Abstract
Purpose
The human epigenome is profoundly altered in cancers, with a characteristic loss of methylation in repetitive regions and concomitant accumulation of gene-promoter methylation. The degree to which these processes are coordinated is unclear so we investigated both in head and neck squamous cell carcinomas.
Experimental Design
Global methylation was measured using the luminometric methylation assay (LUMA), and pyrosequencing of LINE-1Hs and AluYb8 repetitive elements in a series of 138 tumors. We also measured methylation of over 27,000 CpG loci with the Illumina HumanMethylation27 microarray (n=91).
Results
LINE-1 methylation was significantly associated with LUMA and Infinium loci methylation (Spearman’s rho=0.52/rho=0.56, both p<0.001), but not that of AluYb8. Methylation of LINE-1, AluYb8, and Infinium loci differed by tumor site (each Kruskal-Wallis p<0.05). Also, LINE-1 and LUMA methylation were associated with HPV16 E6 serology (each Mann-Whitney p<0.05). Comparing LINE-1 methylation to gene-associated methylation, we identified a distinct subset of CpG loci with significant hypermethylation associated with LINE-1 hypomethylation. An investigation of sequence features for these CpG loci revealed that they were significantly less likely to reside in repetitive elements (GSEA p<0.02), enriched in CpG islands (p<0.001), and were proximal to transcription factor binding sites (p<0.05). We validated the top CpG loci that had significant hypermethylation associated with LINE-1 hypomethylation (at EVI2A, IFRD1, KLHL6, and PTPRCAP) by pyrosequencing independent tumors.
Conclusions
These data indicate that global hypomethylation and gene-specific methylation processes are associated in a sequence-dependent manner, and that clinical characteristics and exposures leading to HNSCC may be influencing these processes.
Introduction
Epigenetic regulation is central to the biological function of all cells. Alteration to the patterns of CpG dinucleotide DNA methylation, particularly at sites with the ability to affect gene function (such as gene promoter regions), are commonplace in human diseases. Epigenetic hallmarks of malignant tumors, compared to their analogous normal tissues, include both decreases in global DNA methylation (hypomethylation - commonly assessed in DNA repeat regions) and concomitant increases (hypermethylation) in gene-associated (or locus-specific) methylation. Gene-associated hypermethylation is often associated with phenotypic silencing of tumor suppressor genes in cancer. In fact, gene promoter hypermethylation events during the pathogenesis of neoplasia may be as common, if not more frequent than the well recognized genetic mechanisms of gene inactivation, like mutation and loss of heterozygosity (1).
Head and neck squamous cell carcinomas (HNSCCs) are prevalent throughout the world, representing the sixth most common malignancy overall and the eighth most common in U.S. males (excluding non-melanoma skin cancers) (2). The major etiologic risk factors are alcohol use, tobacco use, and human papillomavirus type 16 (HPV16) infection. Altered epigenetic states have been described in these tumors, including hypermethylation of promoters associated with known tumor suppressor genes CDKN2A, CDH1, DAPK1, RASSF1, and MGMT (3–5). However, further research is needed to understand the epigenetic basis of this deadly disease.
In addition to gene-specific hypermethylation, loss of repetitive element methylation is a common feature of human cancers. Repetitive elements, a majority of which consist of long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) such as AluYb8 are maintained in a highly methylated state across normal tissues. Methylation of repetitive and retrotransposable elements are thought to play a role in maintenance of genomic stability (due to their high copy number and correlation to total genomic 5-methylcytosine methylation (6)) and are often used as a surrogate for levels of global methylation. Loss of repetitive element methylation in cancer has been shown to activate expression of proto-oncogenes, increase mutational events, and engender chromosomal instability (7, 8). Previous studies have indicated that hypomethylation of repetitive elements is widespread in HNSCC (9). In addition, there is evidence that the degree of hypomethylation is related to HPV16 infection and lifestyle factors (10, 11) as well as the genomic context of the measured loci (12).
We measured different markers of global DNA methylation and gene-associated methylation in HNSCCs to further elucidate the epigenetic basis of this disease. These data were integrated to reveal potential relationships between clinicopathologic/lifestyle variables, global methylation levels, and gene-directed methylation patterns.
Materials and Methods
Study Population/Ethics
The study population was previously described (13). Briefly, samples from incident cases of HNSCC were microscopically examined and histologically confirmed to be >70% tumor content by the study pathologist. Patients were enrolled upon providing written, informed consent. This study was approved by the Brown University institutional review board. Clinical information was collected and HPV16 status was assessed using a multiplex serology assay to detect antibodies against the HPV16 E6 protein according to previously published methods (14). In total, 138 fresh-frozen tumor specimens from head and neck sites (excluding nasopharyngeal carcinomas) were subjected to methylation analysis. Fresh frozen normal (non-diseased) head and neck tissues (n=18) taken at autopsy from the oral cavity, pharynx, and larynx were provided by the National Disease Research Interchange In addition, peripheral blood, drawn from 213 cancer-free individuals living in New England (enrolled as part of a population-based study of bladder cancer) was studied as a comparison convenience sample; details of this population are described in (15).
DNA Isolation and Genome-wide Methylation Measurements
DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) and 1 μg of genomic DNA was sodium bisulfite-modified with the EZ DNA Methylation Kit (Zymo Research, Orange, CA) as per the manufacturer’s instructions.
LINE-1 methylation was measured in all tumor and blood samples according to the procedures described by Bollati et al. (16).
AluYb8 subfamily methylation was measured as outlined by Choi et al. (17) with minor modifications. PCR was performed on 40ng of modified DNA per 25ul reaction under the following conditions: denaturation 94°C, 2 minutes/94°C for 30 seconds (cycling); annealing 58°C, 30 seconds; extension 70°C, 30 seconds; 50 cycles. The first five of six CpG sites were used in analysis of AluYb8 methylation. Pyrosequencing reactions were performed in triplicate and bisulfite conversion efficiency was monitored using internal non-CpG cytosine residues using the PyroMark Q96 MD system. For LINE-1 and AluYb8, methylation at each CpG position was calculated by taking the percent of methylated signal divided by the sum of the methylated and unmethylated signals and reported as the mean over all positions.
The LUMA procedure (18) was carried out according to the modifications described in (19) with the nucleotide dispensation order (GTGTCACATGTGTG) to compensate for background signal from lower quality DNA. Methylation was estimated from the 1-(HpaII/MspI) ratios. Since this procedure requires a significant amount of DNA, we selected 50 HNSCCs for which there was a sufficient amount of substrate. Restriction enzymes (HpaII, MspI and EcoRI-HF) were purchased from New England Biolabs (Ipswich, MA).
Illumina Infinium HumanMethylation27 Microarray
High-resolution methylation analyses of HNSCCs (n=91), normal tissue (n=18), and control bloods (n=213) were conducted on the Illumina Infinium HumanMethylation27 microarray platform (San Diego, CA). This BeadChip assay measures methylation (20), given as a β value ranging from zero to one, at over 27,000 CpG loci. Arrays were processed at the UCSF Institute for Human Genetics, Genomics Core Facility according to the manufacturer’s protocol. Data were assembled in BeadStudio without normalization, as instructed by Illumina. Array control probes were used to assess sample performance. Specifically, the multivariate characteristics of array control probes based on fitted mean vector and variance-covariance matrix (Mahalanobis distance) was used to screen outliers. Sex chromosome loci (n=1092) were excluded to avoid gender-specific methylation bias, resulting in a final dataset that consisted of 26,486 autosomal loci associated with 13,890 genes. Sequence context information such as CpG island status (as defined by (21)) and transcription factor binding site (TFBS) proximity was extracted from UCSC Genome Table Browser (NCBI36/hg18 assembly) with “repeatmasker v3.2.7” and “TFBS Conserved” (TFBS Z-score > 2) tracks.
Statistical Analysis
Methylation data were analyzed in R statistical software package v2.8.1 (http://www.r-project.org). Global methylation comparisons were performed by computing Spearman’s rank correlation among the tumors measured for LINE-1, AluYb8, LUMA, and mean per-sample Infinium array methylation. An omnibus p-value was assigned by comparing the observed correlation estimate with the corresponding null distribution obtained by permutation (10,000 permutations). We tested for univariate associations with epidemiologic factors as predictors of global methylation using a permutation test (10,000 permutations) with the Kruskal-Wallis or Mann-Whitney U test statistic for categorical predictors and Spearman correlations for continuous data. Multivariable regression models were used to control for the HNSCC risk factors risk factors age, HPV16 serology, site, stage, drinking, and smoking.
LINE-1 methylation was first measured in a core set of 138 HNSCCs. Microarray analysis was next performed on 91 HNSCCs (one 96-well plate; limited by resources) which had the most complete clinical/exposure covariate information available. Among these 91 samples, 5 failed AluYb8 pyrosequencing, leaving 86 tumors. Finally, as each LUMA assay requires a substantial amount of DNA, and sufficient substrate was only available to allow for analysis of 50 tumors. In multivariable models, the number of samples reflects those with complete data for all variables in the model.
To discern the relationships between LINE-1 and array CpGs with coordinate methylation, we used recursively partitioned mixture modeling (RPMM), a computationally efficient likelihood-based method of hierarchical clustering (22), to cluster autosomal CpGs into 16 classes based on their methylation pattern. Although metric (nonparametric) hierarchical clustering is a well-characterized method, it does not scale to tens of thousands of observations, and appears to have poorer clustering consistency in this context (see Supplementary Text). Individual CpG-specific array values (β’s) were subsequently averaged together, within each class, to form 16 average methylation values (per individual, one for each class). This dimensionality reduction approach combines data from CpGs with similar patterns, thereby attenuating biochemical noise and reducing multiple comparisons (23). We subsequently determined the methylation class-specific Spearman correlation coefficients with LINE-1 methylation after stratifying the tumors by site. The null distribution of the multivariate 16-dimensional correlation statistic was obtained by randomly permuting the LINE-1 values with respect to their corresponding methylation profiles 10,000 times. An omnibus test of significance was performed by comparing the observed maximum absolute value over the 16 correlations with the corresponding quantity over the permutation distribution.
To investigate the CpG-specific correlations with LINE-1 methylation, we stratified the tumors by site and calculated Spearman correlations at all autosomal array loci. P-values were corrected for multiple comparisons (using the qvalue package in R) and significant (q<0.05) loci were grouped by correlation sign (+/−). To interrogate these loci for their representation among genomic sequence elements, we utilized Gene Set Enrichment Analysis (GSEA) (24). GSEA allows for evaluation of overrepresentation among loci associated with genomic sequence elements among the rank-ordered list of CpGs correlated with LINE-1 methylation. The enrichment analysis uses a Kolmogorov-Smirnov-like statistic to estimate the proportion of loci at the top of the ranked list compared to all loci examined (n=26,486). For this implementation, the procedure was implemented in R using the R function obtained from the Broad Institute GSEA web site, with Pearson correlations and a permutation null distribution generated with 1000 permutations.
Array Validation
Pyrosequencing assays for the top 5 CpG loci negatively associated with LINE-1 were designed, and four of these five assays (associated with the genes EVI2A, IFRD1, KLHL6, and PTPRCAP) were amenable to sequencing. Pyrosequencing was performed both on our original oral/pharyngeal tumors (n=59) and in an independent set of 48 paraffin-embedded HNSCCs. Sample collection and extraction of substrate is described elsewhere (3). Spearman correlations were again calculated and significance determined via permutation test. Primer sequences are listed in Supplementary Table 1.
Results
Global measures of methylation are correlated
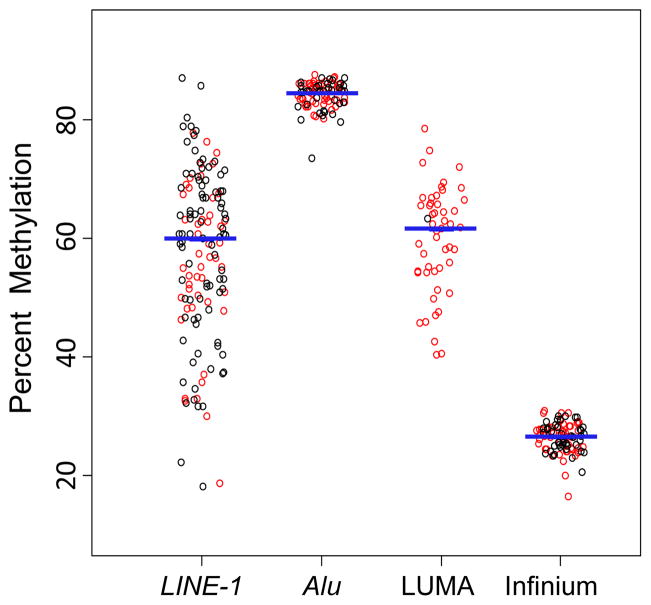
To examine the nature and distribution of global methylation in a case-series of HNSCCs (summarized in Table 1), we bisulfite pyrosequenced LINE-1 and AluYb8 consensus repetitive element sequences; measured CCGG simple sequence repeat methylation using the LUminometric Methylation Assay (LUMA) and measured locus-specific DNA methylation with Illumina’s Infinium microarray (26,486 autosomal CpG loci). The data for each global methylation indicator is shown in Figure 1. Specifically, the tumor-to-tumor variability in methylation was greatest for LINE-1 (median 60%, interquartile range (IQR) 18.3%) and LUMA (median 62%, IQR 11.4%). Conversely, AluYb8 (median 85%, IQR 3.0%) and mean Infinium methylation (median 27%, IQR 3.3%) values were much more tightly distributed.
Table 1.
Clinicopathologic Characteristics of Study Participants
| All Cases (n=138) | |
|---|---|
| Gender, n (%) | |
| Female | 38 (28) |
| Male | 100 (72) |
| Age at Diagnosis (years) | |
| Range | 32–91 |
| Mean (SD) | 61 (12.0) |
| HPV16 E6 Serology, n (%)* | |
| Positive | 12 (12) |
| Negative | 92 (88) |
| Tumor Site, n (%) † | |
| Oral | 76 (64) |
| Pharynx | 24 (20) |
| Larynx | 19 (16) |
| Clinical Stage, n (%) ‡ | |
| I | 9 (7) |
| II | 27 (21) |
| III | 24 (19) |
| IV | 67 (53) |
| Lifetime Drink-Years of Consumption, n § | |
| Range | 0–61 |
| Mean (SD) | 26 (20) |
| Never-drinkers, n | 34 |
| Lifetime Pack-Years Smoked, n || | |
| Range | 0–135 |
| Mean (SD) | 35 (31) |
| Never-smokers, n | 20 |
Thirty-four samples missing serology data
Nineteen samples missing site data
Eleven tumors missing stage data
Twenty-two patients missing self-reported drinking data
Seventeen patients missing self-reported smoking data
Figure 1.
Summary HNSCCs. Red data points indicate tumors common across measurement methods. Black circles represent samples that were not used for all methods, however all tumors were subsets of those measured in the assay were n=138 (levels.
Next, the association between markers of global methylation was investigated. All correlations between markers of global methylation (except between AluYb8 and LUMA) were statistically significant (Supplementary Table 2, also Supplementary Figure 1). AluYb8 methylation was least associated with all other indicators of global methylation. Despite the differences in measurement procedures and nature of the loci examined, the strongest correlation was between LINE-1 methylation and mean array CpG methylation (Spearman correlation=0.56). In addition, there were clear relationships between LUMA and LINE-1 methylation as well as between LUMA and Infinium methylation, suggestive of a link between global- and gene-targeted methylation.
Global methylation measures are associated with tumor site and HPV16 seropositivity
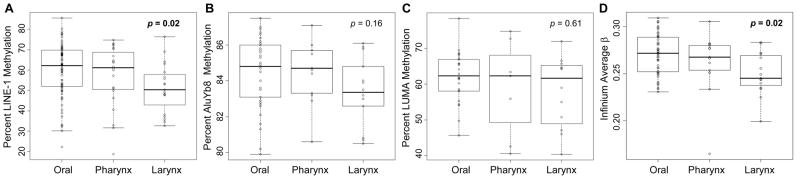
Because LINE-1 methylation has previously been shown to be modified by environmental factors in a number of diseases (11, 16) and we observed associations between LINE-1 and other measures of global methylation, we hypothesized that clinical and exposure characteristics may alter global methylation indicators. Univariate analysis revealed a significant relationship between tumor site and both LINE-1 methylation and average array CpG methylation (each Kruskal-Wallis p<0.03) but not with other global methylation measures (Figure 2). In a multivariable regression model controlled for potential confounders, LINE-1, AluYb8, and average array CpG methylation were significantly associated with tumor site (all three p<0.05). For each global methylation measure, laryngeal tumors had decreased methylation compared to pharyngeal or oral tumors (Table 2).
Figure 2.
Tumors of different anatomical origin have distinct global methylation profiles. P-values in bold are considered significant by Kruskal-Wallis test (p<0.05). Plots show the effect of tumor site-stratification on percent methylation of (A) LINE-1, (B) AluYb8, (C) LUMA, and (D) averaged Infinium array loci.
Table 2.
Multivariable Regression for Global Measures of Methylation on Patient Characteristics*
| Predictors | L1Hs (n=81)
|
AluYb8 (n=39)
|
Infinium (n=40)
|
LUMA (n=28)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | P | Estimate | 95% CI | P | Estimate | 95% CI | P | Estimate | 95% CI | P | |
| Age | 0.2 | (0.0,0.5) | 0.06 | −0.01 | (−0.08,0.06) | 0.78 | ~0 | (0.0,0.0) | 0.52 | −0.03 | (−0.45,0.40) | 0.90 |
| HPV16 E6 Serology | ||||||||||||
| Negative | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref |
| Positive | 9.8 | (0.8,18.8) | 0.04 | 1.00 | (−1.2,3.3) | 0.38 | 0.02 | (−0.01,0.04) | 0.16 | 21.5 | (5.3,37.8) | 0.02 |
| Tumor Site | ||||||||||||
| Oral | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref |
| Pharynx | −6 | (−13.6,1.7) | 0.13 | −0.29 | (−2.0,1.4) | 0.75 | −0.02 | (−0.03, −0.01) | 0.04 | −6.8 | (−16.4,2.8) | 0.18 |
| Larynx | −14.2 | (−22.5, −5.8) | <0.01 | −1.98 | (−3.7, −0.3) | 0.03 | −0.04 | (−0.06, −0.02) | <0.01 | −7.8 | (−16.8,1.2) | 0.11 |
| Stage | ||||||||||||
| I & II | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref | ref |
| III & IV | −5.7 | (−11.8,0.5) | 0.07 | −0.23 | (−1.8,1.3) | 0.78 | 0.01 | (−0.01,0.03) | 0.25 | −1.6 | (−10.5,7.2) | 0.72 |
| Lifetime Drink-Years of Consumption | 0.05 | (−0.01,0.21) | 0.50 | 0.04 | (−0.02,0.09) | 0.21 | ~0 | (0.0,0.0) | 0.79 | 0.20 | (−0.1,0.5) | 0.28 |
| Lifetime Pack-Years Smoked | −0.01 | (−0.10,0.08) | 0.80 | −0.01 | (−0.03,0.02) | 0.56 | ~0 | (0.0,0.0) | 0.28 | −0.04 | (−0.16,0.09) | 0.56 |
Models controlled for all variables listed. CI = Confidence Interval. Results in bold are considered significant where the 95% CI does not cross zero.
HPV16 infection is an established risk factor for HNSCC and was evaluated for association with global methylation measures. In the adjusted regression model HPV16 E6 antibody seropositivity was significantly associated with an increase in LINE-1 (p<0.05) and LUMA methylation (p<0.03) (Table 2). Although the prevalence of HPV-positive tumors differed by site (highest in pharyngeal tumors), we did not have sufficient data for an analysis stratified by additional tumor subsites (Supplementary Table 3). There were no significant associations between any of the global methylation measures and gender, race, patient age at diagnosis, combined TNM stage, body mass index, or smoking intensity/duration. Because the number of samples for each model differed due to missing covariate data, we performed a subset analysis and determined that all tumor subsets were representative of the overall HNSCC population (data not shown).
CpG cluster methylation patterns are correlated with global methylation measures in a sequence-dependent manner
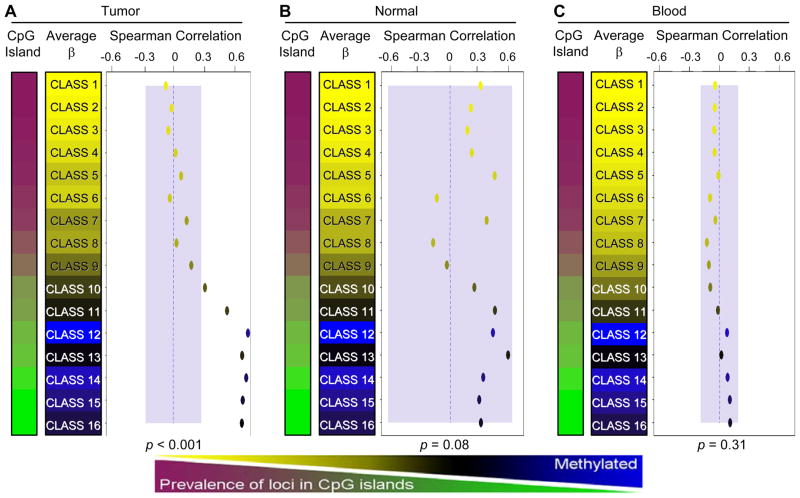
As the data suggest a general correlation between global measures of methylation, we next focused on the relationship between LINE-1 methylation and array CpG methylation using recursively partitioned mixture modeling (RPMM). RPMM has been previously described in detail (22) and applied in numerous DNA methylation array analyses (3, 23, 25). In contrast to computationally inefficient hierarchical clustering methods, this approach robustly classifies loci (or samples) into biologically informed groups whose methylation patterns can be examined for associations with covariates. All array autosomal CpG loci were clustered into 16 methylation classes (median group n=1763 CpGs, range=763–2821 CpGs). For each class, we computed the Spearman correlation between average class methylation and LINE-1 methylation (Figure 3A). Among the 16 CpG methylation classes, mean methylation was significantly positively correlated with LINE-1 methylation in the 7 classes that had a lower CpG island locus prevalence (Supplementary Figure 2). No negative correlations between CpG class methylation and LINE-1 methylation were observed by class. To determine if the observed positive correlations are unique to HNSCCs, an identical analysis was performed in normal head and neck tissues (n=18) and a convenience sample of DNA extracted from peripheral blood of healthy individuals (n=213). In both of these tissue types, no RPMM CpG methylation classes were significantly associated with LINE-1 methylation (Figure 3B & 3C). Because LINE-1 retrotransposons are highly methylated in the normal genome, we compared LINE-1 methylation of all 138 HNSCCs to that of the 18 normal head and neck tissues. This analysis revealed that 126 (92%) tumors had less LINE-1 methylation than the least LINE-1-methylated normal sample.
Figure 3.
Classes of array CpG loci are associated with global methylation levels in tumors but not in normal tissues. RPMM is used to cluster loci with similar methylation β values into sixteen methylation classes. Spearman’s correlation coefficients are calculated by comparing the mean methylation of class member loci for each sample to LINE-1 methylation level and plotted by class. Correlation points are colored according to their mean class methylation (as indicated in the color sidebars). Classes were ordered according to the percentage of their member loci mapping to CpG island regions. A separate RPMM model was applied to the data for each tissue type: (A) HNSCCs, (B) non-diseased head and neck tissues, and (C) normal peripheral blood. Blue shaded regions represent the 95% confidence limits of the observed maximums correlation from 10,000 random permutations (representing the null distribution). Therefore, correlations lying outside of these regions are considered statistically significant. Permutation test omnibus p-values are displayed at the bottom of each panel. Dotted blue lines indicate the zero correlation axis.
LINE-1-associated loci are enriched for sequence-dependent elements
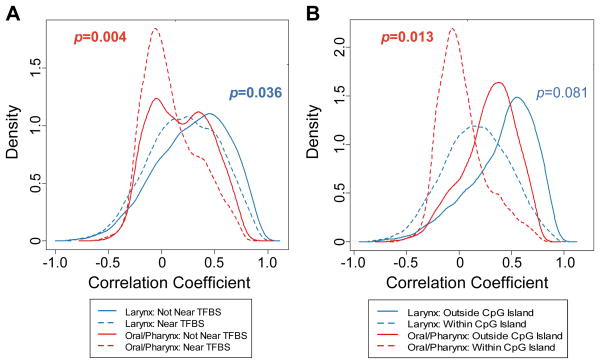
We next sought to identify which individual loci were significantly correlated with LINE-1 hypomethylation. Because LINE-1 methylation varied by tumor site, we stratified the tumors (larynx or oral/pharynx) then calculated Spearman correlations between LINE-1 methylation and all autosomal array CpG loci in a locus-by-locus fashion for each tumor site. Following correction for multiple comparisons, 3029 CpG loci (11%) and 7367 CpG loci (28%) were significantly correlated with LINE-1 methylation (q<0.05) in the laryngeal and oral/pharyngeal analyses, respectively. Among the top 2000 most highly associated loci for both sites, about 97% were positively correlated with tumor LINE-1 methylation levels (hypomethylated CpG loci and hypomethylated LINE-1); a small group of CpGs (37 loci (2%) in the oral cavity/pharynx and 60 loci (3%) in the larynx) were negatively correlated with LINE-1 methylation (hypermethylated CpG and hypomethylated LINE-1; see Supplementary Table 4 as compared to Supplementary Table 5). Our previous analyses have indicated an interaction between epigenetic and genetic alterations in cancer (26, 27). We therefore investigated the hypothesis that the genomic sequence context influences selection for de novo methylation in the LINE-1-negatively correlated CpG loci using Gene Set Enrichment Analysis (GSEA, i.e. determination of the proportion of LINE-1-negatively correlated CpG loci are found at the top of a ranked correlation scores among all loci). These CpGs were significantly more prevalent in CpG islands for oral/pharyngeal tumors (p<0.02), near transcription factor binding sites (TFBSs; both sites p<0.05, Figure 4), and significantly depleted within repetitive elements (p<0.05, Supplementary Figure 3) among all the loci analyzed by microarray.
Figure 4.
Methylation varies with the presence of local structural elements. LINE-1-discordantly methylated loci are enriched for (A) proximity to transcription factor binding sites and (B) CpG islands. Tumor data (for 61 oral/pharyngeal and 15 laryngeal tumors) are stratified by site and the distributions of loci, based on the Spearman correlations between CpG methylation and LINE-1 methylation, are plotted according to their respective kernel density estimations. GSEA permutation test p-values indicated in bold represent significant relationships (p<0.05). Dotted lines indicate the distribution of loci located within the sequence element, while solid lines indicate the distribution of loci located outside of the sequence element.
Bisulfite pyrosequencing was used for array validation of the top oral/pharyngeal CpG loci (within the EVI2A, IFRD1, KLHL6, and PTPRCAP gene regions) negatively correlated with LINE-1 methylation. Tumors from our original fresh-frozen oral/pharyngeal population, where DNA remained (n=59 out of 100), and tumors from an independent set of HNSCCs (n=48), representing all tumor sites, were pyrosequenced. In this validation, all 4 of the CpG loci examined were significantly negatively correlated with LINE-1 methylation (all permutation test p<0.01, Supplementary Figure 4). To ensure the statistical rigor of this novel method for identification of molecularly-selected loci, a random CpG (associated with CD22) was pyrosequenced for the independent validation tumor set, and no significant correlation with LINE-1 methylation was observed (rho=−0.03, p=0.84). We also investigated the possibility that clinical stage might be a significant predictor of methylation in these CpGs, but no such association was found.
Lastly, we determined whether the CpGs with discordant LINE-1 methylation in tumors were also discordant in all normal head and neck tissues and peripheral blood samples using Infinium array CpG methylation at EVI2A, IFRD1, KLHL6, and PTPRCAP. Consistent with this being a tumor-specific phenomenon, among all 4 CpGs, no significant negative correlations were observed in normal head and neck tissues or peripheral blood samples (minimum rho=−0.24, lowest p=0.32).
Discussion
Malignant transformation is a complex process characterized by the accumulation of genetic and epigenetic abnormalities. The patterns of DNA methylation alterations in cancer include both genome-wide hypomethylation and gene-specific hypermethylation. Gene silencing associated with hypermethylation can prevent transcription of critical proteins, however the consequences of hypomethylation of repetitive DNA sequences are less clear. Furthermore, the nature of the relationship between locus-specific methylation and global DNA methylation alterations is underexplored. Here, we extensively examined both global methylation and gene-specific methylation in HNSCC. We showed that different markers of global methylation are correlated but that LINE-1, LUMA, and mean array methylation were more often associated with clinical characteristics of HNSCC than AluYb8 methylation. We also investigated the relationship between global and CpG-specific methylation, and found that LINE-1 hypomethylation was correlated with significantly increased CpG methylation at only a small number of loci. GSEA analysis indicated that these CpG loci were enriched in CpG islands and TFBSs but depleted in repetitive elements, indicating that genomic context may play a role in this coordination.
Methylation of CCGG repeat DNA (as measured by LUMA), LINE-1, and AluYb8 are commonly used as indicators of global methylation. We observed these to be generally correlated, but with different degrees of variance, consistent with previous comparisons between LINE-1 and other global markers in normal blood (16, 28) and various human cancers (9, 10, 17, 29–38). The basis for their differential variance is not well understood, though unequal genomic distributions (39), evolutionary age of retrotransposable elements, and sequence context may all contribute (40). Our observations stress the need to consider inter-assay differences when comparing and interpreting global methylation measures.
Methylation of LINE-1 by pyrosequencing has previously been measured in HNSCC (38). Univariate analyses in that study identified decreased methylation in HNSCC tumors compared to non-diseased tissue of the aerodigestive tract, and tumor-only associations with tobacco, alcohol, and stage. Consistent with this, we identified a (nonsignificant) decrease in methylation for high stage tumors. Tumor stage was not a significant predictor of LINE-1 methylation in the Smith et al. (38) multivariable analysis, but our combined studies suggest a potential association of global hypomethylation with more advanced disease. This association has been noted in other cancers (41).
We have identified hypermethylated CpG loci that are significantly negatively correlated with LINE-1 methylation in HNSCC. The fact that locus-specific methylation increases while global methylation decreases suggests that these CpGs are actively selected to maintain the tumor phenotype through DNA methylation. We next explored the possibility that genomic sequence context may influence the coordination between global and gene-specific methylation. Highlighting the influence of genome architecture on gene-specific methylation, Estécio et al. more generally examined the link between repetitive elements and gene-specific methylation in bladder cancer (42). They demonstrated that the predisposition for de novo methylation of CpG Islands is associated with the absence of transposable elements. Consistent with their findings, we have shown that hypermethylated CpGs negatively correlated with LINE-1 methylation are more likely to be in CpG islands. Furthermore, these CpGs were less likely to reside in transposable elements and more likely to be located near transcription factor binding sites. We did not identify an association with specific transcription factors prevalent these regions, but due to the unique and critical functions that transcription factors provide, the intersection of gene-specific methylation, repeat methylation, and transcription factor presence represents an intriguing area for further exploration. One study has described an overall enrichment for DR2 retinoic acid response elements (RARE) within AluS repetitive elements, yet this may be explained by the spontaneous deamination of a single CpG within AluS elements to generate a RARE. (43). Further research using larger tumor populations and additional cancer types may reveal additional insight into these interactions.
Among the top CpGs whose methylation was negatively correlated with LINE-1 methylation, many of the associated genes are frequently dysregulated or epigenetically repressed in cancers. Our locus-by-locus analysis identified CpGs associated with CDKN2A (Supplementary Table 4), a well known tumor suppressor gene commonly silenced in HNSCC (4) and other cancers. Additionally, CpGs associated with two genes known to be hypermethylated in breast tumors were also identified: SIM1 and GHSR (Supplementary Table 4). SIM1 methylation has been identified as a biomarker for early-stage cancers (44), while methylation of the GHSR gene distinguishes infiltrating ductal carcinoma from normal and benign tissues with a high sensitivity and specificity (45). To our knowledge, these genes have not been explored previously in HNSCC and further studies are required to clarify their role in cellular transformation. Among the genes that were validated by pyrosequencing (commonly altered in the oral cavity/pharynx or also in the larynx), only the tumor-suppressor IFRD1 has a well-defined role, specifically in lung carcinogenesis (46).
There were a surprisingly small number of CpG loci whose methylation was negatively correlated with that of LINE-1 among the top 2000 significantly-altered CpGs. One potential mechanism to explain this discrepancy involves a recent modification to the maintenance CpG methylation theory (in mother to daughter cells). In this proposed model, DNMT3A/DNMT3B (classically considered to be de novo methyltransferases) act cooperatively to maintain DNA methylation marks in CpG islands and repeat regions, whereas DNMT1 (the classic maintenance DNA methyltransferase) maintains non-CpG island methylation (47). A follow-up study by Sharma et al. (48) demonstrated a self-regulatory mechanism between global methylation and expression of DNMT3A/DNMT3B, whereby a reduced global methylation results in a diminished capacity for aberrant de novo methylation due to the degradation of DNMT3A/DNMT3B enzymes. Applying this concept to cancer studies (in the context of genome-wide hypomethylation), hypermethylation would be expected to occur primarily at gene loci within dynamic regulatory trafficking centers where the potential for protein stabilization is highest, such as those inside of TFBSs, CpG islands, and outside of repeat elements (Figure 4 and Supplementary Figure 3).
Collectively, our data support the hypothesis that gene regulation is coordinated through global and locus-specific methylation at certain sequence elements important for cellular differentiation and carcinogenesis. Importantly, our locus-by-locus comparison of CpG-specific methylation with LINE-1 methylation provides a novel route to identify selectively hypermethylated genes in cancers. Further studies in additional tumor types are necessary to clarify the mechanistic basis for and to expand upon these findings.
Supplementary Material
Acknowledgments
This study was supported by the Flight Attendant Medical Research Institute (CJM) and the National Institutes of Health (KTK: CA078609, CA100679).
The authors would like to thank Devin C. Koestler, Charlotte S. Wilhelm-Benartzi, and William P. Accomando Jr.
Footnotes
The authors declare no potential conflicts of interest.
Microarray data from this study have been contributed to the NCBI Gene Expression Omnibus under the accession number GSE25093 (http://www.ncbi.nlm.nih.gov/geo).
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Marsit CJ, Christensen BC, Houseman EA, et al. Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis. 2009;30:416–22. doi: 10.1093/carcin/bgp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–6. doi: 10.1038/sj.onc.1205528. [DOI] [PubMed] [Google Scholar]
- 5.Dikshit RP, Gillio-Tos A, Brennan P, et al. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer. 2007;110:1745–51. doi: 10.1002/cncr.22975. [DOI] [PubMed] [Google Scholar]
- 6.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–23. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr. 2002;132:2424S–9S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 9.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 10.Richards KL, Zhang B, Baggerly KA, et al. Genome-wide hypomethylation in head and neck is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furniss CS, Marsit CJ, Houseman EA, Eddy K, Kelsey KT. Line region hypomethylation is associated with lifestyle and differs by human papillomavirus status in head and neck squamous cell carcinomas. Cancer Epidemiol Biomarkers Prev. 2008;17:966–71. doi: 10.1158/1055-9965.EPI-07-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008;36:5704–12. doi: 10.1093/nar/gkn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119:1761–6. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 14.Waterboer T, Sehr P, Michael KM, et al. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin Chem. 2005;51:1845–53. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]
- 15.Wilhelm CS, Kelsey KT, Butler R, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16:1682–9. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 17.Choi SH, Worswick S, Byun HM, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125:723–9. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karimi M, Johansson S, Stach D, et al. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–95. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 21.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houseman EA, Christensen BC, Yeh RF, et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilhelm-Benartzi CS, Koestler DC, Houseman EA, et al. DNA methylation profiles delineate etiologic heterogeneity and clinically important subgroups of bladder cancer. Carcinogenesis. 2010;31:1972–6. doi: 10.1093/carcin/bgq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poage GM, Christensen BC, Houseman EA, et al. Genetic and epigenetic somatic alterations in head and neck squamous cell carcinomas are globally coordinated but not locally targeted. PLoS One. 2010;5:e9651. doi: 10.1371/journal.pone.0009651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen BC, Houseman EA, Poage GM, et al. Integrated profiling reveals a global correlation between epigenetic and genetic alterations in mesothelioma. Cancer Res. 2010;70:5686–94. doi: 10.1158/0008-5472.CAN-10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jintaridth P, Mutirangura A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00146.2009. [DOI] [PubMed] [Google Scholar]
- 29.Daskalos A, Nikolaidis G, Xinarianos G, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124:81–7. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 30.Cho NY, Kim BH, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–77. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 31.Choi IS, Esteci MR, Nagano Y, et al. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod Pathol. 2007;20:802–10. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- 32.Lee JJ, Geli J, Larsson C, et al. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. Int J Oncol. 2008;33:861–9. [PubMed] [Google Scholar]
- 33.Geli J, Kiss N, Karimi M, et al. Global and regional CpG methylation in pheochromocytomas and abdominal paragangliomas: association to malignant behavior. Clin Cancer Res. 2008;14:2551–9. doi: 10.1158/1078-0432.CCR-07-1867. [DOI] [PubMed] [Google Scholar]
- 34.Deneberg S, Grovdal M, Karimi M, et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia. 2010;24:932–41. doi: 10.1038/leu.2010.41. [DOI] [PubMed] [Google Scholar]
- 35.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 37.Estecio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007;121:1724–8. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 41.Florl AR, Steinhoff C, Muller M, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br JCancer. 2004;91:985–94. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Estecio MR, Gallegos J, Vallot C, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010 doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto K, Fukutomi T, Akashi-Tanaka S, et al. Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer. 2005;116:407–14. doi: 10.1002/ijc.21054. [DOI] [PubMed] [Google Scholar]
- 45.Ordway JM, Budiman MA, Korshunova Y, et al. Identification of novel high-freuency DNA methylation changes in breast cancer. PLoS One. 2007;2:e1314. doi: 10.1371/journal.pone.0001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latif F, Duh FM, Bader S, et al. The human homolog of the rodent immediate earlyresponse genes, PC4 and TIS7, resides in the lung cancer tumor suppressor gene region on chromosome 3p21. Hum Genet. 1997;99:334–41. doi: 10.1007/s004390050368. [DOI] [PubMed] [Google Scholar]
- 47.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance. PLoS Genet. 2011;7:e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.