Abstract
Purpose
Chk1 inhibitors, such as AZD7762 are in clinical development in combination with cytotoxic agents for the treatment of solid tumors, including pancreatic cancers. To maximize the likelihood of their clinical success, it is essential to optimize drug scheduling as well as pharmacodynamic biomarkers in preclinical models.
Experimental Design
We tested multiple schedules of administration of gemcitabine and AZD7762 on the survival of pancreatic cancer cells. Potential pharmacodynamic biomarkers including pChk1, pChk2, pHistone H3, and caspase-3 were evaluated in vitro, followed by assessment of promising candidate biomarkers in vivo. We then went on to determine the contributions of PP2A and DNA damage to the mechanism(s) of induction of the identified biomarker, pS345 Chk1.
Results
AZD7762 given during and after or after gemcitabine administration produced maximum chemosensitization. In vivo, AZD7762 significantly inhibited the growth of pancreatic tumor xenografts in response to gemcitabine. Of the biomarkers assessed, pS345 Chk1 was most consistently increased in response to gemcitabine and AZD7762 in tumors and normal tissues (hair follicles). pS345 Chk1 induction in response to gemcitabine and AZD7762 occurred in the presence of PP2A inhibition and in association with elevated γH2AX, suggesting that DNA damage is an underlying mechanism.
Conclusions
AZD7762 sensitizes pancreatic cancer cells and tumors to gemcitabine in association with induction of pS345 Chk1. Together these data support the clinical investigation of AZD7762 with gemcitabine in pancreatic cancer under a dosing schedule in which gemcitabine is administered concurrent with or prior to AZD7762 and in conjunction with skin biopsies to measure pS345 Chk1.
Keywords: pancreatic cancer, Chk1, chemosensitization, gemcitabine, pharmacodynamic biomarker
Introduction
Gemcitabine is the first line of treatment for patients with pancreatic cancer and is associated with median survivals of approximately 6 and 9 months for metastatic and locally advanced disease, respectively. Many clinical trials have been conducted in an effort to improve upon the efficacy of gemcitabine, yet very few have yielded clinically significant survival advantages (1) (2). Furthermore, even these modest improvements have been accompanied by a substantial increase in toxicity. Thus, a great deal of attention has been focused on the development of molecularly targeted therapies, with the hope of producing improved outcome without increasing toxicity. One such approach has focused on the discovery of small molecule inhibitors targeted to DNA damage response machinery such as Chk1 (checkpoint kinase 1) (for review see (3)). The goal in the development of these types of agents is that they could be used to selectively sensitize cancer cells containing defects in other cell cycle checkpoint proteins, such as p53, to DNA damaging agents (4). Currently, several small molecule Chk1 inhibitors are being developed for clinical use (AZD7762, PF-00477736, LY2606368, and SCH900776) as sensitizers in combination with DNA damaging agents (such as gemcitabine, cisplatin, pemetrexed, and irinotecan) (4–5).
Chk1 is a central mediator of the cellular response to DNA damage. Activation of Chk1 by ATR in response to DNA damage or replication stress results in inhibition of Cdc25 phosphatases, cyclin-Cdk inhibition, and cell cycle arrest (for review see (6)). Chk1 also regulates HRR (homologous recombination repair), as DNA damage-induced HRR is dependent on Chk1-mediated Rad51 phosphorylation (7–8). In addition, Chk1 functions to stabilize stalled replication forks, induce the mitotic spindle checkpoint (in the presence of mitotic spindle defects) (9), and inhibit caspase-3-mediated apoptosis in response to genotoxic stress (10). Previous work from our and other laboratories has shown that inhibition of Chk1 with AZD7762 sensitizes pancreatic cancer cells and xenografts to gemcitabine and radiation through mechanisms involving both inhibition of cell cycle arrest and inhibition of homologous recombination repair (4, 11–12).
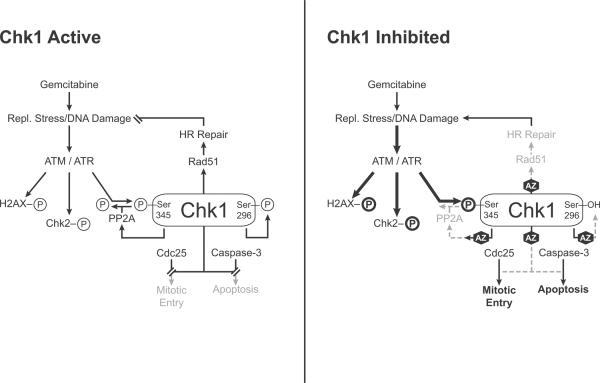
Based on these known functions of Chk1, several possible pharmacodynamic responses would be predicted to be affected by Chk1 inhibition (Fig. 1). We have reported that Chk1 inhibition results in both normal and premature mitotic entry in response to gemcitabine thus resulting in increased phosphorylated histone H3, a marker of mitosis (13). Others have demonstrated that caspase-3 cleavage occurs in response to gemcitabine and Chk1 inhibition (5). In addition, Chk1 inhibition in combination with gemcitabine results in increased DNA damage as evidenced by impairment of homologous recombination repair, ATM-mediated γH2AX induction, as well as Chk1 and Chk2 phosphorylation (11, 14). In response to DNA damage, ATR phosphorylates Chk1 at two established sites, S345 and S317, thus prompting autophosphorylation at S296. We and others observed that pS345 Chk1 (phosphorylated S345 Chk1) is increased in response to Chk1 inhibition (11, 15–16) and there are at least two potential mechanisms through which this may occur. The protein phosphatase, PP2A regulates dephosphorylation of Chk1 and has been reported to be, in part, dependent on Chk1 kinase activity (15). Thus, Chk1 inhibitors could cause an accumulation of pS345 Chk1 as a consequence of PP2A inhibition, occurring secondary to the lack of Chk1 kinase activity. Another possible mechanism for the induction of pS345 Chk1 in response to Chk1 inhibition is through an increase in DNA damage that further amplifies ATR/ATM-mediated Chk1 phosphorylation (17).
Figure 1. Chk1 signaling in response to replication stress and Chk1 inhibition.
In response to replication stress/DNA damage, ATR phosphorylates Chk1 at S345, leading to Chk1 autophosphorylation at S296. Phosphorylation of Chk1 at S345 is counterbalanced by PP2A phosphatase activity, which is in part dependent on Chk1 kinase activity. In addition, ATM mediates phosphorylation of Chk2 and H2AX in response to DNA damage. Active Chk1 inhibits Cdc25 phosphatases, inhibits caspase-3, and induces Rad51 focus formation, ultimately resulting in cell cycle arrest, cell survival, and HRR. In response to replication stress, in the presence of a Chk1 inhibitor, such as AZD7762 (AZ), Chk1 kinase activity and autophosphorylation are inhibited. Inhibition of Chk1 results in Cdc25 phosphatase activity, caspase-3 activation (under some conditions), and inhibition of Rad51 focus formation as well as HRR, ultimately resulting in mitotic entry (marked by pS10 histone H3), apoptosis (under some conditions), and unrepaired DNA damage. As a result of Chk1 inhibition, DNA damage accumulates which in turn amplifies ATM/ATR signaling leading to an increase in phosphorylation of H2AX, Chk1 (S345), and Chk2 (T68). In addition, the increase in pS345 Chk1 is also regulated, in part, by PP2A. Gray color indicates inhibited processes, bold indicates amplified signaling.
In order to maximize the potential clinical efficacy of Chk1 inhibitors, we sought to identify potential pharmacodynamic biomarkers as well as the optimum dosing schedule of gemcitabine and AZD7762. We found that a dosing schedule of gemcitabine followed by AZD7762 was optimal and produced significant gemcitabine-sensitization in both in vivo and in vitro pancreatic tumor models. We then went on to test a panel of potential biomarkers of gemcitabine and AZD7762 activities, and identified pS345 Chk1 as being most consistently increased in response to gemcitabine and AZD7762. We validated pS345 Chk1 as a pharmacodynamic biomarker of gemcitabine and AZD7762 in pancreatic tumor xenografts as well as in normal surrogate tissues. Finally, we determined the contributions of DNA damage and PP2A to the mechanisms of pS345 Chk1 induction in response to gemcitabine and Chk1 inhibition.
Materials and Methods
Cell culture and drug solutions
MiaPaCa-2 cells were obtained from American Type Culture Collection and grown in DMEM supplemented with 10% fetal bovine serum (Invitrogen) and 2 mmol/L L-glutamine (Sigma). Experiments were conducted on exponentially growing cells. Cells were tested for mycoplasma once every 3 months. Gemcitabine (Eli Lilly) was dissolved in PBS. AZD7762 was dissolved in DMSO or 11.3% 2-hydroxypropyl-β-cyclodextrin (Sigma), 0.9% sterile saline for in vitro or in vivo purposes, respectively. Okadaic acid (Sigma) was dissolved in DMSO. Clonogenic survival assays were conducted as previously described (18–19).
Flow cytometry
For γ-H2AX analysis, samples were processed as previously described (20). Samples were analyzed on a FACScan flow cytometer (Becton Dickinsson) with FlowJo software (Tree Star).
Immunoblotting
Cell pellets or pulverized frozen tumors were lysed and immunoblotted as previously described (19). Proteins were detected with pS345 Chk1, pS296 Chk1, pT68 Chk2, pY15 Cdk1, caspase-3, GAPDH (Cell Signaling), Chk2 (Millipore- in vitro; Cell Signaling- in vivo), Cdc25A (F-6, Santa Cruz- in vitro; Cell Signaling- in vivo) Chk1 (Santa Cruz- in vitro; Cell Signaling- in vivo), or pS10 histone H3 (Millipore) antibodies.
Immunohistochemistry
Harvested tumors or tissue sections were fixed in 10% neutral buffered formalin for 24 hours, then embedded in paraffin blocks and sectioned at 5 microns onto slides. Histopathology was conducted using Hematoxylin and Eosin staining and immunohistochemistry using pS345 Chk1 or pS139 H2AX antibodies (Cell Signaling), biotinylated rabbit secondary antibody, SA-HRP complex, and DAB chromogen kit (Ventana). Positive rodent control slides showed strong nuclear staining and negative control slides (Rabbit IgG) showed levels of non-specific staining, if any. Tumors were microscopically evaluated with a 20× objective to assess morphological changes and results were reported by a pathologist. Slide images were produced on an Olympus IX71 microscope with a 60× objective. H-score was determined by assigning a score of 0 – 4, based on the percentage of cells staining positive in a field where 0 = no positive cells, 1 = 1 – 25% positive, 2 = 26 – 50%, 3 = 51 – 75%, and 4 = 76 – 100%, and then multiplying this value by the staining intensity score (1 – 3, where 1 represents weak staining and 3, intense staining). The maximum H-score value is 12.
In vivo studies
Animals were handled according to a protocol approved by the University of Michigan Committee for Use and Care of Animals. MiaPaCa-2 cells or patient derived pancreatic tumor cells (5×106 or 1×106, respectively) were suspended in a 1 : 1 mixture of 10% FBS/RPMI: Matrigel (BD Biosciences) and injected subcutaneously into the flanks of athymic nude or Nodscid mice, respectively. Samples of human pancreatic adenocarcinomas were handled as described previously (21). Treatment was initiated when the average tumor volume reached 100 mm3. For tumor growth delay studies, the tumor size was measured 2 times/week. Tumor volume (TV) was calculated according to the equation: TV = π/6 (ab2), where a and b are the longer and shorter dimensions of the tumor, respectively (19). Measurements were made until the tumor volume increased by approximately a factor of ten. For normal tissue studies, Balb/C or NCr athymic nude mice were utilized.
Combined drug effect analysis
To examine synergy between gemcitabine and AZD7762, survival was determined in response to a fixed ratio of variable concentrations of gemcitabine (50nM–500nM) and AZD7762 (20nM–400nM) and analyzed by the median effect analysis as described previously (22–23).
Statistical analyses
For in vivo tumor growth, tumor volume doubling was determined for each xenograft by identifying the earliest day on which it was at least twice as large as on the first day of treatment. A cubic smoothing spline (SMOOTH.SPLINE function in R) was used to obtain the exact time of doubling, and the Kaplan-Meier method was used to analyze the doubling times derived from the smoothed growth curves. Log rank test (PROC LIFETEST in SAS) was used for comparisons between any two treatment groups. A Student's t-test was used for other analyses.
Results
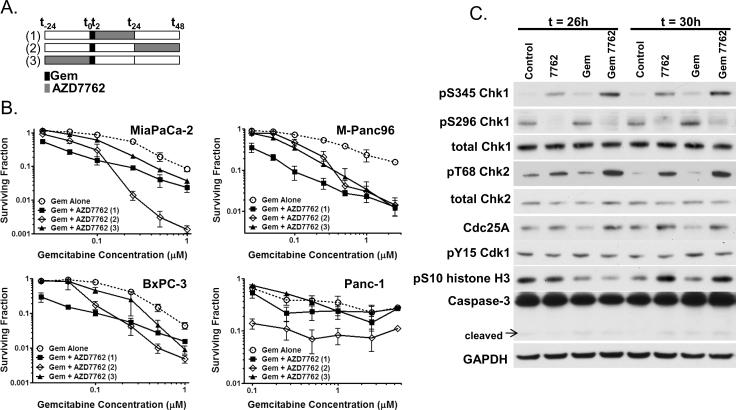
Several recent studies have demonstrated that Chk1 inhibitors sensitize solid tumors to gemcitabine-induced cytotoxicity (4–5, 8, 24). Little has been done, however, to address the issue of optimal scheduling for chemosensitization. We therefore assessed the ability of AZD7762 to sensitize to gemcitabine in a panel of pancreatic cancer cell lines, under three different treatment schedules: AZD7762 during and after (1), 24 hours after (2), or preceding gemcitabine treatment (3) (Fig. 2A). The presumption has been that checkpoint inhibitors should be most effective when given during the time at which cells are arresting at a particular checkpoint. In order to simplify the analysis, we used the maximum dose of AZD7762 which did not produce toxicity by itself (100 nM; Suppl. Fig. 1). We found at low, relatively non-toxic concentrations of gemcitabine that AZD7762 was most effective when present during and immediately following gemcitabine treatment (Schedule 1), producing 4.5 – 6-fold sensitization to a previously nontoxic concentration of gemcitabine (50 nM) (Fig. 2B). At higher concentrations of gemcitabine, AZD7762 was a better chemosensitizer if given 24 hours after gemcitabine treatment (Schedule 2), when the cells were arrested in early S-phase (8). Consistent with the hypothesis that checkpoint inhibition would be most effective when given during cell cycle checkpoint induction, treatment with AZD7762 before gemcitabine (Schedule 3) was the least effective of the schedules tested. Since the greatest extent of gemcitabine sensitization was seen in MiaPaCa-2 cells treated on Schedule 2, we utilized this schedule in our subsequent studies. In order to determine whether AZD7762 and gemcitabine were synergistically affecting cell survival on Schedule 2, we determined the combination indices by median effect analysis (22–23) by using a fixed ratio of AZD7762 and gemcitabine (1:2.5) in MiaPaCa-2 cells. We found that the combination index was significantly less than 1 (P<0.05) at surviving fractions of 0.3 and below (Suppl. Fig. 2) indicating that AZD7762 in combination with gemcitabine produces synergistic cytotoxicity.
Figure 2. AZD7762 sensitizes pancreatic cancer cells to gemcitabine and affects cell cycle checkpoints.
A, Schedule of treatments. B, MiaPaCa-2, M-Panc96, BxPC-3, and Panc-1 cells were treated with gemcitabine (0 – 5 uM) and AZD7762 (100nM) according to the schedule illustrated (A). At the end of the treatment period cells were processed for clonogenic survival. C, MiaPaCa-2 cells were treated with gemcitabine (50 nM) and AZD7762 (100 nM) according to treatment schedule 2. Cells were collected for immunoblotting at t = 26 and 30 hours. The surviving fraction was normalized to the control plating efficiency for `Gem Alone' samples and to the AZD7762 plating efficiency for `Gem + AZD7762' samples for each schedule (B). Data are the mean of n = 3 – 6 independent experiments (B) or are representative of 3 independent experiments (C).
In order to determine potential biomarkers of AZD7762 activity in combination with gemcitabine, we evaluated the known targets of AZD7762 (Chk1 and Chk2), as well as several other potential biomarkers (Fig. 1). In MiaPaCa-2 cells treated on Schedule 2, we found that phosphorylation of Chk1 at S345 was increased in response to gemcitabine or AZD7762 as single agents (Fig. 2C) consistent with activation of the DNA damage response pathway. More importantly the combination of gemcitabine and AZD7762 led to a marked increase in pS345 Chk1. Similarly, the combination of gemcitabine and AZD7762 led to an increase in Chk2 phosphorylation (pT68 Chk2). As anticipated, the ability of Chk1 to undergo autophosphorylation (pS296 Chk1) was inhibited by AZD7762 both in the presence and absence of gemcitabine, indicating that Chk1 kinase activity was inhibited by AZD7762. Consistent with Chk1 activity being inhibited by AZD7762, Cdc25A degradation in response to gemcitabine was inhibited by AZD7762. Phosphorylated Cdk1 (pY15 Cdk1) was minimally affected under these treatment conditions. However, we did observe an increase in the mitotic marker, phosphorylated histone H3 (pS10 histone H3, t=30h), in response to gemcitabine plus AZD7762 relative to gemcitabine alone, indicating abrogation of gemcitabine-mediated cell cycle arrest by AZD7762. In addition, AZD7762 alone produced an increase in phosphorylated histone H3, indicating increased mitotic entry. Finally, since cleaved caspase-3 may be a marker of chemosensitization by Chk1 inhibitors (5), we investigated caspase-3 activation. We did not find that AZD7762 and/or gemcitabine affected caspase-3 activation under the conditions tested, although at later time points with higher concentrations of gemcitabine, we did observe caspase-3 cleavage (data not shown). Based on the magnitude of the effect of gemcitabine and AZD7762 on our panel of potential biomarkers, these data warranted further investigation of pS345 Chk1, pS296 Chk1, and pT68 Chk2.
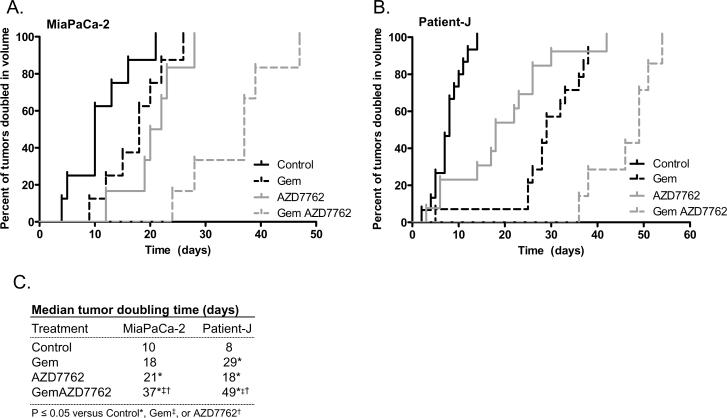
We next tested pancreatic model systems for the in vivo efficacy of AZD7762 as a chemosensitizer. We treated mice bearing MiaPaCa-2-derived subcutaneous xenografts with gemcitabine and AZD7762. Both gemcitabine and AZD7762 demonstrated single agent activity against tumor growth, as evidenced by significant delays in the time to until tumor volume doubling relative to untreated tumors (Δ = 8 and 11 days, gemcitabine and AZD7762, respectively) (Figs. 3A, C, P<0.05). The combination of gemcitabine and AZD7762 was tolerable (Suppl. Table 1) and produced a significant growth delay relative to either gemcitabine or AZD7762 alone (Δ = 19 and 16 days, respectively). Furthermore, in a second in vivo pancreatic tumor model derived from early-passage patient-derived tumors (designated Patient-J), gemcitabine or AZD7762 produced significant tumor growth inhibition evidenced by delays in the time required for tumor volume doubling relative to untreated controls (Δ = 21 and 10 days, respectively) (Figs. 3B–C, P<0.05). The combination of gemcitabine with AZD7762 further delayed tumor growth beyond that induced by gemcitabine or AZD7762 alone (Δ = 20 and 31 days, respectively, P<0.05), which appeared to be a greater than additive effect.
Figure 3. AZD7762 sensitizes pancreatic tumor xenografts to gemcitabine.
A, Mice bearing MiaPaCa-2 xenografts were treated with gemcitabine (120 mg/kg) on days 0, 7, and 14 and AZD7762 (25 mg/kg) on days 0, 1, 7, 8, 14, and 15. B, Alternatively, Patient J-derived xenografts were treated with gemcitabine (60 mg/kg) on days 0, 3, 7, 10, 14, and 17 and AZD7762 (20 mg/kg) on days 0, 1, 3, 4, 7, 8, 10, 11, 14, 15, 17, and 18. Data are expressed as the portion of tumors doubled in volume (A–B) or the median time required for tumor volume doubling (P<0.05) (C). Each treatment group contained 6–8 (A) or 10–20 (B) tumors.
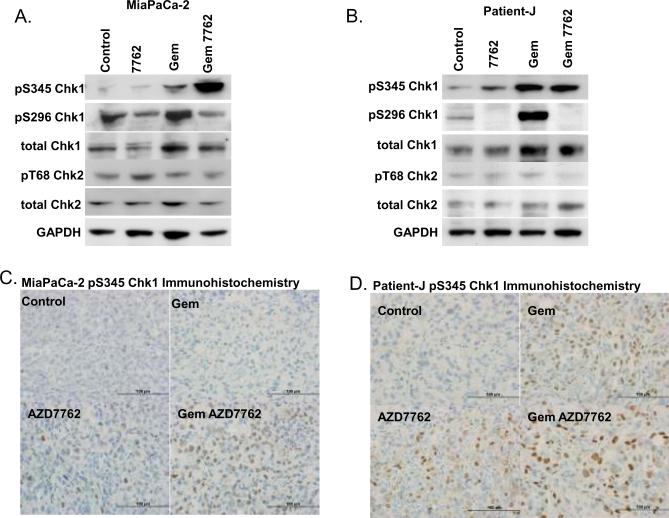
In order to assess potential biomarkers of AZD7762 and gemcitabine activity, we treated mice with gemcitabine and AZD7762, and then monitored pS345 Chk1, pS296 Chk1, pT68 Chk2, and γH2AX, as potential response markers. Consistent with our in vitro observations, we found S345 Chk1 phosphorylation to be increased in response to gemcitabine but to be markedly increased in response to gemcitabine and AZD7762 in MiaPaCa-2 tumors (Fig. 4A, Suppl. Fig. 3). Similarly, the combination of gemcitabine plus AZD7762 increased pS345 Chk1 in Patient-J-derived tumors, however gemcitabine alone produced an equivalent effect on pS345 Chk1 (Fig. 4B, Suppl. Fig. 4). Chk1 autophosphorylation (pS296) was inhibited in MiaPaCa-2 and Patient-J tumors following AZD7762 treatment. In contrast to our in vitro observations, pT68 Chk2 was not affected by gemcitabine and/or AZD7762 under these treatment conditions. Consistent with results obtained by immunoblotting, immunohistochemical detection of pS345 Chk1 revealed increased nuclear staining in response to gemcitabine plus AZD7762, with more subtle effects in response to the single agents (Figs. 4C–D, Suppl. Table 2). pS296 Chk1 immunohistochemistry produced high background staining and results inconsistent with immunoblotting (data not shown) which precluded further investigation of S296 Chk1. In addition, we found γH2AX staining to be increased in the MiaPaCa-2 tumors only in response to gemcitabine plus AZD7762, while γH2AX was increased similarly in response to gemcitabine and AZD7762, either alone or in combination, in Patient-J xenografts (Suppl. Table 2). Taken together these data demonstrate that AZD7762 sensitizes pancreatic tumor xenografts to gemcitabine, a result most consistently marked by an increase pS345 Chk1.
Figure 4. AZD7762 increases pS345 Chk1 in response to gemcitabine.
MiaPaCa-2 xenografts were treated with gemcitabine (120 mg/kg) on day 0 and AZD7762 (25 mg/kg) on days 0 and 1 (A, C). Alternatively, Patient-J-derived xenografts were treated with gemcitabine (60 mg/kg) on day 0 and AZD7762 (20 mg/kg) on days 0 and 1 (B, D). Tumors were harvested on day 1, following the last dose of AZD7762 and prepared for immunoblotting (A–B) or immunohistochemistry (C–D). Data are from a single experiments representative of 3 – 4 tumors per treatment group.
In order to demonstrate target pathway inhibition with AZD7762, we sought to further develop pS345 Chk1 as a pharmacodynamic biomarker for use in future clinical trials. Since obtaining paired pre- and post-treatment biopsies of pancreatic tumors is not usually feasible in patients, we set out to identify an easily attainable normal tissue which might be used as a surrogate for tumor pS345 Chk1 in response to gemcitabine and AZD7762. Thus we treated mice with gemcitabine and AZD7762 and prepared biopsy specimens of hair follicles as well as colon. We found in both hair follicles (Fig. 5A, C) and colon (Fig. 5B) that pS345 Chk1 immunostaining was increased in response to the combination of gemcitabine plus AZD7762, with little to no staining observed in response to gemcitabine or AZD7762 as single agents. Furthermore, the induction of pS345 Chk1 in hair follicles was dependent on gemcitabine and AZD7762 dose. This is in contrast to the pS345 Chk1 staining observed in matched tumor samples which occurred over a range of doses of gemcitabine and AZD7762, as well as in response to gemcitabine alone (Suppl. Fig. 5). These data demonstrate that pS345 Chk1 induction by gemcitabine and AZD7762 can be detected in normal tissues and suggest that pS345 Chk1 in hair follicles is a reliable surrogate for pS345 Chk1 in tumors.
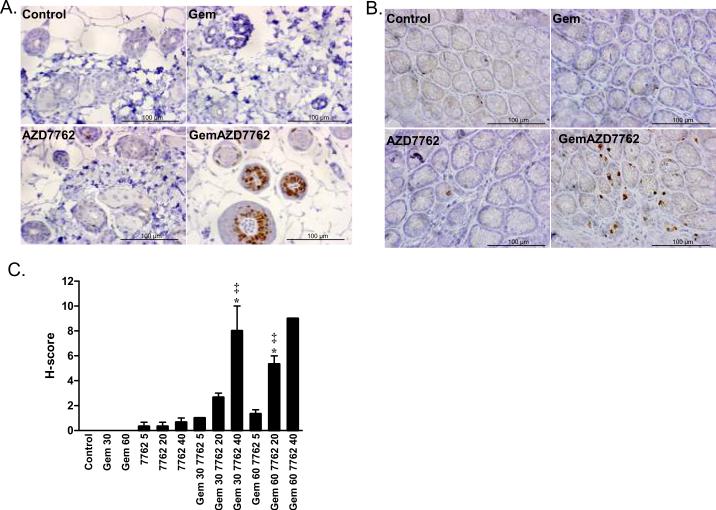
Figure 5. Induction of pS345 Chk1 in hair follicles and colon by gemcitabine and AZD7762.
Mice were treated with gemcitabine (30 – 60 mg/kg) and three hours later with AZD7762 (5 – 40 mg/kg). Three hours post-AZD7762, tissues were harvested and fixed for immunohistochemistry. Images are from single experiment with 60 mg/kg gemcitabine and 20 mg/kg AZD7762, as indicated in hair follicles (A) or colon (B). The mean H-score ± standard error in hair follicles for 2 – 4 animals per treatment condition is shown (C). Statistically significant differences from control* or gemcitabine‡ are indicated (P<0.05) and were determined by one-way ANOVA.
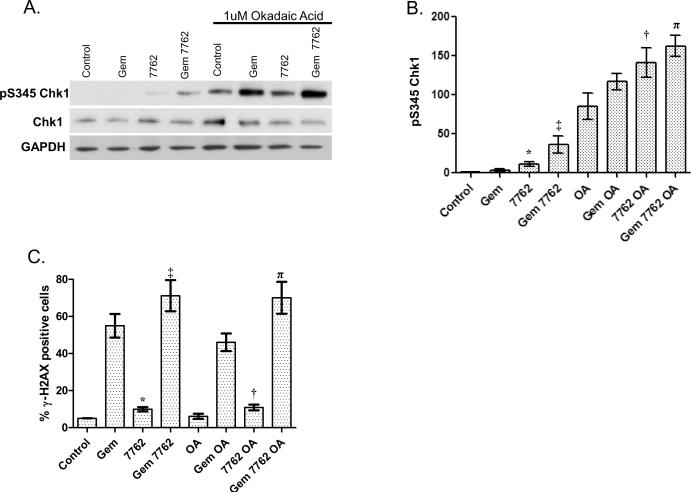
In order to better characterize pS345 Chk1 induction in response to gemcitabine and Chk1 inhibition and thus improve its usefulness as a pharmacodynamic biomarker, we investigated the mechanisms contributing to pS345 Chk1 accumulation. There are at least two possible mechanisms by which this may occur. Chk1 inhibition has been shown to inhibit HRR and cell cycle checkpoints, thus leading to increased DNA damage which could form a feedback loop with ATR/ATM, resulting in further ATR/ATM-mediated phosphorylation of Chk1 at S345 (Fig. 1) (11, 14, 16). Alternatively, Chk1 inhibition has been shown to result in inhibition of the Chk1 phosphatase, PP2A, thus leading to an accumulation of pS345 Chk1(15). In order to distinguish between these two possible mechanisms we treated MiaPaCa-2 cells with okadaic acid, an inhibitor of the PPP family of protein phosphatases including PP2A. We hypothesized that if the increase in pS345 Chk1 in response to AZD7762 were mediated by PP2A, then, in the presence of okadaic acid, AZD7762 would produce no additional effect on pS345 Chk1. Conversely, if the increase in pS345 Chk1 were mediated by increased DNA damage, then, AZD7762 would still increase pS345 Chk1, even in the presence of okadaic acid. We found that baseline pS345 Chk1 was increased in response to okadaic acid (Fig. 6A–B). More interestingly, in the presence of okadaic acid, AZD7762 significantly increased pS345 Chk1 (OA vs. 7762 OA). In addition, in the presence of okadaic acid and gemcitabine, AZD7762 produced a small, but reproducible increase in pS345 Chk1 (Gem OA vs. Gem 7762 OA). Although AZD7762 did increase pS345 Chk1 in the presence of okadaic acid, the magnitude of the effect was less than in the absence of okadaic acid. To further assess the potential role of DNA damage in AZD7762-mediated pS345 Chk1 induction, we analyzed γH2AX, a marker of DNA damage. We found that AZD7762 caused an increase in the percentage of γH2AX positive cells in the presence of okadaic acid, with or without gemcitabine (Fig. 6C). Taken together, these data support the conclusion that, although the primary cause of the increase in pS345 Chk1 in response to AZD7762 with gemcitabine is increased in DNA damage, PP2A inhibition also contributes to the induction.
Figure 6. pS345 Chk1 induction occurs independent of PP2A and correlates with increased DNA damage response.
MiaPaCa-2 cells were treated according to Schedule 2 (Fig. 1A). At t = 24 hours cells were treated with AZD7762 and/or okadaic acid (OA, 1uM) for 1 hour. At the end of treatment, cells were prepared for immunoblotting (A, B) or FACS analysis (C). Data are from a single experiment (A) or are the mean of 3 – 4 independent experiments (B–C). Statistically significant differences (P<0.05) were determined by a paired t-test comparing control versus 7762*, Gem versus Gem 7762‡, OA versus 7762 OA or Gem OA†, versus Gem 7762 OAπ.
Discussion
In this study we demonstrated that AZD7762 sensitizes pancreatic cancer cells and tumors to gemcitabine in a schedule dependent manner, and this correlated directly with pS345 Chk1 induction. The optimal dosing schedules of AZD7762 and gemcitabine were those in which AZD7762 is given during and after (1) or after (2) gemcitabine exposure. We also found that gemcitabine treatment followed by AZD7762 inhibited tumor growth in in vivo pancreatic tumor xenografts. In addition, of the many potential biomarkers we evaluated, pS345 Chk1 was found to be the most robust and reliable biomarker of gemcitabine and AZD7762 activity. Together these data support the clinical investigation of AZD7762 with gemcitabine in pancreatic cancer under a dosing schedule in which gemcitabine is administered concurrent with or prior to AZD7762 and in conjunction with skin biopsies to measure pS345 Chk1 as a pharmacodynamic biomarker of AZD7762 and gemcitabine activity.
Our data demonstrates that S345 Chk1 phosphorylation is elevated in response to gemcitabine and AZD7762 in both tumor and normal tissues. While a response in a normal tissue surrogate does not necessarily equate to a response in a tumor, it is at minimum informative as to whether appropriate concentrations of drug were obtained to achieve target inhibition as well as a biological response. In our present and previously published studies we observed S345 Chk1 phosphorylation in tumor cells over a range of gemcitabine doses and time points (11). In contrast, in normal tissues pS345 Chk1 appears to be a fairly rapid and short lived response that is sensitive to the gemcitabine and AZD7762 doses. These findings suggest that pS345 Chk1 is a much more robust response in tumor than in normal tissue, which is consistent with the selective toxicity against tumors observed in our animal model. The differences between the normal and tumor tissues (both p53 wild type and mutant) could be attributable to multiple other defects present in tumors (K-Ras, p53, and p16) which make them more susceptible to DNA damage by Chk1 inhibition and thus increased pS345 Chk1 (4, 25). Taken together, these data imply that if we observe the induction of pS345 Chk1 in normal tissue, it would likely indicate that pS345 Chk1 is being induced in tumor tissue. In addition, it seems possible that anti-tumor effects could occur even in the absence of normal tissue induction of pS345 Chk1.
There are two potential mechanisms through which pS345 Chk1 may accumulate in response to Chk1 inhibition. Induction of S345 Chk1 phosphorylation in response to Chk1 inhibitors has been shown to be mediated by PP2A, independent of γH2AX induction (Fig. 1)(15). Others have shown that induction of Chk1 phosphorylation and γH2AX in response to Chk1 inhibition is ATR- and ATM- dependent (16–17), suggesting that DNA damage also plays a role in pS345 Chk1 accumulation. Our previous data demonstrated that AZD7762 either alone or in combination with gemcitabine caused an increase in pS345 Chk1 which was accompanied by an increase in γH2AX (11). Thus, we sought to determine the contributions of PP2A and DNA damage to S345 Chk1 phosphorylation in our model system. Since we found that AZD7762 increased pS345 Chk1, even when PP2A was inhibited, an effect associated with induction of γH2AX, we conclude that DNA damage does contribute to pS345 Chk1 induction. However, because the magnitude of the effect of AZD7762 on pS345 Chk1 was greater in the absence of okadaic acid, it is likely that while PP2A inhibition by AZD7762 may also play a role in maintaining pS345 Chk1 levels. While these findings support the model that both PP2A, as well as increased DNA damage, contribute to pS345 Chk1 induction in response to Chk1 inhibition, in the present study it seems that DNA damage is the predominate mechanism of pS345 Chk1 induction. Furthermore, it is likely that the relative contributions of these two mechanisms to pS345 Chk1 accumulation vary in different cell types and under different conditions.
Given the finding that pS345 Chk1 induction in response to Chk1 inhibition is mediated by DNA damage, it seems plausible that γH2AX would also be a biomarker of response to Chk1 inhibition. Certainly, γH2AX has been demonstrated to be a useful pharmacodynamic biomarker of DNA damage and is being used in a number of clinical trials (26). However, in our present study, γH2AX did not demonstrate a clear a relationship with chemosensitization or the likely extent of DNA damage in tumor specimens. It is possible that γH2AX focus formation rather than immunohistochemical staining would have produced a more reliable biomarker of response to Chk1 inhibition. This however, would have required the use of fresh rather than fixed tissue specimens, thus limiting the feasibility for application in future clinical specimens.
Since AZD7762 is an inhibitor of both Chk1 and Chk2, it is possible that Chk2 inhibition may play a role in AZD7762-mediated chemosensitization. Several pieces of evidence however, suggest that sensitization is mediated by Chk1 inhibition. In our own studies and those of others, siRNA-mediated depletion of Chk1 but not Chk2 produced sensitization to gemcitabine as well as other DNA damaging agents (11, 13, 27). In addition, other small molecule Chk inhibitors which are 100-fold more selective for Chk1 over Chk2, such as PD-321852 and PF-00477736, produced chemosensitization (5, 8). On the other hand, there is emerging evidence supporting that Chk2 inhibition may play a role in chemosensitization (28), and small molecule inhibitors selective for Chk2 are being developed for clinical use (6). It will be important in future studies to evaluate the contributions of Chk1 and Chk2 inhibition by assessing the efficacy of selective Chk1 inhibitors.
Although Chk1 inhibitors have been developed with the goal that they could be used to selectively sensitize p53 mutant tumors to DNA damaging agents, reports of single agent activity are beginning to emerge (4, 29–31). In the present study it is noteworthy that we observed single agent activity by AZD7762 with regard to several endpoints including pS345 Chk1, tumor growth, γH2AX, and pS10 histone H3. These observations are supported by our previously published studies demonstrating that AZD7762 alone induces γH2AX, results in more rapid cell cycle progression, inhibits HRR, and delays tumor growth (11). The mechanism underlying this single agent activity is not known but it has been hypothesized that cancer cells which express oncogenes, harbor endogenous DNA damage, and contain defective checkpoint/repair pathways, require Chk1 activity for otherwise unperturbed cell proliferation (17). A better understanding of the single agent activity of Chk1 inhibitors will be important in order to optimize their combination with cytotoxic agents and radiation.
The development of biomarkers, either genetic or pharmacodynamic, is essential to the clinical success of all new molecularly targeted therapies. Our finding that pS345 Chk1 is a pharmacodynamic biomarker of Chk1 inhibition, at least in part mediated by an increase in DNA damage, suggests that pS345 Chk1 could be a useful biomarker for many other novel molecularly targeted agents. Of particular interest, pS345 Chk1 should be investigated as a potential biomarker of response to small molecule inhibitors targeted to DNA damage response and repair pathways such as Chk1, Chk2, and PARP. It will be important in future studies to validate pS345 Chk1 as biomarker of response to other agents which exacerbate DNA damage.
Statement of translational relevance.
One approach to improve the efficacy of current treatment regimens is by incorporating small molecule inhibitors of Chk1 with cytotoxic agents, such as gemcitabine. While Chk1 inhibitors are currently in Phase I clinical trials in multiple advanced malignancies, it is likely that pancreatic cancer will be a focus of upcoming clinical studies. In order to design the most promising clinical trials, issues such as schedule of dosing as well as potential biomarkers of response need to be addressed in preclinical models. In this study, we address both of these key issues, and provide recommendations for clinical trial design which include administration of gemcitabine either concurrent with or followed by Chk1 inhibition, as well as the use of phosphorylated S345 Chk1 as a pharmacodynamic biomarker in skin punch biopsies.
Supplementary Material
Acknowledgements
We would like to thank Barbara Davis, Deborah Lawson, Adam Sheehy, and Graham Betton for their contributions to the immunohistochemical studies presented in this work.
Grant support: This work was funded by NIH Grants R01CA78554, R01CA78554 10S1, R01CA138723, P50CA130810,Cancer Center Core Grant P30 CA046592, and AstraZeneca.
Abbreviations
- AZD7762 or 7762
AstraZeneca Drug 7762
- Chk
checkpoint kinase
- HRR
homologous recombination repair
- pS345 Chk1
phosphorylated S345 Chk1
Footnotes
Disclosure of potential conflicts of interest: L.H.-G., D.M., and J.L.B. are employees of AstraZeneca. S.D.Z. was an employee of AstraZeneca and is currently an employee of Pfizer.
References
- 1.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–6. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 2.Conroy T, Desseigne F, Ychou M, et al. Randomized phase III trial comparing FOLFIRINOX (F: 5FU/leucovorin [LV], irinotecan [I], and oxaliplatin [O]) versus gemcitabine (G) as first-line treatment for metastatic pancreatic adenocarcinoma (MPA): Preplanned interim analysis results of the PRODIGE 4/ACCORD 11 trial. J Clin Oncol. 2010;28:4010. Meeting Abstracts. [Google Scholar]
- 3.Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res. 2009;15:6314–20. doi: 10.1158/1078-0432.CCR-09-0096. [DOI] [PubMed] [Google Scholar]
- 4.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–66. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 5.Blasina A, Hallin J, Chen E, et al. Breaching the DNA damage checkpoint via PF-00477736, a novel small-molecule inhibitor of checkpoint kinase 1. Mol Cancer Ther. 2008;7:2394–404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- 6.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen CS, Hansen LT, Dziegielewski J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 8.Parsels LA, Morgan MA, Tanska DM, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zachos G, Black EJ, Walker M, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–60. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan MA, Parsels LA, Zhao L, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–81. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell JB, Choudhuri R, Fabre K, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–84. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan MA, Parsels LA, Parsels JD, Lawrence TS, Maybaum J. The relationship of premature mitosis to cytotoxicity in response to checkpoint abrogation and antimetabolite treatment. Cell Cycle. 2006;5:1983–8. doi: 10.4161/cc.5.17.3184. [DOI] [PubMed] [Google Scholar]
- 14.McNeely S, Conti C, Sheikh T, et al. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle. 2010;9:995–1004. doi: 10.4161/cc.9.5.10935. [DOI] [PubMed] [Google Scholar]
- 15.Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol. 2006;26:7529–38. doi: 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell C, Park MA, Eulitt P, Yang C, Yacoub A, Dent P. PARP1 modulates the lethality CHK1 inhibitors in carcinoma cells. Mol Pharmacol. 2010 doi: 10.1124/mol.110.067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syljuasen RG, Sorensen CS, Hansen LT, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence TS. Ouabain sensitizes tumor cells but not normal cells to radiation. Int J Radiat Oncol Biol Phys. 1988;15:953–8. doi: 10.1016/0360-3016(88)90132-0. [DOI] [PubMed] [Google Scholar]
- 19.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin Cancer Res. 2008;14:5142–9. doi: 10.1158/1078-0432.CCR-07-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Halicka HD, Darzynkiewicz Z. Detection of histone H2AX phosphorylation on Ser-139 as an indicator of DNA damage (DNA double-strand breaks) Curr Protoc Cytom. 2004;Chapter 7(Unit 7):27. doi: 10.1002/0471142956.cy0727s30. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 22.Morgan MA, Meirovitz A, Davis MA, Kollar LE, Hassan MC, Lawrence TS. Radiotherapy combined with gemcitabine and oxaliplatin in pancreatic cancer cells. Translational Oncology. 2008;1:36–43. doi: 10.1593/tlo.07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou T-C, Talalay P. Quantitative analysis of dose-effective relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances Enzyme Res. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 24.Matthews DJ, Yakes FM, Chen J, et al. Pharmacological Abrogation of S-Phase Checkpoint Enhances the Anti-Tumor Activity of Gemcitabine In Vivo. Cell Cycle. 2007;6 doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- 25.Gilad O, Nabet BY, Ragland RL, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redon CE, Nakamura AJ, Zhang YW, et al. Histone {gamma}H2AX and Poly(ADP-Ribose) as Clinical Pharmacodynamic Biomarkers. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrassa L, Broggini M, Erba E, Damia G. Chk1, but not Chk2, is involved in the cellular response to DNA damaging agents: differential activity in cells expressing or not p53. Cell Cycle. 2004;3:1177–81. [PubMed] [Google Scholar]
- 28.Jobson AG, Lountos GT, Lorenzi PL, et al. Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide] J Pharmacol Exp Ther. 2009;331:816–26. doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Xiao Z, Gu WZ, et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119:2784–94. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 30.Gilad O, Nabet B, Durham AC, Brown EJ. ATR checkpoint kinase suppression in combination iwth oncogenic Ras expression synergistically increases genomic instability and potentiates tumorigenesis in mice. 101st Annual Meeting of the American Association for Cancer Research; 2010. Absract 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrao PT, C. C, Raleigh J, Srinivasa S, Levin W, Johnstone RW. Single agent activity of checkpoint kinase inhibitor PF-477736, in a MYC-driven lymphoma model. 101st Annual Meeting of the American Association for Cancer Research; 2010. Abstract 2499. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.