Abstract
The serial analysis of gene expression (SAGE) method was used to generate a catalog of 53,875 short (14 base) expressed sequence tags from polyadenylated RNA obtained from vastus lateralis muscle of healthy young men. Over 12,000 unique tags were detected. The frequency of occurrence of each tag reflects the relative abundance of the corresponding mRNA. The mRNA species that were detected 10 or more times, each comprising ≥0.02% of the mRNA population, accounted for 64% of the mRNA mass but <10% of the total number of mRNA species detected. Almost all of the abundant tags matched mRNA or EST sequences cataloged in GenBank. Mitochondrial transcripts accounted for ∼20% of the polyadenylated RNA. Transcripts encoding proteins of the myofibrils were the most abundant nuclear-encoded mRNAs. Transcripts encoding ribosomal proteins, and those encoding proteins involved in energy metabolism, also were very abundant. The database can be used as a reference for investigations of alterations in gene expression associated with conditions that influence muscle function, such as muscular dystrophies, aging, and exercise.
The relative abundances of mRNA species can be estimated by determining the proportion of ESTs from a cDNA library that match the mRNA sequence (Audic and Claverie 1997). The abundances of many of the mRNAs expressed in human skeletal muscle have been cataloged according to this method in the Genexpress Index (Houlgatte et al. 1995) and by the Centro di Ricerca Interdepartmentale per la Biotecnologie Innovative (CRIBI) Biotechnology Center (Lanfranchi et al. 1996). Comparison of such catalogs obtained from normal muscle and from various patient groups could elucidate changes in muscle gene expression associated with neuromuscular diseases, myopathies, disuse, or age-related atrophy. However, cataloging enough ESTs to make meaningful comparisons of mRNA abundances between different groups requires sequencing of >104 clones. Moreover, the extent to which even minor differences in the methods used to produce and normalize cDNA libraries influence the proportional representation of different cDNA species is unclear. These problems can be ameliorated to a great extent by the serial analysis of gene expression (SAGE) method (Velculescu et al. 1995), in which each clone contains many short ESTs. This method is extremely useful for quantitating gene expression. For example, the SAGE method was used to identify transcripts differentially expressed in neoplastic cells (Zhang et al. 1997), to detect genes induced by p53 (Madden et al. 1997; Polyak et al. 1997), and to characterize the yeast transcriptome (Velculescu et al. 1997). In this paper, we present an inventory of the SAGE tags obtained from skeletal muscle (vastus lateralis) of healthy young men. This database represents the average of several individuals studied under standard conditions, and describes the mRNA population in a muscle that is frequently biopsied for molecular, biochemical, and histological studies.
RESULTS
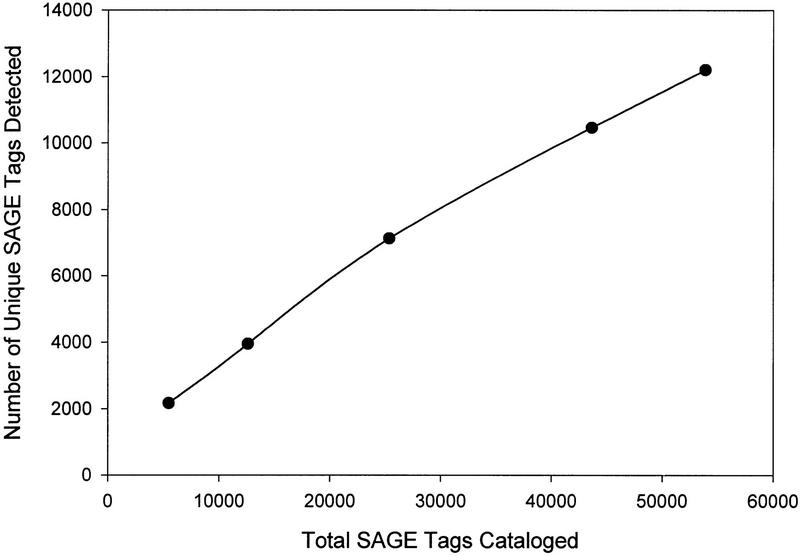
A total of 53,875 SAGE tags was cataloged, representing 12,207 unique SAGE tag species. Figure 1 shows the number of unique tags as a function of the total number cataloged. Most of the tags (8434 or 69%) were detected only once. Another 2553 species (21%) were detected two to four times, and 1220 (10%) were detected five or more times.
Figure 1.
Number of unique SAGE tags detected as a function of the total number of tags cataloged.
Table 1 shows the most abundant SAGE tags, those detected 100 or more times (those detected 50 or more times are available as an online supplement at www.genome.org). The complete list of tags and their abundances is available at the Rochester Muscle Database web site, (http://www.gcrc.rochester.edu/SWindex.html). A longer version of Table 1, including 295 tags that were detected at least 20 times, also can be viewed at this web site. The data files generated by the SAGE software, including a Microsoft Excel database showing matches of SAGE tags with GenBank primate sequences, can be downloaded (instructions at the above web site).
Table 1.
Skeletal Muscle SAGE Tags Detected 100 or More Times in a Sample of 53,875 Total Tags
| Tag (CATG + . . .) | Gene product | GenBank accession no. | Times detected |
|---|---|---|---|
| CCCATCGTCC | cytochrome c oxidase 2a | 7627–40a | 1780 |
| AAGATCAAGA | α actinb | J00068 | 1001 |
| ATCCCCGCCC | creatine kinase M | M14780 | 851 |
| TTCATACACC | NADH dehydrogenase 4/4L | 11491-504 | 847 |
| CTGGAGCCTG | type 1 (slow, cardiac β) myosin heavy chain | X05631 | 830 |
| CCCACCACCC | β-tropomyosin | X06825 | 703 |
| AGCCCTACAA | NADH dehydrogenase 3 | 9709–22 | 702 |
| TACCATCAAT | glyceraldehyde-3-phosphate dehydrogenase | M36164 | 593 |
| GCGACCGTCA | fructose 1,6-diphosphate aldolase a | M21190 | 582 |
| GTTTGGATCT | myoglobin | X00373 | 581 |
| ACCCTTGGCC | NADH dehydrogenase 1 | 3263–76 | 565 |
| GAATGACTGA | myosin heavy chain 2a | Z32858 | 563 |
| TCCTCAACCC | slow twitch troponin C | M37984 | 499 |
| TGATTTCACT | cytochrome c oxidase 3 | 8726–39 | 489 |
| ACTAACACCC | NADH dehydrogenase 2 | 4606–19 | 478 |
| CTAAGACTTC | 16 S rRNA | 2276–89 | 442 |
| TGGGCGGCCT | myosin light chain 2 | M21812 | 409 |
| CCCCGGCCAC | desmin | U59167 | 400 |
| GAGGGCCGGA | fast troponin I | L21715 | 400 |
| ATGGTGCGCC | fast skeletal troponin C | X07898 | 397 |
| AGGCACCTGG | EST similar to slow twitch skeletal muscle troponin I | N87243F17753 | 393 |
| TTCCAATAAA | cardiac myosin light chain 2 | X66141 | 388 |
| GGGGAGGAAC | slow troponin T | M19309 | 364 |
| AAAACATTCT | truncated 16 S rRNA | 1738–51 | 325 |
| AGGATCGAGG | β enolase | X51957 | 324 |
| CACCTAATTG | ATPase 6/8 | 8460–73 | 304 |
| GGAGCCAACT | EST from muscle, similar to troponin T | AA192217 | 287 |
| CAGAGGGTGG | glycogen phosphorylase | X03031 | 285 |
| TAGGTTGTCT | translationally controlled tumor protein | L13806 | 277 |
| AATCAACAAA | myosin light chain 3f | X05451c | 257 |
| CAAGCATCCC | truncated 12 S rRNA | 118–31 | 248 |
| CACTACTCAC | cytochrome b | 14326–39 | 238 |
| CAAGTATAAA | titin | X69490 | 208 |
| CCTCAGGATA | NADH dehydrogenase 5 | 13853–66 | 186 |
| GAGGCTGTGG | phosphoglycerate mutase muscle specific subunit | M18172 | 186 |
| ATTTGAGAAG | cytochrome c oxidase 1 | 6737–50 | 177 |
| GGGCTGGGGT | ribosomal protein L29 | U10248 | 175 |
| GCTTTGCCTC | cytochrome c oxidase 6a | M83308 | 169 |
| TCGAAGCCCC | NADH dehydrogense 4L | 10841–54 | 169 |
| AAGACAGTGG | ribosomal protein L37a | X66699 | 163 |
| TGGGCAAAGC | elongation factor 1γ | Z11531 | 163 |
| AAAGTCATTG | α-tropomyosin | M19713 | 146 |
| TGCCAGAAAA | telethonin | AJ000491 | 142 |
| AGCACCTCCA | elongation factor 2 | Z11692 | 131 |
| TTGGTCCTCT | ribosomal protein L41 | AF026844 | 131 |
| TAATGACAAT | skeletal muscle LIM-protein FHL1 | U60115 | 128 |
| TGAATAAAGT | sarcolipin | U96093 | 124 |
| TTCAATAAAA | acidic ribosomal phosphoprotein P1 | M17886 | 117 |
| TTTACTCAGC | skeletal muscle C-protein (slow myosin binding protein C) | X66276 | 111 |
| X73114 | |||
| CTCATAGCAG | EST similar to translationally controlled tumor protein | AA716547 | 110 |
| TTGGAGATCT | NADH dehydrogenase MLRQ subunit | U94586 | 110 |
| GTTTCAGGTA | Ca2+ ATPase (HK2) | M23115 | 107 |
| AGGGCTTCCA | Wilm’s tumor-related protein QM | M642421 | 103 |
| TCTGCACCTC | S1 (elongation factor-1 α2) | X70940 | 100 |
Complete lists of all tags, in both alphabetical order and order of abundance, are available via the internet at http://www.gcrc.rochester.edu/SWindex.html.
Italics denotes tags matching the mitochondrial genome (HSMITG, GenBank accession no. X93334), and values listed under accession number for these tags indicate the locus within the mitochondrial genome.
Also matches other actin isoforms, but α-actin is much more abundant in skeletal muscle.
Results from present study, Lanfranchi et al. (1996), and Wade et al. (1989) have A in eighth position; X05451 has G in eighth position.
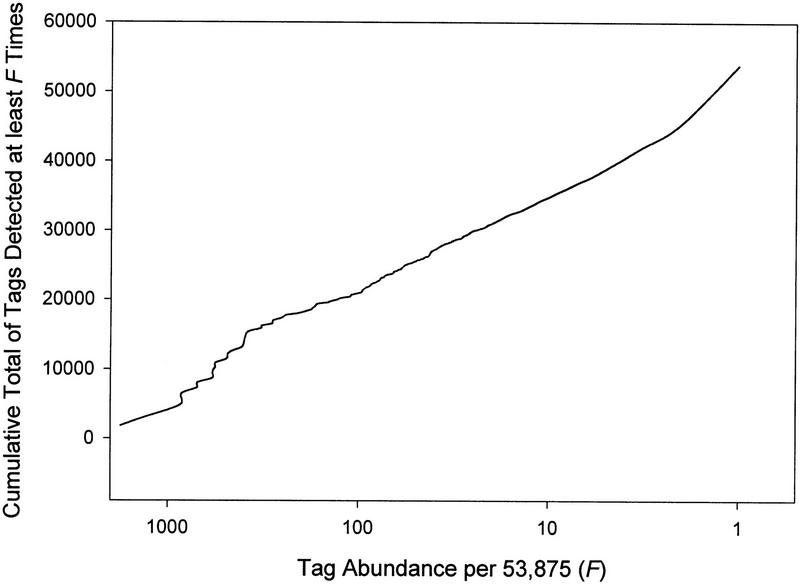
Transcripts detected 50 or more times accounted for 48% of the mRNA population, those detected 20 or more times accounted for 57%, and those detected 10 or more times accounted for 64% (Fig. 2). Transcripts of mitochondrial DNA accounted for 17% of the polyadenylated RNA population. These are minimal estimates for the contribution of the high-abundance transcripts to the total mRNA population, because exclusion of replicate ditags causes some underestimation of the most abundant transcripts (see Methods). Thus, mitochondrial transcripts probably account for 20%–25% of the mRNA population.
Figure 2.
Cumulative number of SAGE tags as a function of tag abundance, in decreasing order of abundance.
Approximately two-thirds of the 264 nonmitochondrial tags that were detected 20 or more times match sequences of mRNAs whose protein products have been identified. Almost all of the abundant tags that did not match known mRNAs did match human ESTs cataloged in GenBank.
Transcripts encoding proteins of the myofibrils were very abundant. Of the 22 nonmitochondrial tags detected >250 times, 13 matched GenBank sequences for mRNAs (or homologous ESTs) encoding myofibrillar proteins. Genes encoding proteins involved in energy metabolism also were highly expressed. In addition to the high concentration of mitochondrial transcripts, 31 of which were detected 20 or more times, 48 nonmitochondrial mRNAs that encode proteins involved in glucose metabolism or ATP production were detected at least 20 times. The mRNAs encoding ribosomal proteins were very abundant. Transcripts of 52 of the ∼80 ribosomal protein genes were detected 20 or more times.
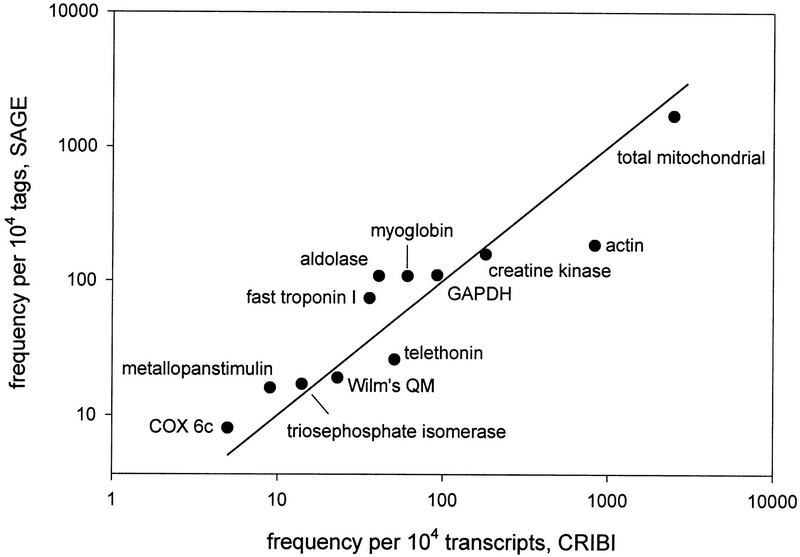
In general, the data presented here are consistent with the CRIBI database (Lanfranchi et al. 1996; Valle et al. 1997), which is the only other database with >104 ESTs from muscle and the only one for which extensive quantitative data have been reported. Both studies indicate a high proportion of mitochondrial transcripts and ribosomal protein mRNAs, a very high abundance of mRNAs encoding contractile proteins, and a high abundance of transcripts encoding several enzymes involved in glucose and energy metabolism. Figure 3 illustrates the similarity of these databases. The transcripts in this figure were chosen to be representative of different levels of expression over a wide range, and were chosen randomly so that there would be no bias toward similarity between databases. The CRIBI catalog has many α- and β-globin transcripts, indicating that a significant amount of blood was present in the tissue sample. The globin mRNAs were present at a much lower level in our sample. Other muscle cDNA libraries described in the literature (Houlgatte et al. 1995) or available via the internet (e.g., Stratagene catalog 937209 at http://www.ncbi.nlm.nih.gov/cgi-bin/UniGene) also support the present data. All catalogs agree that α-actin mRNA is the most abundant nonmitochondrial transcript, and that creatine kinase, myoglobin, β enolase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, myosin heavy chain, and titin mRNAs are among the most abundant transcripts in skeletal muscle. Quantitative information on the less abundant transcripts is not readily available from these other databases, or is not very accurate because of the relatively small number of ESTs that have been cataloged.
Figure 3.
Relation between abundances of selected transcripts in the CRIBI muscle cDNA library (Lanfranchi et al. 1996; Valle et al. 1997) and abundances of the corresponding SAGE tags in the present study. Transcripts were chosen to represent a wide range of abundances (note logarithmic scale) and were not selected according to degree of similarity between databases. The line of identity is shown.
Mitochondrial Transcripts
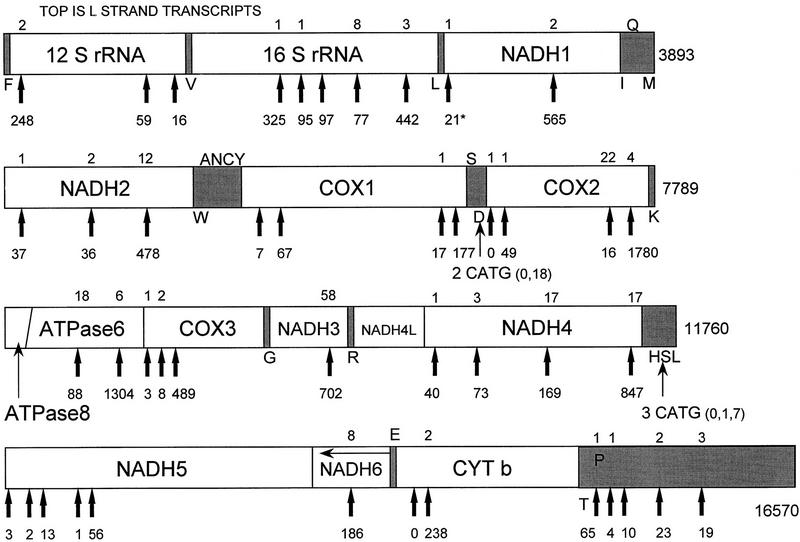
We found 75 different SAGE tags that matched the mitochondrial genome, and these accounted for 17% of the total tags in the catalog. This value is slightly less than the proportion (25%) of mitochondrial transcripts in the CRIBI library (Lanfranchi et al. 1996), but more than the proportion (9%) in the Stratagene muscle library. Of the 9180 total mitochondrial SAGE tags, 8978 (98%) correspond to H strand transcripts. Because the primary mitochondrial transcripts are polycistronic (Clayton 1984), and are sometimes prematurely terminated or spliced at unpredictable locations (Lanfranchi et al. 1996), the analysis of mitochondrial SAGE tags is not straightforward. Figure 4 is a map of the NlaIII restriction sites in the mitochondrial genome and shows the abundances of the SAGE tags corresponding to each of these sites.
Figure 4.
Map of the NlaIII restriction sites in the mitochondrial genome (HSMITG, GenBank Accession no. X93334), and abundances of the SAGE tags corresponding to each of these sites in a sample of 53,875 total tags (excluding replicate ditags). Mitochondrial DNA is circular, but is shown here as several linear pieces to facilitate presentation. Numbers at end of each segment indicate base number in HSMITG, which corresponds to the base sequence of the RNA encoded by the heavy strand (H strand). Numbers under each arrow indicate the frequency of tags corresponding to the H strand transcripts, which include both rRNAs and 12 of the 13 mRNAs (only NADH6 mRNA is encoded by L strand). Numbers above arrows indicate frequency of L strand transcripts corresponding to each site (if no number is shown, no tags matching that site were detected). The presence of a tag indicates that the transcript was polyadenylated somewhere between that NlaIII site and the next one, or between that NlaIII site and the next tRNA (tRNAs are spliced from primary transcript but not polyadenylated). The tRNA genes and control regions are shaded. Single-letter amino acid abbreviations identify location of specific tRNAs (those encoded by H strand indicated below the shaded area, those encoded by the L strand indicated above the shaded area). The NADH1 tag occurring 21 times (asterisk) matches HUMMTCG (GenBank accession no. J01415) rather than HSMITG (G rather than A at base 2740 of HSMITG). [NADH(n)]NADH dehydrogenase (subunit); [COX(n)]cytochrome c oxidase (subunit); (CYT b) cytochrome b.
About 15% of the mitochondrial tags were from rRNA. Because most of the mitochondrial rRNA is not polyadenylated (Clayton 1984), this proportion is not representative of total mitochondrial rRNA abundance. Five separate tags from the 16S rRNA gene were found. The one corresponding to the most 3′ NlaIII restriction site accounted for only 43% of the 16S rRNA tags. Tags from the three NlaIII restriction sites in the 12S rRNA gene were found, but the most 5′ site accounted for most of the tags. Thus, most of the polyadenylated rRNA is not full length.
Thirteen genes of the mitochondrial genome encode proteins, 12 of which are encoded by the H strand (Fig. 4). Most of these are separated from one another by tRNA sequences, which are spliced from the primary transcript but are not polyadenylated. The ATPase subunit 8 and the NADH dehydrogenase subunit 4L genes do not have an NlaIII restriction site. The 3′ ends of these genes overlap with the 5′ ends of the ATPase subunit 6 and NADH dehydrogenase subunit 4 genes, respectively. Separate proteins are produced by frameshifting during translation rather than splicing of the polycistronic mRNA (Clayton 1984). Thus, tags matching the ATPase 6 and NADH dehydrogenase 4 genes correspond to transcripts that can encode two proteins each. About 25% of the tags corresponding to the NADH dehydrogenase 4 gene were not from the most 3′ NlaIII restriction site, and therefore represent transcripts that can encode only NADH dehydrogenase 4L. About 6% of the tags corresponding to the ATPase 6 gene were not from the most 3′ NlaIII restriction site, and represent transcripts that can encode only ATPase 8. The cytochrome c oxidase subunit 3 gene also is contiguous with the ATPase 6 gene, but apparently the primary transcript is cleaved between these genes (Anderson et al. 1981). Nine protein-encoding genes have more than one NlaIII restriction site, and the most 3′ site accounted for most of the tags from all of these genes.
The NADH dehydrogenase subunit 6 mRNA is the only mRNA encoded by the L strand of mitochondrial DNA. It was much less abundant than the mRNAs encoded by the H strand. There were several different tags corresponding to L strand sequences >1000 bases downstream from the NADH dehydrogenase 6 gene. Because there are no tRNAs encoded by the L strand between this gene and the downstream tags, they could theoretically represent NADH6 mRNAs with long 3′ untranslated tails. However, even antisense tRNA sequences may be spliced (Anderson et al. 1981), which would eliminate this possibility.
The most abundant polyadenylated mitochondrial RNA in HeLa cells is a 7S RNA whose function is unknown (Clayton 1984). This RNA does not have an NlaIII restriction site and therefore its abundance in muscle was not assessed.
DISCUSSION
The present inventory should be an accurate reflection of the relative concentrations of the most abundant transcripts in normal adult vastus lateralis muscle. As discussed below, a few abundant transcripts are undetected, but these should account for a very low proportion of the mRNA species. Although there is some uncertainty about the precise abundance of transcripts that were detected infrequently, quantitation is theoretically quite precise for the abundant transcripts (Audic and Claverie 1997).
A limitation of the SAGE method is that any cDNA lacking the required restriction enzyme site is unevaluable. This problem can be overcome by use of different restriction enzymes, but this procedure increases the expense, time, and amount of RNA required. Short transcripts obviously have a lower chance of having a particular restriction site than longer transcripts. If sequences were entirely random, a 2-kb transcript would have a probability of >99.9% of having at least one NlaIII restriction site, a 1-kb transcript a 98% chance, a 0.5-kb transcript an 85% chance, and a 0.25-kb transcript a 62% chance. In this study we did not detect 3 of the 72 most abundant cDNAs in the CRIBI muscle library (Lanfranchi et al. 1996). Two of these are short cDNAs that lack an NlaIII restriction site (cytochrome c oxidase 7a, 341 bases; ribosomal protein S21, 343 bases). The other is an EST whose full mRNA sequence is unknown. There could be under-representation of transcripts in which the most 3′ NlaIII restriction site is far upstream of the polyadenylation site, because of premature termination of cDNA synthesis with very long transcripts.
The production of SAGE tags requires cleavage of the cDNA with a type IIS restriction enzyme (BsmFI in this study) after it is ligated to linkers containing the restriction enzyme recognition site and PCR primer sequences. If, after cleavage with NlaIII, a cDNA fragment contains the sequence GTCCC within 20 bases of the CATG that defines the start of a SAGE tag, that cDNA could be under-represented because the BsmFI digestion can shorten the tag or cleave it from the PCR primer sequence. Only ∼2% of the cDNAs would be expected to be affected by this problem.
We detected 12,207 distinct tags in this study, but not all of these represent distinct transcripts. Some tags, especially those detected only once, represent sequencing and PCR errors. Zhang et al. (1997) estimated that 6.8% of the SAGE tags catalogd in their study might represent sequencing errors. We were careful to catalog only those portions of the raw sequences that appeared to be of high quality, but some errors are unavoidable. A few tags correspond to upstream NlaIII restriction sites in the most abundant cDNAs (see Methods). When mRNA from genetically heterogeneous individuals is pooled, polymorphisms can produce more than one SAGE tag for a particular transcript. Single nucleotide polymorphisms (SNPs) are very common. In 3′ ESTs, one SNP was found for every 750 bases when seven individuals were studied (Wang et al. 1998). On the basis of this frequency, the probability of a SNP occurring within a 14-base SAGE tag is ∼2% when eight individuals are included. A polymorphism could be manifested either by a single nucleotide change in the tag sequence, or a change in the entire sequence if the polymorphism were to eliminate the most downstream NlaIII restriction site or to add a more downstream NlaIII restriction site. If only one person in the group has the SNP, the corresponding tag would generally have a low redundancy, unless the SNP were to occur in a very abundant transcript. We identified only 4 tags with a single-base variation (substitution, insertion, or deletion) of a more abundant tag among the 295 tags detected at least 20 times. However, these potential SNPs have not been verified with DNA from individual subjects, and PCR or sequencing errors cannot be excluded as the cause of such variations.
The absence of a tag from the present catalog does not rule out expression of the corresponding transcript in muscle. On the basis of the RNA/DNA ratio in human muscle (Forsberg et al. 1991), the proportion of RNA recovered as polyadenylated RNA in the present study, the assumption that the average mRNA has a molecular mass of 625 kD (Hastie and Bishop 1976), and the fact that myonuclei account for ∼75% of the nuclei in muscle tissue (Welle et al. 1996), we estimate that there are ∼150,000 molecules of mRNA per myonucleus. With nearly 54,000 tags cataloged, a transcript present at a concentration of 10 copies/myonucleus would have a 3% chance of not being detected, and one present at only 5 copies/myonucleus would have a 17% chance of not being detected.
Mitochondrial tags accounted for ∼20% of the polyadenylated RNA population, even though mitochondrial DNA accounts for <1% of the total DNA. The mitochondrial genome has a very high density of functional genes, and there are hundreds of copies of each mitochondrial gene for each copy of a nuclear gene. Thus, mitochondrial transcripts are not very abundant when expressed per gene rather than per microgram of polyadenylated RNA. As noted by Lanfranchi et al. (1996), the mitochondrial transcripts are frequently polyadenylated at unexpected locations, so many of the polyadenylated RNAs may not be functional. The very low abundance of L strand transcripts relative to H strand transcripts may be due to differential RNA degradation rather than differential transcription (Aloni and Attardi 1971). There was considerable variability in the abundances of the tags matching the 12 H strand mRNA genes, indicating that post-transcriptional processing or differences in mRNA stability must be an important determinant of mitochondrial mRNA concentrations.
Our results are generally similar to the CRIBI (Lanfranchi et al. 1996), Genexpress (Houlgatte et al. 1995), and Stratagene muscle (UniGene library 272 at http://www.ncbi.nlm.nig.gov/cgi-bin/Unigene) databases in terms of the relative concentrations of the high-abundance transcripts in human muscle. The CRIBI data are based on pectoral muscle from a mastectomy patient, the Genexpress data on leg muscle from a 19-year-old woman, and the Stratagene data on muscle from a patient with malignant hyperthermia. Thus, the presence of some quantitative differences (more than fivefold differences when data expressed as percent of total mRNA) among the databases is not too surprising. We used a muscle that is frequently biopsied for histological and biochemical studies, and were careful to control the activity and nutritional status of the donors. We pooled RNA from several donors so that the influence of any individual anomalies should be minimized.
An advantage of using SAGE to quantify gene expression is the reusable nature of the data. Anyone obtaining vastus lateralis muscle from another group of subjects, under similar conditions, and preparing tags according to the same protocol, can make meaningful comparisons with this database. We plan to use this database as a reference for investigating differential gene expression in age-related muscle atrophy and muscular dystrophy. The data will become even more valuable as more of the tags are identified in the course of the sequencing of the human genome. This method provides a systematic approach to searching for transcripts that may be differentially expressed, unlike the random approach offered by differential display. SAGE is more likely than subtractive hybridization methods to detect relatively small differences (e.g., twofold) in the level of expression of abundant transcripts. The main limitation of SAGE is the cost of sequencing enough clones to accurately quantify the low-abundance transcripts. As better sequencing machines become available (Venter et al. 1998), SAGE may become a much more efficient method for quantifying even rare transcripts.
METHODS
Subjects
Muscle biopsies were obtained from eight men, 21–24 years old. All were healthy and had normal neuromuscular function, as determined by physical examination, medical history, and laboratory tests (electrolytes, glucose, liver enzymes, blood count and clotting tests, TSH). We did not include any subjects who participated in unusually strenuous exercise programs or who performed any heavy resistance exercises involving the quadriceps muscles. We asked all subjects to refrain from activities more strenuous than walking for 3 days before the muscle biopsy. All subjects gave written consent after procedures and risks were explained verbally and in a written consent form. The research was approved by the University of Rochester Research Subjects Review Board.
Procedures
Each subject was admitted to the University of Rochester General Clinical Research Center the evening before his muscle biopsy, for standardization of diet and activity. He received a standard meal containing 12 kcal/kg body weight with 10%–15% of energy as protein. No activity more strenuous than walking was permitted. The subject did not eat from 10:00 p.m. until he was given breakfast at 8:00 a.m. the morning of the biopsy. The breakfast contained 7 kcal/kg body weight, with 10%–15% of energy as protein. After breakfast he rested until the muscle biopsy was obtained at ∼9:30 a.m.
The percutaneous needle biopsy of the vastus lateralis muscle was obtained within a few minutes of anesthetizing the skin and muscle with lidocaine. The tissue was frozen in liquid nitrogen as soon as possible after removal from the needle, then stored at −70°C until being used for extraction of RNA.
Production of SAGE Tags
Total RNA was extracted by homogenizing each muscle sample in TriReagent (Molecular Research Center, Cincinnati, OH) with a Polytron. The total RNA yield was assessed by UV absorbance (GeneQuant, Pharmacia, Piscataway, NJ), 10% of each subject’s RNA was set aside, and the rest was included in the combined pool. Polyadenylated RNA was extracted from the pooled RNA with the PolyATract system (Promega, Madison, WI). Total RNA yield was 530 ng/mg tissue. Polyadenylated RNA yield was ∼0.8% of total RNA, or ∼4 ng/mg tissue. According to a slot-blot assay for polyadenylated RNA (Welle et al. 1996), >90% of the mRNA was extracted.
The detailed SAGE protocol (v. 1.0c) and software (v. 1.00) were generously provided by K.W. Kinzler (Johns Hopkins University, Baltimore, MD). The outline of the procedure has been presented elsewhere (Velculescu et al. 1995). The procedure produces ditags, which are two tags joined in the sequence 5′CATG(N)20–24CATG. The first half of the ditag represents the sense sequence of an mRNA, and the second half represents the antisense sequence of an mRNA. We used NlaIII as the anchoring enzyme and BsmFI as the tagging enzyme, so that each tag represents the 10 bases 3′ to the most downstream CAUG in the mRNA. Although the tagging enzyme typically left 11 or 12 bases beyond the NlaIII restriction site, only 10 bases were used in the analysis to ensure that the rare shorter tags would be counted, and to avoid any ambiguity about which side of a ditag the centrally located bases were associated with. The ditags were concatenated, then inserted into a plasmid vector (pZero, Invitrogen, Carlsbad, CA) for cloning and sequencing. Clones were screened by PCR, and those having inserts >500 bases were sequenced. Sequencing reactions were done with the BigDye terminator cycle sequencing kit (Perkin Elmer, Branchberg, NJ), and the reaction products were analyzed with an ABI 377 sequencer (Perkin Elmer) by the University of Rochester Core Nucleic Acid Laboratory.
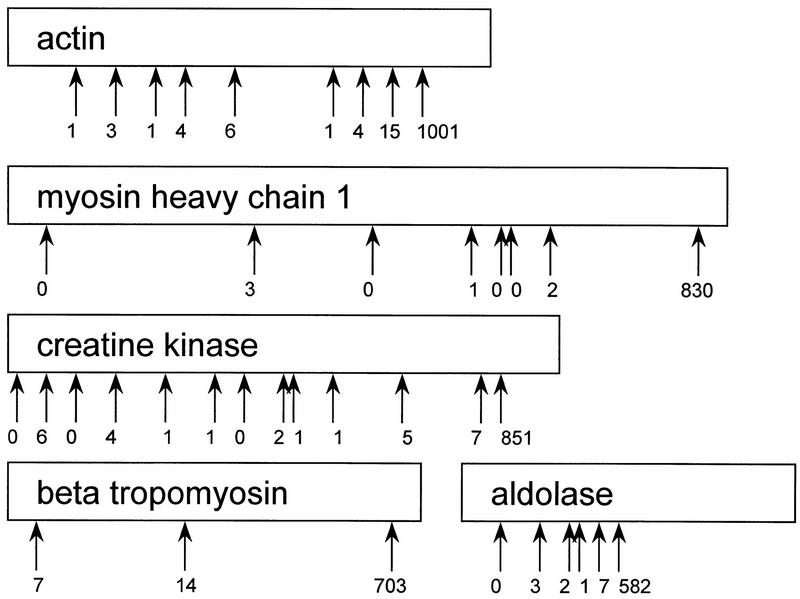
Each transcript should theoretically produce only one species of SAGE tag if the NlaIII digestion of the cDNA is 100% efficient, and if upstream fragments are efficiently removed prior to ligation with linkers and digestion with the tagging enzyme. The SAGE tag sequence should correspond to the bases flanking the NlaIII restriction site closest to the polyadenylation site. The high efficiency of NlaIII digestion and removal of upstream cDNA fragments after NlaIII digestion are demonstrated by the very low proportion (<5%) of SAGE tags that matched sequences upstream of the most 3′ NlaIII restriction site (Fig. 5), except in the case of some mitochondrial transcripts as indicated in Results.
Figure 5.
Map of the NlaIII restriction sites in some of the most abundant nonmitochondrial cDNAs, and abundances of the SAGE tags corresponding to each of these sites.
Data Analysis
Data were analyzed with the SAGE 1.00 software, which automatically detects and counts tags from the sequence files. By chance, the most redundant SAGE tags often are ligated to one another to form the same ditag more than once. So that preferential PCR amplification of certain ditags cannot cause overestimation of the abundance of the constituent SAGE tags, the SAGE software excludes replicate ditags from the catalog. The replicate ditags arose mainly from the most abundant mRNA species, as would be expected by chance. For example, on the basis of the results from the first 14,000 ditags, ∼30 instances of the cytochrome c oxidase 2 tag combining with the actin tag would be expected by chance, and 28 occurrences were detected. About 13 instances of the cardiac β myosin heavy chain tag combining with the actin tag would be expected, and 16 were detected. Table 2 illustrates that exclusion of replicate ditags causes a significant underestimation (up to 34%) of actual tag frequency for only the most abundant species. Underestimation of the most abundant tags due to exclusion of replicates becomes progressively greater as more tags are sequenced. To minimize this problem, we entered the tags into two separate databases with equal numbers of tags in each database, then merged the two databases to produce the counts presented in this paper. The values reported in this paper are those obtained after exclusion of replicate ditags within each database. The values also exclude tags matching linker sequences (1.35% of total) and A10 tags (0.32%).
Table 2.
Effect of Eliminating Replicate Ditags on Tag Counts
| Tag (CATG + . . .) | Identity | Frequency including replicate ditags | Frequency excluding replicate ditags | Decrease after excluding replicates (%) |
|---|---|---|---|---|
| CCCATCGTCC | COX2 | 1348 | 890 | 34 |
| AAGATCAAGA | actin | 756 | 523 | 31 |
| TGATTTCACT | COX3 | 316 | 240 | 24 |
| GGGGAGGAAC | slow troponin T | 205 | 169 | 18 |
| CAAGTATAAA | titin | 134 | 117 | 13 |
| TTTACTCAGC | C protein | 58 | 55 | 5 |
| CCTCTCTGGT | EST F20783 | 34 | 34 | 0 |
| TCTTGTGCAT | LDH A | 10 | 10 | 0 |
| AACAGCTCCC | EST F19683 | 6 | 6 | 0 |
These tag counts do not match those in Table 1 because they represent only half of the data. Because the underestimation of the most abundant tags would have become progressively greater with an increasing number of total tags, two separate databases were created and merged to produce the data in Table 1. Thus, the underestimation for the combined databases does not exceed the percentages shown here. (COX2) Cytochrome c oxidase 2; (COX3) cytochrome c oxidase 3; (LDH A) lactate dehydrogenase A.
Tag sequences were matched with GenBank sequences by use of the advanced BLAST option at the National Center for Biotechnology Information (NCBI) web site (www.ncbi.nlm.nih.gov), and with the database utility of SAGE software version 3.04 (β) after downloading the GenBank primate sequence files from NCBI. We have not listed matches with nonhuman sequences or human non-mRNA sequences, because with only 14 bases many matches are expected by chance. Matches were checked to verify that the tag corresponded to the most downstream NlaIII restriction site.
Acknowledgments
We thank Dr. Kenneth W. Kinzler for providing the detailed SAGE protocol and software. We thank Bharati Shah and Madalina Chirieac for their technical assistance. This project was funded by grants from the National Institutes of Health (AG-13070, AG-10463, RR-00044) and a Paul B. Beeson Physician Faculty Scholar Award to C.A.T. from the Alliance for Aging Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL swelle@ican.net; FAX (716) 760-6236.
REFERENCES
- Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci. 1971;68:1757–1761. doi: 10.1073/pnas.68.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Forsberg AM, Nillson E, Wernerman J, Bergstrom J, Hultman E. Muscle composition in relation to age and sex. Clin Sci. 1991;81:249–256. doi: 10.1042/cs0810249. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Bishop JO. The expression of three abundant classes of messenger RNA in mouse tissues. Cell. 1976;9:761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- Houlgatte R, Mariage-Samson R, Duprat S, Tessier A, Bentolila S, Lamy B, Auffray C. The Genexpress index: A resource for gene discovery and the genic map of the human genome. Genome Res. 1995;5:272–304. doi: 10.1101/gr.5.3.272. [DOI] [PubMed] [Google Scholar]
- Lanfranchi G, Muraro T, Caldara F, Pacchioni B, Pallavicini A, Pandolfo D, Toppo S, Trevisan S, Scarso S, Valle G. Identification of 4370 expressed sequence tags from a 3′-end-specific cDNA library of human skeletal muscle by DNA sequencing and filter hybridization. Genome Res. 1996;6:35–42. doi: 10.1101/gr.6.1.35. [DOI] [PubMed] [Google Scholar]
- Madden SL, Galella EA, Zhu J, Bertelsen AH, Beaudry GA. SAGE transcript profiles for p53-dependent growth regulation. Oncogene. 1997;15:1079–1085. doi: 10.1038/sj.onc.1201091. [DOI] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Valle G, Faulkner G, De Antoni A, Pacchioni B, Pallavicini A, Pandolfo D, Tiso N, Toppo S, Trevisan R, Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DEJ, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO, Hunkapiller M. Shotgun sequencing of the human genome. Science. 1998;280:1540–1542. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- Wade R, Feldman D, Gunning P, Kedes L. Sequence and expresssion of human myosin alkali light chain isoforms. Mol Cell Biochem. 1989;87:119–136. doi: 10.1007/BF00219255. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Welle S, Bhatt K, Thornton C. Polyadenylated RNA, actin mRNA, and myosin heavy chain mRNA in young and old human skeletal muscle. Am J Physiol. 1996;270:E224–E229. doi: 10.1152/ajpendo.1996.270.2.E224. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]