Abstract
HOXA10 encodes a transcription factor required for endometrial receptivity and embryo implantation. The objective of this study was to identify and to characterize those molecular markers regulated by HOXA10 expression. The authors have identified putative HOXA10 target genes identified by microarray analysis employing a murine model of transient HOXA10 expression during the anticipated implantation window. Microarray analysis identified 40 statistically significant genes regulated by HOXA10 overexpression of which 31 genes were downregulated greater than 2-fold over control and 9 genes were upregulated. Cellular ontogenies of differentially expressed genes include cell adhesion molecules, signal transduction factors, and metabolic regulators. Semiquantitative real-time reverse transcriptase polymerase chain reaction confirmed regulation of selected candidate genes. Examples included clusterin (Clu), phoshoglycerate 3-dehydrogenase (3-Pgdh), and tumor-associated calcium signal transducer 2 (Tacstd2). Elucidation of these pathways will allow further characterization of the molecular mechanisms governing endometrial development, which also may function to enhance uterine receptivity.
Keywords: Implantation, HOXA10, 3-phosphoglycerate dehydrogenase, microarray, endometrium
The ability of the blastocyst to implant into a receptive endometrium governs reproductive success. Acute changes within this environment are driven by molecular events essential to the implantation process. Thus, the cascade of signaling events that occur in both fetal and maternal tissues at the time of implantation establishes an appropriate milieu critical to the development and survival of the fetus. Defects in the formation of this network and the inability to sustain this cross talk are believed to result in various pregnancy-associated complications that may manifest throughout the pregnancy.1,2
The homeobox (HOX) genes encode transcription factors that guide embryologic development as well as regulate differential gene expression within the endometrium with each menstrual cycle.3 These regulators, first associated with directing axial patterning4 through activation and repression of downstream targets, since have been demonstrated to be conserved across species and are essential to metazoan existence.5–7 Molecular analysis of HOX gene expression in the reproductive system of a murine model as well as in human cell lines demonstrates that HOX genes are essential to proper development.8 Specifically, HOXA10 gene product is expressed by endometrial glands and stroma throughout the menstrual cycle,9,10 and its level increases maximally during the midsecretory phase at the time of implantation11,12 and remains elevated throughout this phase.
Although there are no known human mutations in HOXA10, women with decreased secretory phase HOXA10 expression have lower implantation rates.13 Animal models generated to harbor selected gene knockouts demonstrate infertility phenotypes and continue to provide insight into the potential biomarkers that are involved in regulating the molecular implantation window. In particular, targeted disruption of the HOXA10 gene in mice results in a transformation of the upper uterine segment into an oviduct-like structure and inhibits implantation, even when embryos are transferred to the grossly unaffected lower uterine segment.13,14 HOXA10-null mice produce normal numbers of embryos that are able to implant in wild-type surrogate mice, whereas wild-type embryos from the surrogate mice cannot implant in the HOXA10-deficient mice.13,15,16
Selective alteration of endometrial HOXA10 expression in mice through the use of liposome-mediated gene transfection dramatically alters implantation.17 Although similar studies have not yet been performed in higher animal models or humans, transfection of a human endometrial adenocarcinoma cell line (Ishikawa cells) with a HOXA10 antisense oligodeoxyribonucleotide also resulted in decreased HOXA10 expression.17 Furthermore, efficient transfection and expression of an Escherichia coli lacZ reporter gene has been accomplished in intact human uteri ex vivo.18,19 Thus, differential maternal expression of endometrial HOXA10 is essential for implantation as this transcription factor regulates the molecular switches that promote the appropriate endometrial receptivity.
Because of the complexity of this system, it is not surprising that HOXA10 interacts with multiple targets, which in conjunction exert signal specificity and are functionally necessary. The purpose of this study was to establish and to define molecular profiles of those downstream targets of HOXA10 essential to the implantation process. Using complimentary DNA (cDNA) microarray technology,we have been able to identify candidate genes differentially expressed in a mouse implantation model where HOXA10 is transiently overexpressed. Identification of these downstream targets may uncover novel mechanisms and signaling cascades that are essential to implantation efficiency.
MATERIALS AND METHODS
Generation of Model System
Plasmid constructs and DNA/liposome preparation
Human HOXA10 cDNA (generous gift of C. Largman, Research 151, Martinez, California) cloned into the pcDNA3.1(+) vector (6.4 kb; Invitrogen, Carlsbad, CA) and the pcDNA3.1(+) vector (5.2 kb) alone have been described previously and have been demonstrated to be expressed in our murine model system.17 Concentrations and ratios of DNA/liposome were titrated previously in vitro and in vivo.17 Briefly, a final concentration of 16 µg/mL of DNA and 40 µg/mL of lipofectamine (a 3:1 [w/w] liposome formulation of the polycationic lipid [DOSPA] and the neutral lipid [DOPE] in membrane-filtered water; Invitrogen) was incubated in Opti-MEM Reduced Serum Media (Invitrogen) to a total volume of 75 µL per animal.
In vivo gene transfection
Nulliparous reproductive age CD1 female and male mice (8–12 weeks old; Charles River, Wilmington, Massachusetts) were mated and examined every 12 hours until the detection of a vaginal plug. Its presence designated day 1 of pregnancy. About 24 to 30 hours after plug detection, the animals were anesthetized with 200 to 400 µL intraperitoneal injection of a mixture of 5% xylazine/10% ketamine in accordance with approved animal care protocols. Laparotomy was performed exposing the uterus. Twenty-five microliters of complexed DNA/liposome mixture (HOXA10 plasmid or empty vector control in equivalent concentration) was injected into the base of each uterine horn using a 28-gauge U-100 insulin syringe. The peritoneum was then reapproximated in a running fashion using 4-0 synthetic, absorbable braided suture. Last, the skin was closed using an interrupted stitch of the same suture.
Procurement of specimen
Forty-eight hours after laparotomy, mice were euthanized in accordance with the Yale Animal Care and Use Committee protocol. The uterus was removed and dissected away from supportive tissues and ovaries.The uteri were minced on ice and placed in 1 mL of Trizol (Invitrogen) and stored at −80°C for total RNA extraction.
Gene Expression Profiling
Isolation of RNA
Total RNA was isolated using Trizol per manufacturer’s protocol. Purified total RNA then was subjected to RNeasy Kit purification (Qiagen, Valencia, CA).The RNA was resuspended in 50 µL of RNAase-free water, and its purity was assessed by both gel electrophoresis and spectrophotometry (A260/A280). All samples demonstrated ratios >1.6 and <1.9. All purified products were stored at −80°C.
Microarray and statistical analysis
Genechip Mouse Expression Set 430 2.0 Array (Affymetrix, Santa Clara, CA) containing more than 39000 transcripts was used as the platform. Data accrued in the microarray experiments (.CEL files) were processed with GeneSpring data analysis software (Agilent Technologies, Santa Clara, CA) to generate a list of genes that demonstrate fold changes in expression that are statistically significant. Raw data containing probe intensities were normalized to background controls within each microchip data set, and the normalized data then underwent statistical analysis. A Student t test was then used to identify those genes whose expression was statistically different between the control and test groups (P < .05). Those selected genes that demonstrated greater than 1.5-fold changes when comparing the control and test arms were retained. Additionally, these genes were queried for ontologic classifications using the GOTree machine Web-based analysis from the Oak Ridge National Laboratory.20 Fold changes in test versus the control samples were calculated and statistical significance determined via Student t test. Data from this microarray series is archived through the MIAMExpress (Accession # E-MEXP-1211).
Validation of Gene Expression Data
Primer pairs for PCR
The primer pairs used in this study were synthesized by the Yale University oligonucleotide synthesis facility. For 18srRNA: forward primer, 5′-tgtggtgttgaggaaagcag; reverse primer, 5′-tcccatccttcacatccttc. For 3-Pgdh: forward primer, 5′-gaccccatctctcctga; reverse primer, 5′-gcacacctttcttgcactga. For Tacstd2: forward primer, 5′-atgtcgggcaatgggctcac; reverse primer, 5′-caggcccaccgagtttacgc. For Clu: forward primer, 5′-agcaggaggtctctgacaatg; reverse primer, 5′-ggcttcctctaaactgttgagc.
Semiquantitative analysis
Analysis of messenger RNAs was performed by real-time reverse-transcription polymerase chain reaction (RT-PCR) using SYBER Green as an intercalator. Briefly, total RNA was quantitated spectrophotometrically and 1 µg of total RNA was used in a 20-µL reaction to generate cDNA via iScript cDNA Synthesis Kit (Biorad, Hercules, CA) employing random hexamer and oligo (dT) primers. Serial dilutions of both primers and substrate were performed to maximize primary efficiency and optimal template dilution. Real-time PCR (MyiQ; Biorad) was performed using the IQ SYBER Green Supermix (Biorad). Each reaction was performed in triplicate. Paired primers (100 nM) were used per 25-µL reaction well. The real-time synthesis protocol consisted of a first cycle 95°C denaturation (10 minutes) followed by 40 cycles at 95°C (30 seconds), annealing at 60°C (45 seconds), and extension at 72°C (45 seconds). A 2% agarose gel was used to confirm expected PCR products. Quantitation was assessed through comparing the number of cycles needed for the fluorescence of PCR products to reach a predefined threshold value. All data were normalized to the nascent expression of the 18s ribosomal RNA subunit through primer amplification. Fold changes in the test arm relative to the control arm were determined21 and a Student t test was used to identify those statistically significant genes.
RESULTS
To identify candidate genes, we employed our mouse model that allows for transient expression of select gene products during the periimplantation period.17 Two animals were used per test group. Endometrium was transiently transfected with the HOXA10 plasmid or the empty vector as a control. High-quality RNA was obtained from each sample. Of the 39 000 representative genes and transcripts available in the microarray slide, 31 unique transcripts were consistently upregulated more than 2-fold. Conversely, 9 transcripts were downregulated. Many of these genes demonstrating altered expression represent commonly occurring gene products within the cell rather than unique regulatory products.
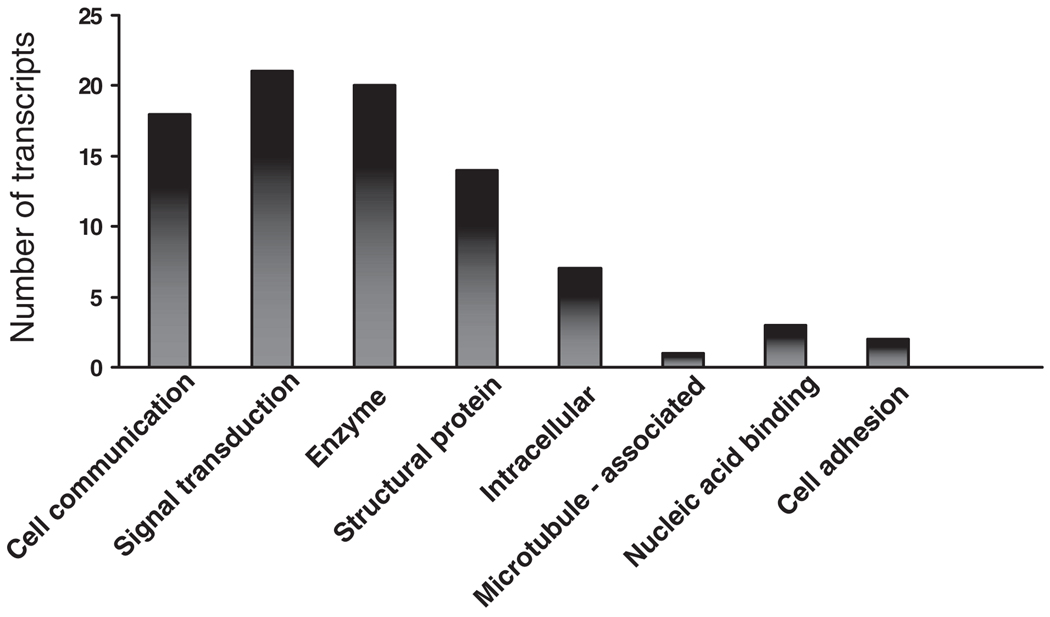
Table 1 provides an overview of genes demonstrating statistically significant differences in expression. Although multiple gene products were altered globally, those categories containing a relatively enriched number of differentially regulated genes include cell communication, signal transduction, enzymatic function, and structural components. Of note, many of these products share multiple ontologic classifications (Figure 1).
Table 1.
Analysis Summary of Statistically Significant, Differentially Expressed Genes
| Fold Change |
Accession Numbera |
Fold Change |
Accession Numbera |
||
|---|---|---|---|---|---|
| Activated leukocyte cell adhesion | −2.3 | 1437466 | Chemokine ligand 12 | 2.45 | 1419262 |
| Keratin complex 1, gene 15 | −77 | 1422667 | Peptidyl arginine deiminase I | −2.36 | 1419323 |
| Fibroblast growth factor | 2.48 | 1418497 | Ig domain-Sema | −2.6 | 1419717 |
| Kit oncogene | 2.65 | 1452514 | Alcohol dehydrogenase | −2.93 | 1421058 |
| Transferrin | −2.16 | 1425546 | Carboxylesterase 3 | −6.8 | 1435031 |
| Aldehyde dehydrogenase | −2.48 | 1418572 | Adenylate kinase 3 | 2.77 | 1440866 |
| Tryptophan 2,3-dioxygenase | 8 | 1419093 | Keratin complex 2 | −3.2 | 1422784 |
| Branched chain aminotransferase | −2 | 1430111 | Tumor-associated Ca2+ signal transducer | −4.4 | 1423323 |
| Flavin-containing monooxygenase | −2 | 1435459 | Glycine decarboxylase | −3 | 1416049 |
| Fatty-acid coenzyme A | −2 | 1451828 | Diacylglycerol O-acyltransferase | −5.49 | 1419504 |
| Topoisomerase | 2 | 1454694 | UDP N-acetyl-galactosamine | −2.2 | 1434399 |
| Fatty-acid binding protein 5 | −2.4 | 1416022 | Clusterin | −2.02 | 1437458 |
| 3-Phosphoglycerate dehydrogenase | −2.66 | 1454714 | Pbx/knotted 1 homeobox 2 | 2.54 | 1433907 |
| RAB17, RAS oncogene | −2.53 | 1422178 | Basic transcription element binding protein | 2.09 | 1436763 |
| Ca2+ channel | −2.5 | 1422178 | Fibroblast growth factor 13 | 2.48 | 1418497 |
| GABA receptor | −5 | 1424647 | Lipocalin | −3.52 | 1427747 |
| Aquaporin | −3.4 | 1425382 | Flavin-containing monooxygenase 2 | −2 | 1435371 |
| Endothelin receptor type A | 2.3 | 145169 | Fatty-acid binding protein | −2.4 | 1416022 |
| Glycine decarboxylase | −3.07 | 1416049 | Ras homolog gene family | −4.4 | 142976 |
| Chemokine ligand 6 | −2.26 | 1417266 | Lymphocyte Ag complex 6 | −5 | 1416930 |
The accession number is a unique identifier given to each gene product represented on the Affymetrix genechip.
Figure 1.
Cellular ontogeny. Ontology of the gene products that are differentially expressed during periimplantation in animals overexpressing HOXA10 when compared to sham controls. Note that one gene may have multiple ontologic classifications.
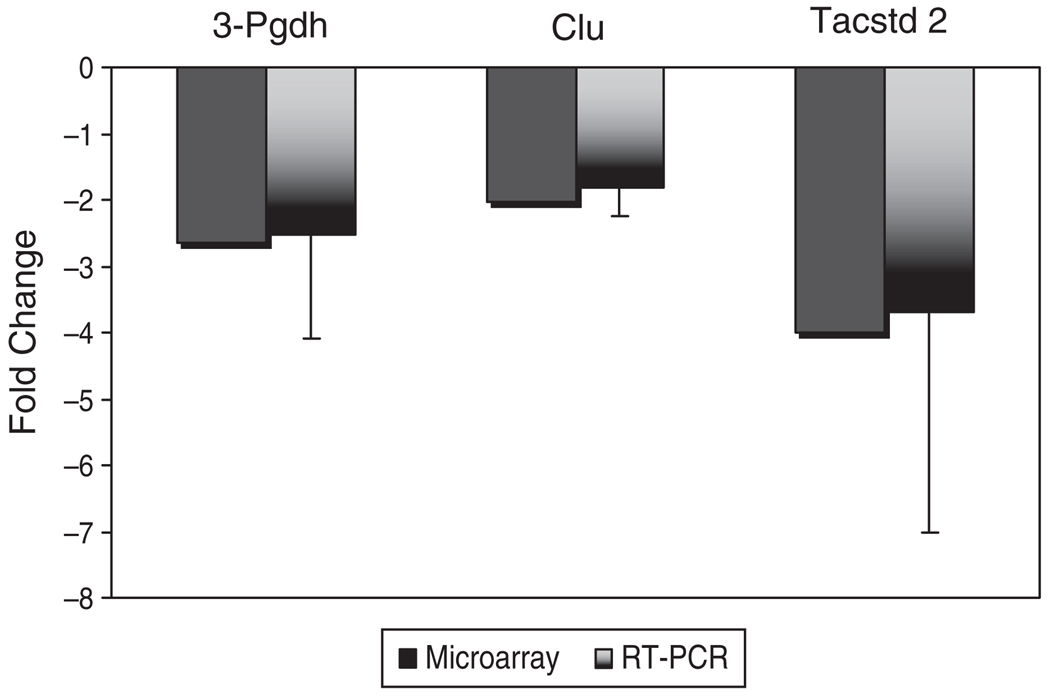
Analysis by real-time RT-PCR validated changes in gene expression obtained from microarray screening. Three genes demonstrated to be expressed differentially in the microarray experiment, which had not previously been identified in the uterus, were verified. These genes included 3-phosphoglyerate dehydrogenase (3-Pgdh), clusterin (Clu), and tumor-associated calcium signal transducer (Tacstd2; Figure 2).
Figure 2.
Validation of cDNA microarray screen. Represented are the fold changes noted in the microarray that correlate to those changes demonstrated by real-time PCR. 18sR RNA was used as the housekeeping control in the real-time assays.
DISCUSSION
Employing a well-established murine model, we sought to identify those genes regulated by HOXA10 that may play a decisive role in uterine receptivity. Relative overexpression of HOXA10 during the implantation window demonstrated allowed us to screen for candidate genes and then validate their differential expression by real-time RT-PCR. There was good correlation between levels of expression of 3-Pgdh, Clu, and Tacstd-2 and all were downregulated. Although many of the genes detailed in Table 1 have been characterized elsewhere,22–24 the genes defined in this study have not been validated previously.
HOXA10 is known to be upregulated by progesterone in uterine stroma15 just prior to blastocyst adhesion and its levels are sustained during decidualization.13,25 These characteristics have made HOXA10 an obvious marker for experimentation in an attempt to elucidate molecular mechanisms of implantation. The severe infertility phenotype observed in HOXA10-deficient female mice is multifactorial, suggesting that HOXA10 has pleiotropic effects and regulates multiple pathways. In addition to the structural developmental defects, local immunity, endometrial proliferation, and differentiation are all altered. These mice demonstrate polyclonal T-cell proliferation in a milieu accustomed to progesterone-mediated immunologic suppression.26 Furthermore, they are incapable of promoting and sustaining stromal cell proliferation and have altered expression of cyclin-dependent kinase.26 They are unable to appropriately regulate several molecules necessary for endometrial differentiation and receptivity.27,28
Proteomic profiling also has been used to investigate the role of HOXA10 in stromal cell activity. Employing the same HOXA10-deficient mouse strain, proteomic analysis similarly revealed altered expression of many proteins. Most notably, these data identified 3-Pgdh as a downstream target of HOXA10, although confirmatory analysis was not performed.29
We acknowledge the protean role of HOXA10 in both Mullerian morphogenesis and menstrual cyclicity. We wished to isolate our study to the periimplantation window and were concerned that genetic perturbations during fetal development in the absence of HOXA10 may influence the ability to study isolated uterine receptivity. It is for this reason that we choose to use a transient-transfection model where exogenous HOXA10 is expressed only briefly just prior to blastocyst adhesion rather than the knockout model.
Furthermore, to define the molecules essential to the implantation process, we procured tissue after transfection and just prior to the anticipated time of embryo implantation. Mice were transiently transfected on day 2 and sacrificed at the time of heightened uterine receptivity. It should be noted that the data were pooled among each test group. Those animals overexpressing HOXA10 plasmid were compared with animals harboring the empty-vector controls. Previously, this laboratory has demonstrated that there exists no detriment from the transient transfection of vector alone in this model system.17 Thus, differences between the overexpressing HOXA10 test group and the sham control group can be ascribed to the presence of the exogenous HOXA10 transcript. Variances between our results and others may be secondary to differences in the development of the uterus in the knockout model, whereas our data are generated by altered maternal HOXA10 expression in the setting of normal embryonic development.
Nonetheless, 3 distinct genes are demonstrated to be affected by levels of HOXA10 expression. It is interesting that these genes are found to be downregulated. The sex steroids may act in concert to regulate gene expression. Estradiol levels and estradiol receptor activity fall temporarily on day 2 and then surge on day 4.Transcription of these targeted genes may be estrogen sensitive and thus downregulated during this time.30 By day 3, corpus lutea–synthesized progesterone initiates stromal cell proliferation and its increasing presence may cause indirect downregulation of gene expression26 through direct HOXA10 regulation. It has been established that homeobox genes are pleotropic transcriptional modulators; HOXA10, specifically, can act as a repressor31,32 as well as an activator33 of transcription. Therefore, it is not surprising that the gene expression profiles validated through this screen demonstrate transcriptional repression of presumed downstream targets of HOXA10 when it is overexpressed.
One of the most notable genes derived through our study and validated here is 3-Pgdh. It is a key enzyme involved in l-serine biosynthesis and is crucial in regulating cellular metabolism in mammalian tissues.34,35 Differential expression of 3-Pgdh gene products in response to sex steroids and selective estrogen-receptor modulators may also be noted in breast cancer cells.36 Interestingly, the mouse 3-Pgdh gene contains a 5′ consensus Sp1 transcription factor recognition sequence and the gene itself shares 71% homology to its human counterpart.37 Data from gel-shift assay as well as cell-culture reporter assay support that 3-PGDH expression is driven by the transcription factor Sp1 in a variety of cells.37 Sp1 expression in endometrial stromal tissue is greatest in the secretory phase of the human menstrual cycle38 and is expressed in a tissue-specific and cell-specific manner. Remarkably, in endometrial culture experiments, expression of HOXA10 appears to be driven by Sp1.38 Both observations implicate that HOXA10 and Sp1 may be involved in a well-regulated mechanism of tightly controlling 3-Pgdh expression within the endometrium during the implantation window, and it is reasonable to postulate that both Sp1 and HOXA10 may regulate 3-Pgdh/3-PGDH expression.
Sex steroids drive uterine development and adult endometrial differentiation in the menstrual cycle. Much of the knowledge detailing the window of implantation is derived from embryo transfer and single-gene knockout experiments in rodents. However, little is known about the molecules that govern this process. Data presented here contribute to defining the molecular signature of uterine receptivity during early mouse implantation. Further characterization of these genes and their role in implantation may assist in elucidating the complex molecular and physiologic events that govern reproductive success.
ACKNOWLEDGMENTS
The authors would like to thank Aiping Lin at the W.M. Keck Foundation Biostatistics Resource at Yale University for expert assistance with the microarray data and Dr Hongling Du for scientific advice.
This study was supported by grants NIH R01 HD038667 and U54 HD052668.
Footnotes
REFERENCES
- 1.Aplin JD. The cell biological basis of human implantation. Best Pract Res Clin Obstet Gynaecol. 2000;14:757–764. doi: 10.1053/beog.2000.0116. [DOI] [PubMed] [Google Scholar]
- 2.Red-Horse K, Zhou Y, Genbacev O, et al. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest. 2004;114:744–754. doi: 10.1172/JCI22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor H, Vanden H, Igarashi P. A conserved HOX axis in the mouse and human reproductive system: late establishment and persistent expression of the HOXA cluster gene. Biol Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 4.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 5.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 6.McGinnis W, Levine M, Hafen E, Kuroiwa A, Gehring W. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984;37:403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- 7.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:227–256. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 8.Block K, Kardana A, Igarashi P, Taylor HS. In utero diethylstilbestrol (DES) exposure alters HOX gene expression in the developing mullerian system. FASEB J. 2000;14:1101–1108. doi: 10.1096/fasebj.14.9.1101. [DOI] [PubMed] [Google Scholar]
- 9.Taylor HS, Igarashi P, Olive D, Arici A. Sex steroids mediate HOXA11 expression in the human peri-implantation endometrium. J Clin Endocrinol Metab. 1999;84:1129–1135. doi: 10.1210/jcem.84.3.5573. [DOI] [PubMed] [Google Scholar]
- 10.Taylor HS, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J Clin Invest. 1998;101:1379–1384. doi: 10.1172/JCI1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor HS, Vandenheuvel GB, Igarshi P. A conserved Hox axis in the mouse and human female reproductive system—late establishment and persistent adult expression of the Hoxa cluster genes. Biol Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 12.Sarno JL, Kliman HJ, Taylor HS. HOXA10, Pbx2, and Meis1 protein expression in the human endometrium: formation of multimeric complexes on HOXA10 target genes. J Clin Endocrinol Metab. 2005;90:522–528. doi: 10.1210/jc.2004-0817. [DOI] [PubMed] [Google Scholar]
- 13.Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122:2687–2696. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- 14.Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14:1328–1331. doi: 10.1093/humrep/14.5.1328. [DOI] [PubMed] [Google Scholar]
- 15.Satokata I, Benson G, Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374:460–463. doi: 10.1038/374460a0. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh-Li HM, Witte DP, Weinstein M, et al. Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development. 1995;121:1373–1385. doi: 10.1242/dev.121.5.1373. [DOI] [PubMed] [Google Scholar]
- 17.Bagot CN, Troy PJ, Taylor HS. Alteration of maternal Hoxa10 expression by in vivo gene transfection affects implantation. Gene Ther. 2000;7:1378–1384. doi: 10.1038/sj.gt.3301245. [DOI] [PubMed] [Google Scholar]
- 18.Daftary GS, Taylor HS. Reproductive tract gene transfer. Fertil Steril. 2003;80:475–484. doi: 10.1016/s0015-0282(03)00970-1. [DOI] [PubMed] [Google Scholar]
- 19.Daftary GS, Taylor HS. EMX2 gene expression in the female reproductive tract and aberrant expression in the endometrium of patients with endometriosis. J Clin Endocrinol Metab. 2004;89:2390–2396. doi: 10.1210/jc.2003-031389. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of intersting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Aronow B, Richardson B, Handwerger S. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key gene function categories. Physiol Genomics. 2001;6:103–116. doi: 10.1152/physiolgenomics.2001.6.2.105. [DOI] [PubMed] [Google Scholar]
- 23.Kao LC, Tulac S, Lobo S, et al. Global gene profiling in human endometrium during the window of implantation. Endocrinology. 2002;143:2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- 24.Reece J, Das S, Paria B, et al. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J Biol Chem. 2001;276:44137–44145. doi: 10.1074/jbc.M107563200. [DOI] [PubMed] [Google Scholar]
- 25.Lim H, Ma L, Ma WG, Maas RL, Dey SK. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol Endocrinol. 1999;13:1005–1017. doi: 10.1210/mend.13.6.0284. [DOI] [PubMed] [Google Scholar]
- 26.Yao M, Lim H, Schust D, et al. Gene expression profiling reveal progesterone-mediated cell cycle and immunoregulatory roles of Hoxa-10 in the preimplantation uterus. Mol Endocrinol. 2003;17:610–627. doi: 10.1210/me.2002-0290. [DOI] [PubMed] [Google Scholar]
- 27.Daftary GS, Taylor HS. Pleiotropic effects of Hoxa10 on the functional development of peri-implantation endometrium. Mol Reprod Dev. 2004;67:8–14. doi: 10.1002/mrd.20013. [DOI] [PubMed] [Google Scholar]
- 28.Bagot CN, Kliman HJ, Taylor HS. Maternal Hoxa10 is required for pinopod formation in the development of mouse uterine receptivity to embryo implantation. Dev Dyn. 2001;222:538–544. doi: 10.1002/dvdy.1209. [DOI] [PubMed] [Google Scholar]
- 29.Daikoku T, Tranguch S, Friedman DB, Das SK, Smith DF, Dey SK. Proteomic analysis identifies immunophilin FK506 binding protein 4 (FKBP52) as a downstream target of Hoxa10 in the periimplantation mouse uterus. Mol Endocrinol. 2005;19:683–697. doi: 10.1210/me.2004-0332. [DOI] [PubMed] [Google Scholar]
- 30.Carson DD, Bagchi I, Dey SK, et al. Embryo implantation. Dev Biol. 2000;223:217–237. doi: 10.1006/dbio.2000.9767. [DOI] [PubMed] [Google Scholar]
- 31.Troy PJ, Daftary GS, Bagot CN, Taylor HS. Transcriptional repression of peri-implantation EMX2 expression in mammalian reproduction by HOXA10. Mol Cell Biol. 2003;23:1–13. doi: 10.1128/MCB.23.1.1-13.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daftary GS, Taylor HS. Endocrine regulation of HOX genes. Endocr Rev. 2004;27:331–355. doi: 10.1210/er.2005-0018. [DOI] [PubMed] [Google Scholar]
- 33.Daftary GS, Troy PJ, Bagot CN, Young SL, Taylor HS. Direct regulation of Beta3-integrin subunit gene expression by HOXA10 endometrial cells. Mol Endocrinol. 2002;16:571–579. doi: 10.1210/mend.16.3.0792. [DOI] [PubMed] [Google Scholar]
- 34.Cho HM, Jun DY, Bae MA, Ahn JD, Kim YH. Nucleotide sequence and differential expression of the human 3-phosphoglycerate dehydrogenase gene. Gene. 2000;245:193–201. doi: 10.1016/s0378-1119(00)00009-3. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida K, Furuya S, Osuka S, et al. Targeted disruption of the mouse 3-phosphoglyerate dehydrogenase gene causes severe neurodevelopmental defects and results in embryonic lethality. J Biol Chem. 2004;279:3573–3577. doi: 10.1074/jbc.C300507200. [DOI] [PubMed] [Google Scholar]
- 36.Al-Dhaheri MH, Shah YM, Basrur V, Pind S, Rowan BG. Identification of novel proteins induced by estradiol, 4-hydroxytamoxifen and acolbifene in T47D breast cancer cells. Steroids. 2006;71:966–978. doi: 10.1016/j.steroids.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Mitoma J, Furuya S, Shimizu M, et al. Mouse 3-phosphoglycerate dehydrogenase gene: genomic organization, chromosomal localization, and promoter analysis. Gene. 2004;334:15–22. doi: 10.1016/j.gene.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Martin R, Taylor MB, Krikun G, Lockwood C, Akbas GE, Taylor HS. Differential cell-specific modulation of HOXA10 by estrogen and specificity protein 1 response elements. J Clin Endocrinol Metab. 2007;92:1920–1926. doi: 10.1210/jc.2006-1694. [DOI] [PubMed] [Google Scholar]