | 16193 | Il6 | interleukin 6 | |
| 11.0 | 6.0 | 20210 | Saa3 | serum amyloid A 3 | |
| – | 10.5 | 16176 | Il1b | interleukin 1 beta | |
| 10.0 | – | 20208 | Saa1 | serum amyloid A 1 | |
| * | 9.9 | 20202 | S100a9 | S100 calcium binding protein A9 (calgranulin B) | |
| 4.2 | 9.9 | 56644 | Clec7a | C-type lectin domain family 7, member a | |
| 9.4 | – | 12628 | Cfh | complement component factor hb | |
| 8.7 | 20208 | Saa1 | serum amyloid A 1 | ||
| 8.2 | – | 55985 | Cxcl13 | chemokine (C-X-C motif) ligand 13b | |
| 8.0 | – | 20302 | Ccl3 | chemokine (C-C motif) ligand 3 | |
| 4.3 | 7.4 | 12475 | Cd14 | CD14 antigen | |
| – | 7.2 | 20344 | Selp | selectin, plateletb | |
| – | 6.7 | 12655 | Chi3l3 | chitinase 3-like 3 | |
| 6.1 | – | 17857 | Mx1 | myxovirus (influenza virus) resistance 1 | |
| – | 6.1 | 12986 | Csf3r | colony stimulating factor 3 receptor (granulocyte) | |
| 4.2 | 5.9 | 14825 | Cxcl1 | chemokine (C-X-C motif) ligand 1a b, b | |
| 5.1 | 5.8 | 20750 | Spp1 | secreted phosphoprotein 1 | |
| 4.4 | 5.8 | 20296 | Ccl2 | chemokine (C-C motif) ligand 2 | |
| 4.3 | 5.7 | 12768 | Ccr1 | chemokine (C-C motif) receptor 1b | |
| 5.7 | – | 12167 | Bmpr1b | bone morphogenetic protein receptor, type 1B | |
| 5.2 | 5.2 | 193740 | Hspa1a | heat shock protein 1A | |
| 5.0 | – | 12274 | C6 | complement component 6 | |
| 4.9 | – | 12273 | C5ar1 | complement component 5a receptor 1b | |
| 4.9 | – | 20306 | Ccl7 | chemokine (C-C motif) ligand 7 | |
| 4.8 | – | 20293 | Ccl12 | chemokine (C-C motif) ligand 12 | |
| 4.3 | 4.7 | 15481 | Hspa8 | heat shock protein 8 | |
| 4.6 | – | 21897 | Tlr1 | toll-like receptor 1 | |
| 4.6 | – | 20303 | Ccl4 | chemokine (C-C motif) ligand 4 | |
| – | 4.6 | 15945 | Cxcl10 | chemokine (C-X-C motif) ligand 10 | |
| 4.6 | – | 15951 | Ifi204 | interferon activated gene 204 | |
| 4.1 | 4.5 | 15525 | Hspa4 | heat shock protein 4 | |
| 4.5 | – | 53606 | Isg15 | ISG15 ubiquitin-like modifier | |
| 4.3 | – | 16644 | Kng1 | kininogen 1 | |
| 4.3 | – | 12266 | C3 | complement component 3 | |
| 4.2 | – | 13197 | Gadd45a | growth arrest and DNA-damage-inducible 45 alpha | |
| Low | 4.2 | – | 79221 | Hdac9 | histone deacetylase 9 |
| Down-regulated Genes | |||||
| Laser vs. | Adjacent | Entrez Gene | Entrez Gene | ||
| |
Control |
vs. Control |
Gene ID |
Symbol |
Gene Name |
| High | 0.07 | – | 11891 | Rab27a | RAB27A, member RAS oncogene family |
| 0.11 | – | 56637 | Gsk3b | glycogen synthase kinase 3 beta | |
| 0.13 | – | 17840 | Mup1 | major urinary protein 1 | |
| 0.15 | – | 110067 | Tcrg | T-cell receptor gamma chain | |
| 0.16 | – | 22589 | Atrx | alpha thalassemia∕mental retardation | |
| syndrome X-linked homolog (human) | |||||
| 0.16 | – | 12443 | Ccnd1 | cyclin D1 | |
| 0.17 | – | 71514 | Sfpq | splicing factor proline∕glutamine rich | |
| (polypyrimidine tract binding protein associated) | |||||
| 0.17 | – | 18829 | Ccl21a | chemokine (C-C motif) ligand 21A | |
| 0.17 | – | 20298 | Ccl21b | chemokine (C-C motif) ligand 21B | |
| 0.17 | – | 65956 | Ccl21c | chemokine (C-C motif) ligand 21C (leucine) | |
| 0.18 | – | 30939 | Pttg1 | pituitary tumor-transforming 1 | |
| 0.18 | – | 17840 | Mup1 | major urinary protein 1a | |
| 0.18 | – | 51869 | Rif1 | Rap1 interacting factor 1 homolog (yeast) | |
| 0.18 | – | 269582 | Clspn | claspin homolog (Xenopus laevis) | |
| 0.20 | – | 20538 | Slc6a2 | solute carrier family 6 (neurotransmitter | |
| transporter, noradrenalin), member 2 | |||||
| 0.20 | – | 22589 | Atrx | alpha thalassemia∕mental retardation syndrome | |
| X-linked homolog (human)a | |||||
| 0.20 | – | 22594 | Xrcc1 | X-ray repair complementing defective | |
| repair in Chinese hamster cells 1 | |||||
| 0.21 | – | 19883 | Rora | RAR-related orphan receptor alpha | |
| 0.21 | – | 19687 | Rfc1 | replication factor C (activator 1) 1 | |
| 0.22 | – | 674070 | LOC674070 | similar to Ig heavy chain V region 1B43 precursor | |
| 0.22 | – | 12189 | Brca1 | breast cancer 1 | |
| 0.22 | – | 17174 | Masp1 | mannan-binding lectin serine peptidase 1b | |
| 0.21 | – | 11920 | Atm | ataxia telangiectasia mutated homolog (human) | |
| 0.24 | – | 54450 | Il1f5 | interleukin 1 family, member 5 (delta) | |
| 0.24 | – | 20299 | Ccl22 | chemokine (C-C motif) ligand 22 | |
| 0.24 | – | 19361 | Rad51 | RAD51 homolog (S. cerevisiae) | |
| 0.25 | – | 19883 | Rora | RAR-related orphan receptor alpha | |
| Low | 0.25 | – | 30939 | Pttg1 | pituitary tumor-transforming 1a |
duplicate Gene ID, different gene chip probe
less than a 4-fold difference from a different probe for the same gene at another location on the gene chip
bold type indicates the fold change that is used to designate the rank on the table
Entrez Gene: gene-centered information at NCBI version v.33, official Gene ID, symbol, and name
In summary, 145 genes were up-regulated by at least 4-fold in the laser spot and 69 genes were 4-fold up-regulated in the region adjacent to the laser spot. Several of these genes are associated with cytoprotection and also for tissue response to thermal stress. Chemokines, the most common up-regulated genes identified, are in the tissue response category since they are secreted and are likely not involved in cytoprotection. Comparison of the thermally damaged laser spot with the thermally stressed adjacent region revealed differences in gene expression over time. The differential expression in these two regions suggests that there are genes that cannot be activated in regions of high thermal stress but are activated in adjacent regions. This may indicate that transcriptional regulation is differentially affected by heat or that there are factors that act as thermometers and produce a graded response depending on the temperature and duration of heating. The fact that this severe damage at this site did not compromise generalized transcriptional machinery, since selected genes were increased, supports the idea of a graded response corresponding to the extent of stress. A full complement of stress response genes was observed in this region at later time points indicating that they may be essential for cell survival and repair, and may be due to an influx of cells from less damaged regions.
In addition to revealing features of the acute response to thermal injury and possible pathways of tissue repair, the genes in these data sets provide possible regulatory regions for use in creating reporter genes with improved or differential responses to cellular stress and tissue damage. These may be useful in the creation of next generation transgenic reporter mice that would reveal damage to tissues during surgery or other injuries and serve as indicators of the healing response. Thermal control of gene expression can be accomplished by placing promoters from these thermally regulated genes in front of therapeutic genes thus creating genetic constructs whose expression can be controlled from a distance using heat deposition from lasers, focused ultrasound, or other tools. Moreover, these genes and their encoded proteins could be used as therapeutic targets for improving wound healing and repairing burned tissues.
Acknowledgments
Thanks to Elizabeth T. Zuo from the Stanford Protein and Nucleic Acid Facility for performing the microarray processing. This research was funded by a grant from the U.S. Department of Defense, the Medical Free Electron Laser Program administered by the Air Force Office of Sponsored Research, Grant No. FA9550–04-1–0045.
References
- Dasu M. R. K., Barrow R. E., and Herndon D. N., “Gene profiling in muscle of severely burned children: Age- and sex-dependent changes,” J. Surg. Res. 123, 144–152 (2005). 10.1016/j.jss.2004.07.248 [DOI] [PubMed] [Google Scholar]
- Sonna L. A., Gaffin S. L., Pratt R. E., Cullivan M. L., Angel K. C., and Lilly C. M., “Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells,'' J. Appl. Physiol. 92, 2208–2220 (2002). 10.1152/japplphysiol.01002.2001 [DOI] [PubMed] [Google Scholar]
- Hang H. Y., He L. S., and Fox M. H., “Cell-cycle variation of Hsp70 levels in Hela-cells at 37-degrees-C and after a heat-shock,” J. Cell. Physiol. 165, 367–375 (1995). 10.1002/jcp.1041650218 [DOI] [PubMed] [Google Scholar]
- Jerome V., Vourch C., Baulieu E. E., and Catelli M. G., “Cell-cycle regulation of the chicken Hsp90-alpha expression,” Exp. Cell Res. 205, 44–51 (1993). 10.1006/excr.1993.1056 [DOI] [PubMed] [Google Scholar]
- Milarski K. L. and Morimoto R. I., “Expression of human Hsp70 during the synthetic phase of the cell-cycle,” Proc. Natl. Acad. Sci. U.S.A. 83, 9517–9521 (1986). 10.1073/pnas.83.24.9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diller K. R., “Stress protein expression kinetics,” Annu. Rev. Biomed. Eng. 8, 403–424 (2006). 10.1146/annurev.bioeng.7.060804.100449 [DOI] [PubMed] [Google Scholar]
- Assefa Z., Garmyn M., Bouillon R., Merlevede W., Vandenheede J. R., and Agostinis P., “Differential stimulation of ERK and JNK activities by ultraviolet B irradiation and epidermal growth factor in human keratinocytes,” J. Invest. Dermatol. 108, 886–891 (1997). 10.1111/1523-1747.ep12292595 [DOI] [PubMed] [Google Scholar]
- S,Beissert and Schwarz T., “Mechanisms involved in ultraviolet light-induced immunosuppression,” J. Investig. Dermatol. Symp. Proc. 4, 61–64 (1999). [DOI] [PubMed] [Google Scholar]
- Herrlich P., Sachsenmaier C., Radlerpohl A., Gebel S., Blattner C., and Rahmsdorf H. J., “The mammalian uv response – mechanism of DNA-damage induced gene-expression,” J. Adv. Enzyme Regul. 34, 381–395 (1994). 10.1016/0065-2571(94)90024-8 [DOI] [PubMed] [Google Scholar]
- Ullrich A. and Schlessinger J., “Signal transduction by receptors with tyrosine kinase-activity,” Cell 61, 203–212 (1990). 10.1016/0092-8674(90)90801-K [DOI] [PubMed] [Google Scholar]
- Zhuang L. H., Wang B. H., and Sauder D. N., “Molecular mechanism of ultraviolet-induced keratinocyte apoptosis,” J. Interferon Cytokine Res. 20, 445–454 (2000). 10.1089/10799900050023852 [DOI] [PubMed] [Google Scholar]
- Rylander M. N., Diller K. R., Wang S. H., and Aggarwal S. J., “Correlation of HSP70 expression and cell viability following thermal stimulation of bovine aortic endothelial cells,” ASME J. Biomech. Eng. 127, 751–757 (2005). 10.1115/1.1993661 [DOI] [PubMed] [Google Scholar]
- Rylander M. N., Feng Y., Bass J., and Diller K. R., “Thermally induced injury and heat-shock protein expression in cells and tissues,” Ann. N.Y. Acad. Sci. 1066, 222–242 (2005). 10.1196/annals.1363.009 [DOI] [PubMed] [Google Scholar]
- Dinh H. K. B., Zhao B. T., Schuschereba S. T., Merrill G., and Bowman P. D., “Gene expression profiling of the response to thermal injury in human cells,” Physiol. Genomics 7, 3–13 (2001). [DOI] [PubMed] [Google Scholar]
- Beckham J. T., Wilmink G. J., Opalenik S. R., Mackanos M. A., Abraham A. A., Takahashi K., Contag C. H., Takahashi T., and Jansen E. D., “Microarray analysis of cellular thermotolerance,” Lasers Surg. Med. 42, 752–765 (2010). 10.1002/lsm.20983 [DOI] [PubMed] [Google Scholar]
- Wilmink G. J., Roth C. L., Ibey B. L., Ketchum N., Bernhard J., Cerna C. Z., and Roach W. P., “Identification of microRNAs associated with hyperthermia-induced cellular stress response,” Cell Stress Chaperones 15, 1027–1038 (2010). 10.1007/s12192-010-0189-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N. J. and Davidson J. M., “Migration inhibitory factor-related protein (MRP)8 and MRP14 are differentially expressed in free-electron laser and scalpel incisions,” Wound Repair Regen. 12, 327–336 (2004). 10.1111/j.1067-1927.2004.012313.x [DOI] [PubMed] [Google Scholar]
- Enk C. D., Shahar I., Amariglio N., Rechavi G., Kaminski N., and Hochberg M., “Gene expression profiling of in vivo UVB-irradiated human epidermis,” Photodermatol. Photoimmunol. Photomed. 20, 129–137 (2004). 10.1111/j.1600-0781.2004.00097.x [DOI] [PubMed] [Google Scholar]
- Sonna L. A., Fujita J., Gaffin S. L., and Lilly C. M., “Invited review: effects of heat and cold stress on mammalian gene expression,” J. Appl. Physiol. 92, 1725–1742 (2002). 10.1152/japplphysiol.01143.2001 [DOI] [PubMed] [Google Scholar]
- Sonna L. A., Wenger C. B., Flinn S., Sheldon H. K., Sawka M. N., and Lilly C. M., “Exertional heat injury and gene expression changes: a DNA microarray analysis study,'' J. Appl. Physiol. 96, 1943–1953 (2004). 10.1152/japplphysiol.00886.2003 [DOI] [PubMed] [Google Scholar]
- Bowman P. D., Schuschereba S. T., Lawlor D. F., Gilligan G. R., Mata J. R., and DeBaere D. R., “Survival of human epidermal keratinocytes after short-duration high temperature: synthesis of HSP70 and IL-8,” Am. J. Physiol.: Cell Physiol. 272, C1988–C1994 (1997). [DOI] [PubMed] [Google Scholar]
- O’Connell-Rodwell C. E., Mackanos M. A., Simanovskii D., Cao Y. A., Bachmann M. H., Schwettman H. A., and Contag C. H., “In vivo analysis of heat-shock-protein-70 induction following pulsed laser iradiation in a transgenic reporter mouse,” J. Biomed. Opt. 13, 030501 (2008). 10.1117/1.2904665 [DOI] [PubMed] [Google Scholar]
- Mackanos M. A., and Contag C. H., “In vivo analysis of hsp70 induction following pulse duration dependent laser irradiation in a transgenic reporter mouse,” (submitted).
- Deisseroth K., “Optogenetics,'' Nat. Methods 8, 26–29 (2011). 10.1038/nmeth.f.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V., Zhang F., Ramakrishnan C., Mattis J., Prakash R., Diester I., Goshen I., Thompson K. R., and Deisseroth K., “Molecular and cellular approaches for diversifying and extending optogenetics,” Cell 141, 154–165 (2010). 10.1016/j.cell.2010.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose R. and Werner S., “Wound-healing studies in transgenic and knockout mice,” Mol. Biotechnol. 28, 147–166 (2004). 10.1385/MB:28:2:147 [DOI] [PubMed] [Google Scholar]
- Korn T., Oukka M., Kuchroo V., and Bettelli E., “Th17 cells: effector T cells with inflammatory properties,” Semin Immunol. 19, 362–371 (2007). 10.1016/j.smim.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivamani R. K., Garcia M. S., and Isseroff R. R., “Wound re-epithelialization: modulating keratinocyte migration in wound healing,” Front. Biosci. 12, 2249–2268 (2007). 10.2741/2277 [DOI] [PubMed] [Google Scholar]
- Romagnani P., Lasagni L., Annunziato F., Serio M., and Romagnani S., “CXC chemokines: the regulatory link between inflammation and angiogenesis,” Mod. Trends Immunol. 25, 201–209 (2004). 10.1016/j.it.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Balsara R. D. and Ploplis V. A., “Plasminogen activator inhibitor-1: the double-edged sword in apoptosis,” Thromb. Haemostasis 100, 1029–1036 (2008). 10.1160/TH08-07-042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. W., Olman M., and Benveniste E. N., “CXCL12-mediated induction of plasminogen activator inhibitor-1 expression in human CXCR4 positive astroglioma cells,” Biol. Pharm. Bull. 32, 573–577 (2009). 10.1248/bpb.32.573 [DOI] [PubMed] [Google Scholar]
- Ghazizadeh M., Tosa M., Shimizu H., Hyakusoku H., and Kawanami O., “Functional implications of the Il6 signaling pathway in keloid pathogenesis,” J. Invest. Dermatol. 127, 98–105 (2007). 10.1038/sj.jid.5700564 [DOI] [PubMed] [Google Scholar]