Abstract
Replication of plus-strand RNA viruses depends on lipids present in cellular membranes. Recent genome-wide screens have revealed that eight phospholipid biosynthesis genes affected the replication of Tomato bushy stunt virus (TBSV) in yeast model host. To test the importance of phospholipids in TBSV replication, we studied one of the identified genes, namely INO2, which forms a heterodimer with Ino4, and is a transcription activator involved in regulation of phospholipid biosynthesis. Deletion of INO2, or double deletion of INO2/INO4, reduced TBSV replication and inhibited the activity of the viral replicase complex. In addition, the stability of the viral replication protein is decreased as well as the localization pattern of the viral protein changed dramatically in ino2Δ ino4Δ yeast. Over-expression of Opi1, a repressor of Ino2 and phospholipid biosynthesis, also inhibited TBSV RNA accumulation. In contrast, over-expression of Ino2 stimulated TBSV RNA accumulation. We also observed an inhibitory effect on Flock house virus (FHV) replication and the reduced stability of the FHV replication protein in ino2Δ ino4Δ yeast. These data are consistent with the important role of phospholipids in RNA virus replication.
INTRODUCTION
The host cell’s organellar membranes are efficiently subverted by plus-stranded (+)RNA viruses for their replication (Miller and Krijnse-Locker, 2008). High concentrations of membrane-bound viral proteins and co-opted host proteins lead to the formation of “viral replication organelles” that provide protection against cellular nucleases and proteases (Ahlquist et al., 2003; Denison, 2008; Miller and Krijnse-Locker, 2008; Novoa et al., 2005; Pogany et al., 2008). In addition, the membrane lipids and proteins could also serve as scaffolds for the assembly of the viral replicase complex or they can facilitate the targeting of the viral replication proteins to a particular microdomain in the membrane. Moreover, the subcellular membranes may provide critical lipid or protein cofactors to regulate the function of the viral replicase. Indeed, dynamic remodeling/deforming membranes to give rise to unique structures, called spherules (i.e., invaginations of lipid membranes), is a characteristic feature for many (+)RNA viruses (Barajas, Jiang, and Nagy, 2009; den Boon, Diaz, and Ahlquist, 2010; Kopek et al., 2007; McCartney et al., 2005; Miller and Krijnse-Locker, 2008; Schwartz et al., 2002). These viral-induced spherules serve as sites of viral RNA replication. Importantly, (+)RNA viruses also induce membrane proliferation that requires new lipid biosynthesis. Indeed, several genome-wide screens identified lipid biosynthesis/metabolism genes affecting (+)RNA virus replication (Cherry et al., 2005; Krishnan et al., 2008; Kushner et al., 2003; Panavas et al., 2005b; Serviene et al., 2006).
The best characterized examples of virus-induced modification of cellular lipid metabolism include the recruitment of host enzymes such as PI4PKIIIß, which is involved in phosphatidylinositol-4-phosphate (PI4P) synthesis, to modify the lipid composition of membranes during poliovirus replication (Belov and Ehrenfeld, 2007; Belov et al., 2007; Hsu et al., 2010; Sasvari and Nagy, 2010). Hepatitis C virus (HCV) modulates phospholipid biosynthesis by recruiting PI4PKIIIα, which is also involved in PI4P synthesis, to facilitate the formation of the “membranous-web” (cellular vesicles), which serves as the site of HCV RNA replication (Berger et al., 2009). Dengue virus co-opts FASN, a major rate-limiting enzyme in fatty acid biosynthesis, by retargeting it to the ER membrane, the site of dengue virus replication (Heaton et al., 2010). Another example is Drosophila C virus (picorna-like virus), whose replication was blocked by depletion of the HLH106 regulator of fatty acid metabolism and fatty acid synthase (Cherry et al., 2006). Mutation of Ole1p, which affects the amount of unsaturated fatty acids, reduced the activity of the BMV replicase, and possibly altered the binding of the BMV 1a replication protein to the membrane due to a reduced ratio of unsaturated fatty acids in S. cerevisiae (Lee and Ahlquist, 2003). Infection with West Nile virus (WNV) was shown to result in redistribution of cholesterol from the plasma membrane to the sites of virus replication (Mackenzie, Khromykh, and Parton, 2007). Yet other change in lipid metabolism is induced by Dengue virus, which promotes autophagy and beta-oxidation of lipids released from lipid droplets to generate extra ATP needed for virus replication (Heaton and Randall, 2010).
Tomato bushy stunt virus (TBSV), a tombusvirus, is among the most advanced model RNA viruses regarding characterization of host factors (Nagy, 2008; Nagy and Pogany, 2010). Among the five proteins encoded by the TBSV genome, only p33 replication co-factor, which is an RNA chaperone, and the p92pol RNA-dependent RNA polymerase (RdRp) are essential for TBSV RNA replication (Stork et al., 2011; White and Nagy, 2004). p33 and p92pol are integral membrane proteins with a topography facing the cytosolic surface of the peroxisomes or occasionally ER, the sites of replicase complex formation and viral RNA replication (Jonczyk et al., 2007; McCartney et al., 2005; Pathak, Sasvari, and Nagy, 2008).
Electron microscopic images of cells replicating tombusviruses demonstrated extensive remodeling of membranes (Barajas, Jiang, and Nagy, 2009; McCartney et al., 2005; Navarro et al., 2006). Moreover, genome-wide screens in S. cerevisiae identified 14 host genes involved in lipid biosynthesis and metabolism affecting tombusvirus replication and recombination, suggesting that tombusviruses depend on active lipid biosynthesis (Jiang et al., 2006; Panavas et al., 2005b; Serviene et al., 2006; Serviene et al., 2005). The identified lipid biosynthesis/metabolism genes included 8 genes affecting phospholipid biosynthesis, 4 genes affecting fatty acid biosynthesis/metabolism and 2 genes affecting sterol synthesis (Nagy, 2008). These finding suggest that lipids are likely involved, directly or indirectly, in TBSV replication in yeast. Accordingly, previous studies showed that sterols are critical for TBSV replication (Sharma, Sasvari, and Nagy, 2010).
In order to demonstrate the roles of phospholipids in TBSV RNA replication, we selected INO2 for additional in-depth studies from the pool of identified genes in the genome-wide screens with TBSV (Jiang et al., 2006; Nagy, 2008; Panavas et al., 2005b). Ino2 is a basic helix- loop-helix transcription activator for phospholipid synthesis genes (Block-Alper et al., 2002). The phospholipid biosynthesis in S. cerevisiae is based on biochemical pathways conserved in higher eukaryotes (Carman and Han, 2009; Daum et al., 1998). The expression of phospholipid biosynthesis genes is controlled at the mRNA transcription and stability steps. The expression of many genes involved in phospholipid biosynthesis is controlled by a cis-acting DNA sequence (UASINO); the transcription activator Ino2, which forms a heterodimer with Ino4; and a repressor, named Opi1 (Fig. 1A) (Carman and Han, 2009; Wagner et al., 1999; Wagner et al., 2001). When low amount of phosphatidic acid (PA, a precursor of phospholipids) is present in the ER membrane, then Opi1 is released from the ER membrane and after its translocation into the nucleus, Opi1 binds to Ino2 and represses the mRNA transcription of the phospholipid biosynthesis genes (Fig. 1A) (Carman and Han, 2009; Schaaf et al., 2008; Young et al., 2010).
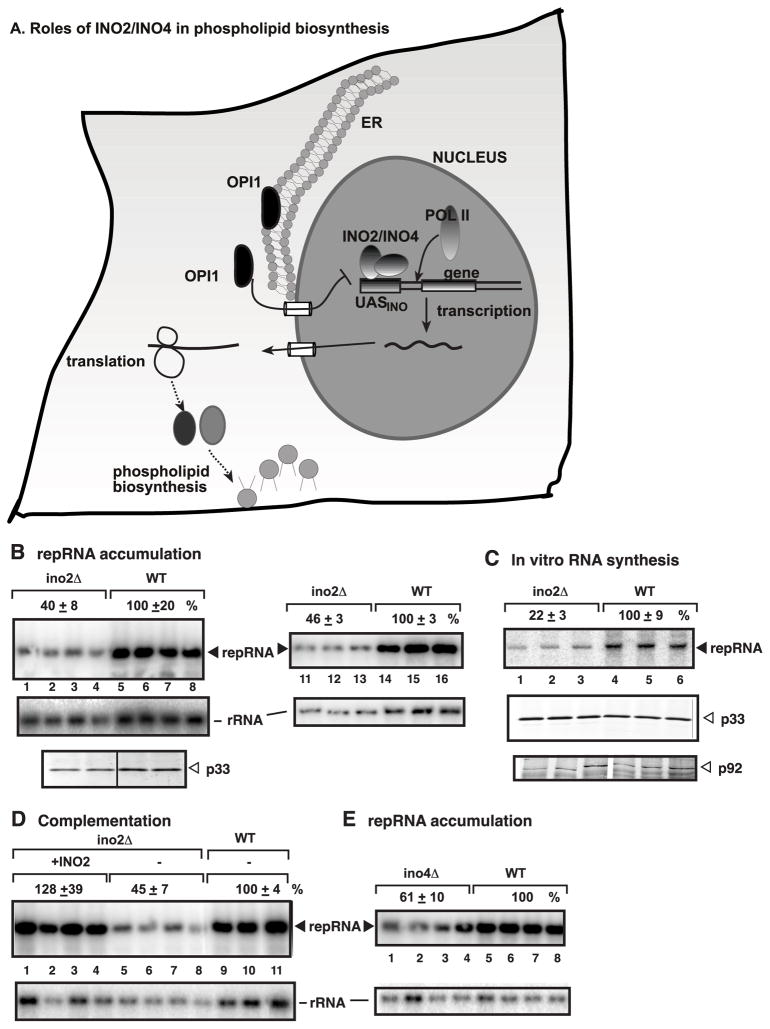
Fig 1.
Deletion of INO2 and INO4 inhibits TBSV repRNA accumulation in yeast. (A) Schematic representation of the regulation of expression of phospholipid biosynthesis genes by Ino2/Ino4 transcription activators and Opi1 repressor. (B) Top panel: Northern blot analysis with a 3' end specific probe was used to detect the accumulation level of the TBSV repRNA in ino2Δ or wt (BY4741) yeast. To launch TBSV repRNA replication, we expressed both 6xHis-p33 and 6xHis-p92 from the ADH1 promoter as well as DI-72(+) repRNA from the galactose-inducible GAL1 promoter from plasmids. Yeast cells were cultured for 24 hours at 23ºC in 2% galactose SC-ULH− media. The accumulation levels of repRNA were calculated using Imagequant software. Middle panel: Northern blot analysis to probe ribosomal rRNA, which was used as a loading control. Bottom panel: Western blot analysis of p33 accumulation using anti-His antibody. (C) Decreased tombusvirus replicase activity in ino2Δ yeast. An in vitro replicase activity assay was performed with membrane-enriched preparations obtained from ino2Δ or wt yeasts grown as in Panel B. The membrane-enriched fraction contains the tombusvirus replicase bound to the endogenous repRNA template that is used during the in vitro replicase assay in the presence of 32P-UTP and the other unlabeled rNTPs. Note that the in vitro activities of the tombusviral replicase were normalized based on p33 (middle panel) levels. Bottom panel shows Western blot analysis of p92 present in the membrane-enriched replicase preparations. (D) Northern blot analysis of TBSV repRNA in ino2Δ yeast complemented with Ino2 expression from the low copy pYC-INO2 plasmid (lanes 1-4), or wt yeast (with the empty pYC plasmid). (E) Northern blot analysis of TBSV repRNA in ino4Δ yeast. See further details in Panel B.
Our approach to test the effect of phospholipid biosynthesis regulators on TBSV RNA replication is justified by the data from the genome-wide screens that INO2 and additional transcription regulators of the phospholipid biosynthesis genes [called SAGA complex and HAT (histone acetyltransferase) complex] have been identified, while single structural genes for phospholipid biosynthesis were not (Jiang et al., 2006; Nagy, 2008; Panavas et al., 2005b). This is expected because two parallel pathways, the de novo and the Kennedy, exist to produce phospholipids in yeast (Nohturfft and Zhang, 2009). Thus, single deletion of structural genes for phospholipid biosynthesis could be partially complemented under the conditions we performed the genome-wide screens (Jiang et al., 2006; Panavas et al., 2005b). However, deletion of INO2 affects both pathways (Carman and Han, 2009; Nohturfft and Zhang, 2009). Accordingly, we demonstrate in this paper that co-deletion of INO2-INO4 leads to a reduced level of TBSV repRNA accumulation in yeast. The tombusvirus replicase complexes isolated from ino2Δino4Δ yeast show poor activity, suggesting that phospholipids are important for the assembly/activity of the tombusvirus replicase. In addition, we demonstrate altered cellular localization of the tombusvirus replication protein in ino2Δino4Δ yeast. Moreover, to expand our findings to other RNA viruses, we also show that the replication of Flock house virus is inhibited in ino2Δino4Δ yeast. Thus, the emerging picture from the current work is that phospholipid biosynthesis is required for efficient replication of some (+)RNA viruses.
RESULTS
Deletion of Ino2 and Ino4 transcription activators in yeast reduces TBSV RNA accumulation and inhibits the viral replicase activity in vitro
To confirm the role of phospholipid biosynthesis in TBSV replication, we tested replication of the TBSV replicon (rep)RNA, which is an efficiently replicating surrogate RNA template derived from the TBSV genomic (g)RNA (Panavas and Nagy, 2003; White and Morris, 1994), in ino2Δ yeast, co-expressing the p33 and p92pol replication proteins and DI-72 repRNA from plasmids. Northern blot analysis revealed ~60% less efficient replication of DI-72 repRNA in ino2Δ yeast in comparison with the parental yeast (Fig. 1B, lanes 1-4 versus 5–8), confirming the data from the previous genome-wide screen (Panavas et al., 2005b). The expression level of p33 also decreased by ~50% in ino2Δ yeast (Fig. 1B), which could be one of the reasons for the reduced accumulation level of the TBSV repRNA in ino2Δ yeast. To test if the activity of the tombusvirus replicase complex was similar in ino2Δ and wt yeast strains, we isolated the membrane-bound tombusvirus replicase from these yeast strains and tested the in vitro template activity on the co-purified endogenous template RNA. These experiments revealed that the tombusvirus replicase showed only 22% activity when obtained from ino2Δ yeast, although the amount of p33/p92pol was adjusted to comparable levels in the membrane fractions (Fig. 1C). The replicase-based in vitro data suggest that the reduced phospholipid synthesis in ino2Δ yeast inhibits the relative activity of the tombusvirus replicase. Thus, phospholipids are likely important for tombusvirus replication.
Additional control experiments showed that plasmid-born INO2 could complement the negative effect of INO2 deletion on TBSV RNA accumulation (Fig. 1D, lanes 1–4). Moreover, deletion of INO4 also reduced TBSV replication significantly (Fig. 1E). This is not surprising since Ino4 is a transcription activator that forms a heterodimer with the Ino2 to control the transcription of phospholipid biosynthesis genes (Carman and Han, 2009; Nohturfft and Zhang, 2009).
Since inhibition of TBSV replication was consistently stronger in ino2Δ yeast than in ino4Δ yeast, we decided to test TBSV replication in a double-deletion yeast strain (ino2Δino4Δ). TBSV repRNA replication was even more debilitated in the double-deletion yeast strain (down to ~25% level Fig. 2A). In addition, we observed consistently reduced levels of p33/p92pol in the double-deletion yeast (Fig. 2A), although both replication proteins were expressed from the constitutive ADH1 promoter. To exclude the possibility that the reduced p33/p92pol levels were due to inhibition of ADH1 promoter-driven transcription, we also tested the accumulation of p33/p92pol when expressed from GAL1 galactose inducible/glucose suppressible promoter (Fig. 2B) or from the copper-inducible CUP1 promoter (Fig. 2C). Interestingly, repRNA replication as well as p33/p92pol levels were also reduced in these yeasts, suggesting that viral RNA replication and reduction in replication protein levels is not promoter-specific (Fig. 2A-C). Testing the tombusvirus replicase activity in the membrane-enriched fraction obtained from ino2Δ ino4Δ yeast revealed that the normalized template activity (after adjustment of p33 to comparable levels in each sample) of the tombusvirus replicase on the endogenous template was low (Fig. 3, lanes 1–2 versus 3–4). Altogether, these data suggest that repRNA accumulation, the normalized activity of the tombusvirus replicase and the level of p33/p92pol decreased markedly in ino2Δino4Δ yeast.
Fig. 2.
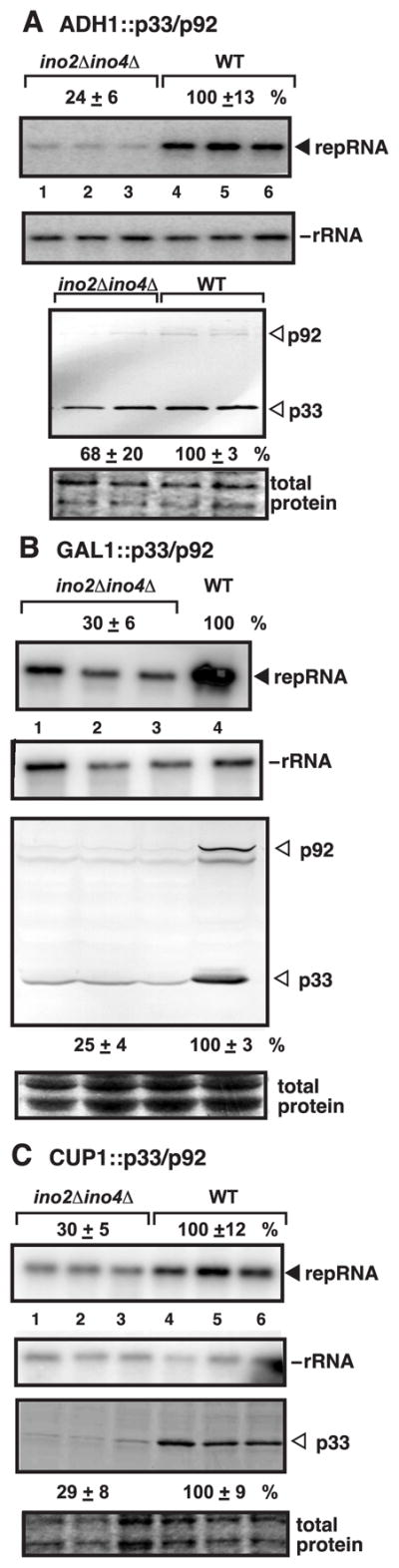
Reduced TBSV repRNA accumulation in ino2Δ ino4Δ yeast. (A) Northern blot analysis of TBSV repRNA in ino2Δ ino4Δ or wt yeast. To launch TBSV repRNA replication, we expressed 6xHis-p33 and 6xHis-p92 from the ADH1 promoter and DI-72(+) repRNA from the galactose-inducible GAL1 promoter. Yeast cells were cultured for 24 hours at 23ºC in 2% galactose SC- ULH− media containing 2mg/L inositol. See further details in Fig. 1B. (B) To induce TBSV repRNA replication, we expressed 6xHis-p33, 6xHis-p92, and DI-72(+) repRNA from the GAL1 promoter. See further details in Fig. 1B. (C) To induce TBSV repRNA replication, we expressed 6xHis-p33 and 6xHis-p92 from the copper-inducible CUP1 promoter and DI-72(+) repRNA from GAL1 promoter. See further details in Fig. 1B.
Fig. 3.
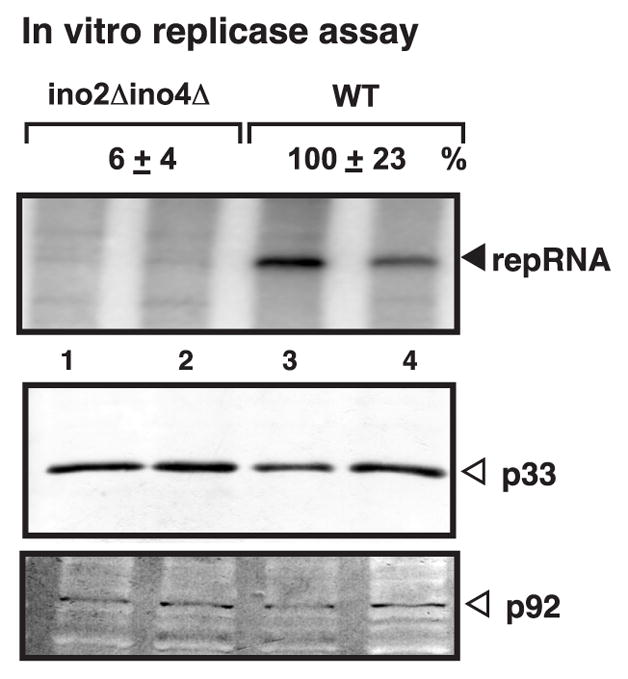
Reduced tombusvirus replicase activity in ino2Δ ino4Δ yeast. TBSV repRNA replication was induced by expressing 6xHis-p33 and 6xHis-p92 from the ADH1 promoter and DI-72(+) repRNA from GAL1 promoter in yeast for 24 hours at 23ºC in 2% galactose SC-ULH- media containing 2mg/L inositol. See further details in Fig. 1C.
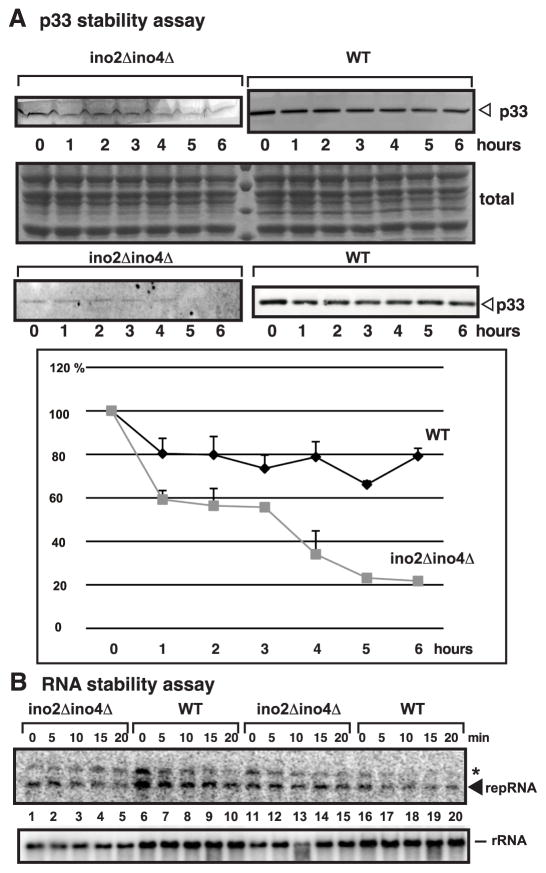
Reduced stability of p33 replication protein in ino2Δino4Δ yeast
To test the stability of p33 replication protein when phospholipid biosynthesis is inhibited, we first expressed p33 from GAL1 promoter for 6 hours, followed by adding cycloheximide (to inhibit new protein synthesis), followed by measuring protein levels (Fig. 4A). We found that the half-life of p33 decreased ~ 2-fold to 3.5 hours in ino2Δino4Δ yeast from more than 6 hours in wt yeast (Fig. 4A). Interestingly, the stability of the repRNA did not change in ino2Δino4Δ yeast (Fig. 4B), suggesting that the primary effect of reduced phospholipid synthesis is on the replication proteins and not on the stability of repRNA.
Fig. 4.
Reduced half-life of p33 replication protein in ino2Δ ino4Δ yeast. (A) Yeast was pre- grown at 29 ºC for 12 hours in SC-U− with 2% glucose, followed by replacing the media with SC-U− with 2% galactose for 6 hours to induce the expression of p33, followed by addition of cycloheximide (100 μg/ml). Samples were collected at the shown time points. The amount of p33 was estimated via Western blotting based on anti-His antibody and ECL-Plus. The images were analyzed by a phosphorimager and quantitated via Imagequant. The experiments were repeated three times (two repeats are shown). The error bars represent the upper half of standard error. (B) Yeast was pre-grown at 29 ºC for 12 hours in SC-U− with 2% glucose, followed by replacing the media with SC-U− with 2% galactose containing 78mg/L inositol for 12 hours at 23 ºC to induce the expression of DI-72(+) repRNA, followed by replacing the media with SC-U− with 2% glucose. Samples were collected at the shown time points. The amount of DI-72(+) repRNA was estimated by Northern blotting. * marks the full-length transcripts (uncleaved) carrying nonviral sequences at the 3’ end, while the repRNA (which was quantified) carries the authentic TBSV 3’ end due to cleavage of the 3’ extension by a ribozyme and is pointed at by a solid arrowhead.
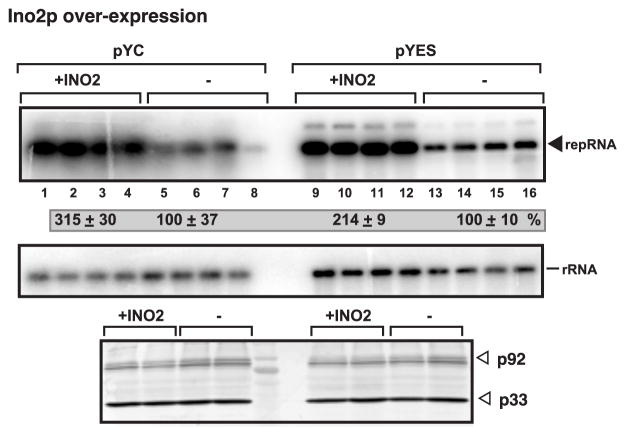
Over-expression of Ino2 increases TBSV RNA accumulation in yeast
Over-expression of Ino2 has been shown to increase the phospholipid synthesis in yeast (Carman and Han, 2009; Nohturfft and Zhang, 2009). We found that over-expression of Ino2 from either a low (Fig. 5, lanes 1–4) or high (lanes 9–12) copy number plasmids led to 2-3-fold increase in TBSV RNA accumulation. The level of p33/p92pol replication proteins did not change in the over-expression strains, suggesting that the increased RNA accumulation was likely due to increased TBSV RNA replication.
Fig. 5.
Over-expression of Ino2 enhances TBSV repRNA replication in yeast. Top panel: Northern blot analysis of TBSV repRNA in BY4741 yeast over-expressing Ino2 from the GAL1 promoter from the low copy pYC-INO2 plasmid (+INO2, lanes 1–4), or from the high copy pYES-INO2 (+INO2, lanes 9–12). The control yeast carried either the empty pYC or pYES plasmids as shown. The expression of Ino2 started 15 hours before launching TBSV repRNA replication, which started by expressing 6xHis-p33 and 6xHis-p92 from the CUP1 promoter and DI-72(+) repRNA from the GAL1 promoter, and continued to the end of the experiment (24 hours of TBSV replication at 29 ºC). We omitted inositol from the growth media. In addition, we used 100 μM BCS for 15 hours before inducing TBSV replication (to prevent leaky transcription from the CUP1 promoter). See further details in Fig. 1B. Bottom panel shows the Western blot analysis of p33 and p92 replication proteins using anti-His antibody.
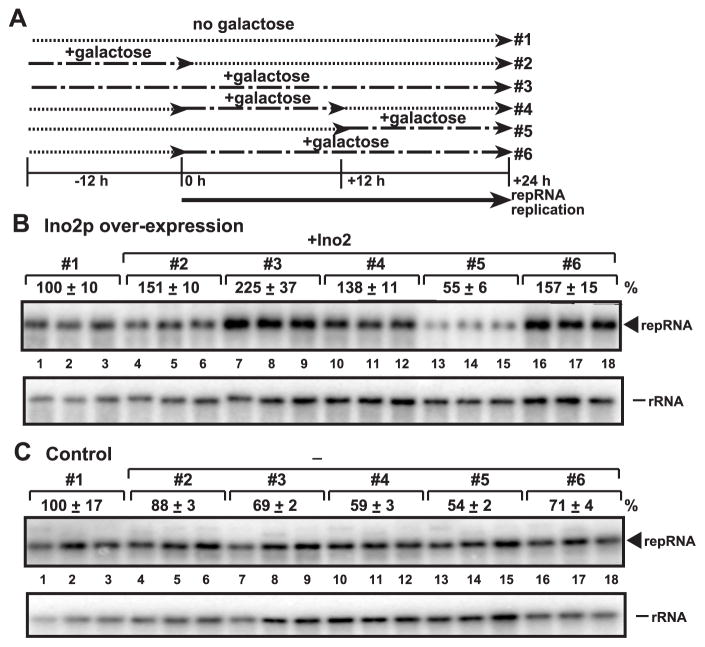
To test when TBSV is the most dependent on phospholipid biosynthesis, we devised a scheme for controlled over-expression of Ino2, and thus regulated increase in phospholipid biosynthesis, followed by measuring TBSV RNA accumulation 24 hours after inducing repRNA replication (see Fig. 6A for the experimental scheme). The control experiments included the same expression plasmid and inducer (galactose), but without the INO2 open reading frame (Fig. 6C), which leads to the over-expression of a small peptide. We found that Ino2 over-expression increased TBSV repRNA accumulation to the largest extent when it was over-expressed all the time (3x increase, from 69% to 225%, compare treatment #3 in Fig. 6B-C, lanes 7–9). We also observed ~2-fold increase in TBSV RNA accumulation when Ino2 was over-expressed for 12 h either before the induction of TBSV repRNA accumulation (from 88% to 151%, compare treatment #2, Fig. 6B-C, lanes 4–6) or the first half of TBSV replication period (treatment #4, lanes 10–12). Similarly, over-expression of Ino2 for 24 h during the entire TBSV replication period led to ~2x increase in TBSV RNA accumulation (compare treatment #6, Fig. 6B-C, lanes 16–18). In contrast, over-expression of Ino2 for 12 h during the second half of TBSV replication period (compare treatment #5, Fig. 6B-C, lanes 13–15) did not alter TBSV RNA accumulation. Altogether, these data are consistent with the model that TBSV replication is the most dependent on phospholipid biosynthesis at the early stage of the replication cycle and less dependent at the latter stage. However, it is important to note that since we over-expressed Ino2 transcription activator, the actual effect on phospholipid levels in yeast cells could take place couple of hours after the induction of Ino2.
Fig. 6.
Over-expression of Ino2 facilitates TBSV repRNA accumulation the most effectively when expressed continuously in yeast. (A) The scheme of Ino2 over-expression from the GAL1 promoter from the low copy pYC-INO2 plasmid relative to initiation of repRNA replication. repRNA replication took place for 24 hours at 29 ºC before RNA analysis. (B) Northern blot analysis of TBSV repRNA in yeast samples over-expressing Ino2 as shown schematically in panel A. We omitted inositol from the growth media, which always contained 2% raffinose plus 0 or 2% galactose as shown in panel A. In addition, we used 100 μM BCS for 12 hours before inducing TBSV replication (to prevent leaky transcription from the CUP1 promoter). Note that treatment #1 (no Ino2 is expressed) is the same as in panel C and it is chosen as 100% to allow comparison between the two panels. (C) Northern blotting shows the level of TBSV repRNA accumulation when a small peptide was over-expressed from the GAL1 promoter in pYC as shown schematically in panel A. The accumulation level of DI-72(+) repRNA (shown in percentage) was normalized based on 18S rRNA.
Over-expression of Opi1, a repressor of phospholipid synthesis, decreases TBSV RNA accumulation in yeast
To further show that TBSV replication depends on active phospholipid biosynthesis, we over-expressed Opi1, which is a repressor of Ino2-Ino4 complex (Carman and Han, 2009; Nohturfft and Zhang, 2009; Wagner et al., 1999). TBSV repRNA accumulation decreased by 2.5-fold in yeast over-expressing Opi1 (Fig. 7, lanes 1–4). The amount of p33 and p92pol replication proteins also decreased in this yeast (Fig. 7). Thus, the overall effect of over-expression of Opi1 was very similar to the situation seen with ino2Δino4Δ yeast (Fig. 2), suggesting that the phospholipid biosynthesis pathway is important in tombusvirus replication.
Fig. 7.
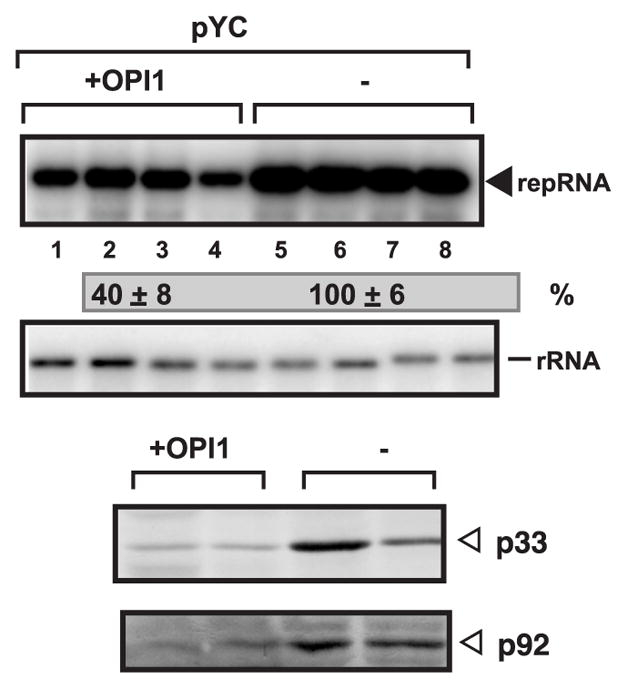
Over-expression of Opi1 repressor inhibits TBSV repRNA replication in yeast. Top panel: Northern blot analysis of TBSV repRNA in BY4741 yeast over-expressing Opi1 from the GAL1 promoter from the low copy pYC-OPI1 plasmid (+OPI1, lanes 1–4). The control yeast carried the empty pYC plasmid (lanes 5–8). The expression of Opi1 started 15 hours before launching TBSV repRNA replication, which started by expressing 6xHis-p33 and 6xHis-p92 from the CUP1 promoter and DI-72(+) repRNA from the GAL1 promoter, and continued to the end of the experiment (24 hours of TBSV replication at 29 ºC). We used 100 μM BCS in the media for 15 hours before inducing TBSV replication (to prevent leaky transcription from the CUP1 promoter). See further details in Fig. 1B. Bottom panel shows the Western blot analysis of p33 and p92 replication proteins using anti-His antibody.
Phospholipid synthesis is required for proper subcellular localization of tombusvirus replication proteins
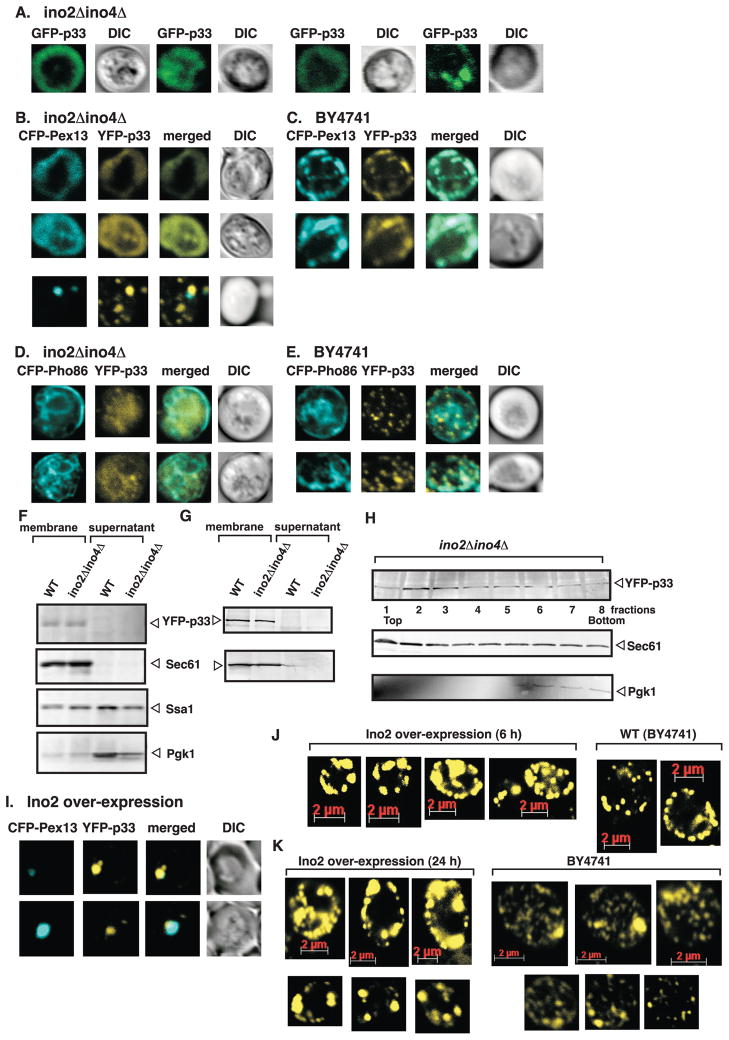
To monitor the subcellular localization of the tombusvirus replication protein in ino2Δino4Δ yeast, first we used GFP-tagged p33 and confocal microscopy. Interestingly, the large majority of yeast cells showed unusual diffused pattern for GFP-p33 in ino2Δino4Δ yeast (Fig. 8A), which is in contrast with the punctate structures formed in wt BY4741 yeast (not shown and Fig. 8C, E, J-K) (Jonczyk et al., 2007; Panavas et al., 2005a). A smaller fraction of yeast cells showed GFP-p33 as part of punctate structures, as expected from cells replicating the TBSV repRNA (Fig. 8A).
Fig. 8.
Phospholipid synthesis is essential for the proper subcellular localization of tombusvirus p33 replication protein in yeast. (A) Confocal laser microscopy analysis of subcellular distribution of GFP-tagged p33 in ino2Δ ino4Δ yeast. (B-C) YFP-tagged p33 co-expressed with CFP-tagged Pex13, a peroxisomal marker protein, in ino2Δ ino4Δ yeast and in wt BY4741. (D-E) Subcellular localization of YFP-p33 co-expressed with CFP-Pho86, an ER marker protein, in ino2Δ ino4Δ yeast and in wt BY4741, respectively. Throughout the experiments GFP- or YFP- tagged p33 was expressed from GAL1 promoter and the marker proteins, CFP-Pex13 and CFP-Pho86 from the ADH1 promoter. (F) Western blot analysis of p33 (6xHis-tagged) after fractionation from wt (BY4741) and ino2Δ ino4Δ yeast cells. Sec61 ER protein, Ssa1 both cytosolic and membranous protein, and Pgk1p cytosolic protein were used as controls and detected with specific antibodies. (G) Western blot analysis of p33 after fractionation from wt (BY4741) and ino2Δ ino4Δ yeast cells. Note that the membrane fraction was treated with alkaline to remove peripheral proteins from the membrane as described in Materials and Methods. (H) Western blot analysis of p33 from wt and ino2Δ ino4Δ yeast cells after membrane flotation. The top fractions contain the membrane-associated proteins, while the bottom fractions contain soluble or aggregated proteins. See further details in panel F. (I) Confocal laser microscopy analysis of subcellular localization of YFP-p33 co-expressed with CFP-Pex13 peroxisomal marker protein at 6 h time point in yeast over-expressing Ino2. (J) and (K) Localization of YFP-p33 at 24 h time point, in yeast over-expressing Ino2 or in wt (control) background as shown. The bottom row of images are shown at lower magnification of yeast cells to illustrate the presence of ~1-4 large punctate structures in yeast over-expressing Ino2, while smaller punctate structures form in the control BY4741 yeast transformed with the empty vector.
To define the subcellular location of p33 in ino2Δino4Δ yeast, we co-expressed YFP-p33 with CFP-Pex13, which is a peroxisomal marker protein (Jonczyk et al., 2007; Panavas et al., 2005a). Surprisingly, both YFP-p33 and CFP-Pex13 showed the unusual diffused pattern in the majority of yeast cells (Fig. 8B), which is dramatically different from the usual punctate structures and co-localization as seen in the wt BY4741 strain (Fig. 8C). In a small fraction of cells (~25%), in which YFP-p33 and CFP-Pex13 showed punctate structures (Fig. 8B, bottom panel), their co-localization was detectable, suggesting that in a few cells tombusvirus p33 might be able to get transported to proper subcellular location, while it is mislocalized in the majority of ino2Δino4Δ yeast cells.
To further test the subcellular location of p33 in ino2Δino4Δ yeast, we co-expressed YFP-p33 with CFP-Pho86, which is an ER marker protein (Jonczyk et al., 2007; Panavas et al., 2005a). We did not observe co-localization of p33 and the ER marker protein in ino2Δino4Δ yeast (Fig. 8D), while we detected some co-localization of YFP-p33 with CFP-Pho86 in wt BY4741 yeast (Fig. 8E). Most of the punctate structures formed by YFP-p33 were located in the proximity of ER, but not co-localized with the ER marker, as shown previously (Jonczyk et al., 2007; Panavas et al., 2005a). This pattern is typical for tombusviruses, which is interpreted as peroxisomal location.
We also used cell-fractionation experiments to test if YFP-p33 is still membrane-bound in ino2Δino4Δ yeast. The obtained data from independent sets of experiments revealed that YFP-p33 sedimented with the membrane-containing fraction, not the supernatant containing the soluble proteins (Fig. 8F). Treating the membrane-fraction with alkaline to remove peripheral membrane-bound proteins did not remove p33, indicating that p33 is an integral membrane-protein in ino2Δino4Δ yeast (Fig. 8G). Membrane flotation experiments confirmed that p33 was the most abundant in the top fractions 2 and 3, which contains most of the membrane-bound proteins, such as Sec61p (Fig. 8H, lanes 2-3). Altogether, these data suggest that most YFP-p33 is likely membrane-associated in ino2Δino4Δ yeast.
Over-expression of Ino2 in BY4741 strain resulted in YFP-p33 pattern with frequent formation of large punctate structures both at 6 h and 24 h time points (Fig. 8J-K), which were co-localized with Pex13 peroxisomal marker at the 6h time point (Fig. 8I). In comparison, YFP-p33 is distributed to a large number, but smaller punctate structures in the wt yeast (Fig. 8J-K). Since Ino2 over-expression simulates TBSV repRNA accumulation, it is likely that the large punctate structures in Ino2 over-expressing cells are increasingly active in viral RNA synthesis.
Phospholipid synthesis is needed for replication of Flock house virus
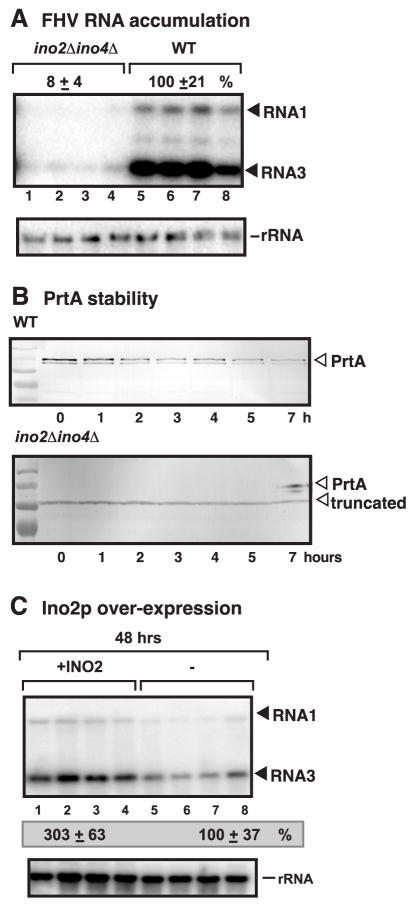
To test if the reduced phospholipid synthesis in ino2Δino4Δ yeast could also affect the replication of another RNA virus, we chose Flock house virus (FHV) (Castorena, Stapleford, and Miller, 2010; Kopek et al., 2010; Odegard, Banerjee, and Johnson, 2010; Pogany et al., 2010; Price, Ahlquist, and Ball, 2002), which is an insect virus distantly related to TBSV, with both viruses belonging to the Flavivirus-supergroup among (+)RNA viruses. The accumulation of FHV RNA1 and the subgenomic RNA3 (produced from RNA1 during replication) was reduced dramatically in ino2Δino4Δ yeast (Fig. 9A). Interestingly, an N-terminally truncated protein A (PrtA) replication protein accumulated in ino2Δino4Δ yeast (Fig. 9B), suggesting that the FHV replication protein is unstable in presence of reduced amounts of phospholipids. Time-course experiments showed that the full-length FHV PrtA was barely detectable even at the beginning of the protein stability experiments (Fig 9B).
Fig 9.
Deletion of INO2 and INO4 inhibits FHV RNA accumulation in yeast. (A) Top panel: Northern blot analysis with a 3' end specific probe was used to detect the accumulation level of the FHV RNA1 and RNA3 in ino2Δ ino4Δ or wt (BY4741) yeast. To launch FHV RNA replication, we expressed FHV RNA1 from the copper-inducible CUP1 promoter from a plasmid. Yeast cells were cultured for 48 hours at 29ºC in 2% galactose SC-H− media containing 2mg/L inositol. The accumulation levels of repRNA were calculated using Imagequant software. The accumulation level of FHV RNA3 (shown in percentage) was normalized based on 18S rRNA. Middle panel: Northern blot analysis to probe rRNA, which was used as a loading control. (B) Western blot analysis of protein A (prtA) accumulation using anti-FLAG antibody. Samples were collected at 0, 1, 2, 3, 4, 5 and 7 hours. Note that sample 7 (bottom panel) also contains a trace amount of full-length prtA, depicted by an arrowhead, as a size control to illustrate the difference in protein A products. (C) Over-expression of Ino2 facilitates FHV RNA accumulation in yeast. Ino2 was over-expressed from the GAL1 promoter from the low copy pYC-INO2 plasmid. FHV replication took place for 48 hours at 29 ºC before RNA analysis. The accumulation level of FHV RNA3 (shown in percentage) was normalized based on 18S rRNA. We omitted inositol from the growth media, which always contained 2% raffinose.
Over-expression of Ino2 in wt BY4741 yeast resulted in ~3-fold increase in FHV RNA3 accumulation (Fig. 9C, lanes 1-4 versus 5-8). Thus, similar to TBSV, FHV replication is also dependent on phospholipid synthesis in yeast.
DISCUSSION
Phospholipids are major components of cellular membranes, affecting the size, shape and rigidity of cells and intracellular organelles (Nohturfft and Zhang, 2009). Replication of various RNA viruses, which hijack subcellular membranes to induce the formation of viral replication organelles (den Boon, Diaz, and Ahlquist, 2010; Miller and Krijnse-Locker, 2008; Novoa et al., 2005), likely depends on phospholipids. Indeed, all the genome-wide screens performed with (+)RNA viruses have led to the identification of a number of host genes affecting lipid biosynthesis or metabolism (Cherry et al., 2005; Krishnan et al., 2008; Kushner et al., 2003; Li et al., 2009). Similarly, genome-wide screens with TBSV identified at least 14 host genes affecting phospholipid, sterol and fatty acid biosynthesis/metabolism (Jiang et al., 2006; Panavas et al., 2005b). The abundance of the identified host lipid synthesis genes suggests that lipids are important for TBSV replication. Indeed, we have shown previously that sterols affect TBSV replication in yeast, in plants and in vitro (Sharma, Sasvari, and Nagy, 2010).
The role of phospholipids in TBSV replication is supported by several pieces of evidence presented in this paper. First, single deletions of INO2 and INO4 or the double-deletion of INO2-INO4 inhibited TBSV replication in yeast (Figs. 1-2). INO2 and INO4 are activators of phospholipid biosynthesis genes, and, thus, their deletions are known to reduce phospholipid levels and prevent membrane proliferation (Block-Alper et al., 2002; Schuck et al., 2009). The ino2Δ ino4Δ yeast is viable due to the base-level phospholipid biosynthesis that occurs in the absence of Ino2/Ino4 transcription activators. We found that ino2Δ ino4Δ yeast still can support low level of TBSV replication (Figs. 1-2), likely due to the presence of some phospholipids in the cellular membranes. Second, over-expression of Opi1, which is a repressor of phospholipid biosynthesis by binding to Ino2 (Wagner et al., 1999; Wagner et al., 2001), also inhibited TBSV repRNA accumulation in yeast (Fig. 7). Third, over-expression of Ino2 increased TBSV RNA accumulation (Figs. 5–6).
The follow-up experiments revealed that the reduced phospholipid levels affected many steps/processes during TBSV replication. For example, the in vitro activity of the tombusvirus replicase in the membrane-enriched fraction from ino2Δ ino4Δ yeast was poor when compared with a similar preparation from wt yeast (Fig. 3). Since we adjusted the preparations to have comparable amounts of tombusvirus replication proteins, the differences in the template activity of these replicase preparations are likely due to either poor assembly of the replicase complex or the low activity of the replicase in ino2Δ ino4Δ yeast. Thus, phospholipids affect the activity of the replicase to make viral RNA products.
Another characteristic of TBSV replication in ino2Δ ino4Δ yeast is the reduced stability of the tombusvirus replication protein (Fig. 4A). This reduced stability could be due to incorrect localization of the replication proteins in ino2Δ ino4Δ yeast. Instead of the usual punctate structures formed by p33 on the peroxisomal or ER membranes, p33 shows diffused distribution in ino2Δ ino4Δ yeast, albeit most of the p33 proteins seem to be still associated with membranes based on cell-fractionation and treatment of membranes with alkaline (Fig. 8). Based on these data, we propose that the tombusvirus replication proteins are not targeted to the proper subcellular locations in ino2Δ ino4Δ yeast. This interferes with the assembly of the viral replicase, resulting in reduced replicase activity and possibly faster turnover of the replication protein in ino2Δ ino4Δ yeast. On the contrary, the intracellular targeting of p33 and the assembly of the replicase might be facilitated by over-expression of Ino2, which resulted in enlarged punctate structures in yeast (Fig. 8H-I).
FHV replication also occured at a reduced level in ino2Δ ino4Δ yeast (Fig. 9). This confirms previous findings that phospholipids are important for FHV replication (Castorena, Stapleford, and Miller, 2010). A new finding is the occurrence of a truncated prtA replication protein in ino2Δ ino4Δ yeast, suggesting that phospholipids could be important to protect prtA from cleavage by cellular proteinases. We did not observe similar abundant truncated products of p33 or p92pol in ino2Δ ino4Δ yeast, but this could be due to faster degradation of p33 or p92pol that rapidly removes the putative truncated protein products. Overall, the replication proteins of both viruses seem to require phospholipids for enhanced stability.
Although the data shown here support strongly the roles of phospholipids in TBSV and FHV RNA replication, we cannot yet pinpoint the critical phospholipids, since INO2/INO4 transcription activators affect the production of many phospholipids in yeast (Carman and Han, 2009). A more detailed work on FHV demonstrated that genes involved in the production of phosphatidylcholine are critical for FHV replication (Castorena, Stapleford, and Miller, 2010). Additional experiments will be needed to identify the critical phospholipids for TBSV replication.
MATERIALS AND METHODS
Yeast strains and expression plasmids
Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and single-gene deletion strain ino2Δ and ino4Δ strains were obtained from Open Biosystems (Huntville, AL). The following yeast expression plasmids have been generated before: pHisGBK-His33 (Panaviene et al., 2004); pGAD-His92 (Panaviene et al., 2004); pYC-DI72sat (Panavas and Nagy, 2003); pHisGBK-CUP1::His33-ADH::DI72 (Mendu et al., 2010); pESC-His-p33-DI-72 (Jonczyk et al., 2007); pGBK-His33/DI-72 (Jiang et al., 2006); pYES-HisGFP-33 (Panavas et al., 2005a); pex13-CFP (Panavas et al., 2005a); pho86-CFP (Panavas et al., 2005a); pGAD-His92-Cup1 and pHisGBKHis33Cup1/GAL-DI (provided by K. Pathak). pESC-His-Cup-FHV-RNA1-TRSVRz was provided by J. Pogany.
To create double deletion strain ino2Δino4Δ, we used homologous recombination in ino2Δ strain by replacing INO4 ORF with hphNT1 (Hygromycin resistance) gene. The hphNT1 ORF was PCR amplified from pFA6a-HPH (Euroscarf) using the following primers: #3693 (CGAAGGAGTTAAGAGGGCGGCTTGAACTAAAAAGAGAAAAGCA-cgtacgctgcaggtcga) and #3694 (AGAATTTCTTCGCTTATATTAC-TTACTTTACCCTACTCCTTGatcgatgaattcgagctc). The obtained PCR product was used to transform ino2Δ strain. The new strain ino2Δino4Δ was confirmed with primers #2501 (ATCCACGCCCTCCTACATC) and #3695 (GGGTACCTCCAAATCTGCGAAGGTA).
To express Ino2 in yeast, the full-length ORF of INO2 was cloned into pYES/NT/C (Invitrogen). First, INO2 ORF was amplified by PCR from yeast genomic DNA by using primers #2311 (CAGCGGATCCATGCAACAAGCAACTGGGAACGAATTACT) and #2312 (GACCCTCGAGTCAGGAATCATCCAGTATGT) that were appended with BamHI and XhoI recognition sequences, respectively, to facilitate directional cloning. To express Ino2, we PCR amplified the INO2 ORF and digested with HindIII and XhoI sites and cloned into pYC2/CT low copy vector digested with the same pair of enzymes.
To obtain pYES-His92, which contains GAL1-p92 and URA3 auxotrophic marker, p92 ORF was amplified using primers #788 (GAGGGATCCGAGACCATCAAGAGAATG) and #952 (CCCGCTCGAGTCATGCTACGGCGGAGTCAAGGA) appended with BamHI and XhoI restriction enzyme recognition sites to facilitate directional cloning and then ligated and cloned into pYES vector digested with the same pair of enzymes. Similarly, pYES-His33, which contains GAL1-p33 and URA3 auxotrophic marker, the p33 ORF was amplified using primers #788 and #1403 (gccgCTCGAGCTATTTCACACCAAGGGACTCA) appended with BamHI and XhoI restriction enzyme recognition sites to facilitate directional cloning and then ligated and cloned into pYES vector digested with the same pair of enzymes. Construct BG1805-Opi1-zz carries OPI1 ORF behind the GAL1 promoter and fused to a tandem affinity tag that includes a His-tag and the zz domain of protein A at the C terminus (Li et al., 2008).
TBSV replication assay in yeast
Yeast strains BY4741, ino2 , ino4 and ino2Δ ino4Δ were transformed with pHisGBKHis33 and pGADHis92 (Panaviene et al., 2004) as well as with pYC-DI72 (Panavas and Nagy, 2003). Replication assay was performed by measuring the accumulation of DI-72(+) repRNA relative to the 18S rRNA (Panavas and Nagy, 2003). To precisely regulate the amount and timing of expression of replication proteins p33 and p92 and to measure their subsequent effect on RNA accumulation levels, ino2Δ ino4Δ and BY4741 yeast strains were transformed with the following combination of plasmids: (a) pHisGBKHis33 and pGADHis92 and pYC-DI72; (b) pYES-92 and pESC-His33/Gal-DI; and (c) pGAD-92Cup1, pHisGBKHis33Cup1-GAL-DI. Yeast cells were pre-grown at 29°C with shaking for 15 hrs and then transferred to media containing 2% galactose supplemented with 50μM Cu++. Standard RNA extraction and Northern blot was performed as mentioned in previous publications (Panavas and Nagy, 2003; Panaviene et al., 2004).
In vitro replicase assay using membrane-enriched (ME) fraction of yeast
The procedure used to obtain functional ME fractions was the same developed by us earlier (Panaviene, Panavas, and Nagy, 2005). Briefly, yeast was pre-grown in SC-ULH- medium containing 2% glucose for 24 h at 29 °C with shaking at 250 rpm. Then, yeast cells were transferred to SC-ULH- containing 2% galactose and incubated at 23 °C with shaking at 250 rpm. After 24 h growth, yeast samples were collected by centrifugation at 3000g for 5 min, followed by washing the pellet with 20 mM Tris–HCl, pH 8.0. The pelleted cells were resuspended in 1 ml of 20 mM Tris–HCl, pH 8.0, followed by centrifugation at 21,000g for 1 min. Yeast cells were broken by glass beads in a Genogrinder (Glen Mills Inc., Clifton NJ) for 2 min at 1500 rpm. After mixing with 600 μl chilled extraction buffer (200 mM sorbitol, 50 mM Tris–HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma), the samples were centrifuged at 100g for 5 min at 4 °C. The supernatant was moved to a new microcentrifuge tube, followed by centrifugation at 21,000g for 10 min at 4 °C. The pellet was resuspended in 0.7 ml extraction buffer, resulting in the ME fraction. The replicase assay with the ME fraction was performed in 100 μl volume containing RdRp buffer [40 mM Tris pH 8.0, 10 mM MgCl2, 10 mM DTT, 100 mM potassium glutamate, 0.2μl Rnase inhibitor, 1 mM ATP, CTP, GTP, 0.1 μl radioactive P32-UTP (3000 mCi/mmol ICN) and 50 μl ME fraction. Samples were incubated at 25 °C for 2 h. The reaction was terminated by adding 70 μl SDS/EDTA (1% SDS, 50 mM EDTA pH 8.0) and 100 μl phenol-chloroform (1:1). After standard isopropanol precipitation of the RNA products, the RNA samples were electrophoresed under denaturing conditions (5% PAGE containing 8 M urea) and analyzed by phospho-imaging using a Typhoon (GE) instrument as described (Panaviene, Panavas, and Nagy, 2005).
RNA and protein stability assays
Yeast strains BY4741 and ino2Δ ino4Δ were transformed with pYC2-DI72. The transformed yeast strains were grown at 29°C in SC-U (synthetic complete without uracil) with 2% galactose. After 20 h, the cultures were re-suspended in SC-U supplemented with 2% glucose and collected after indicated time-points. Northern blotting was performed to measure repRNA levels at various time points.
To study the stability of p33 in yeast, BY4741 and ino2Δ ino4Δ were transformed with pYES-33 expressing 6xHis-tagged CNV p33 from the inducible GAL1 promoter. Yeast transformants were cultured overnight in SC U− medium containing 2% glucose at 29°C. Yeast cultures were transferred to U- 2% Galactose medium for 6 hrs 29°C. To study stability of protein A of FHV, BY4741 and ino2Δ ino4Δ strains were transformed with pGAD/CUP/PtnA/C-HA/FLAG construct. After pre-growing the cells in L− media containing 2% glucose, protein A expression was induced with 50μM Cu++ for 12 hrs. Then, cycloheximide was added to a final concentration of 100 μg/ml to inhibit protein synthesis. Equal amounts of yeast cells were collected at given time points after cycloheximide treatment and cell lysates were prepared by the NaOH method as described previously (Sharma, Sasvari, and Nagy, 2010). The total protein samples were analyzed by SDS-PAGE and Western blotting with anti-His and anti-FLAG antibody as described previously (Panaviene, Panavas, and Nagy, 2005; Panaviene et al., 2004).
Complementation assay and over-expression of Ino2p and Opi1p
pYC-INO2 or pYC empty plasmids were transformed into BY4741 and ino2 strains containing pGAD-His92, pGBK-His33/DI-72 (to co-express p33 protein from the constitutive ADH1 promoter and DI-72 repRNA under inducible GAL1 promoter). pYC-INO2, pYC-empty, pYES-INO2 or pYES-empty plasmids were transformed into BY4741 strain containing pGAD-His92-Cup, pHisGBK-Cup1::His33-ADH1::-DI72 (Mendu et al., 2010), which expresses p33 protein from CUP1 promoter and DI-72 repRNA under constitutive ADH1 promoter. Cells were pre-grown at 29°C for 15 hrs in 2 % galactose media followed by supplementation with 50μM Cu++ and further incubation at 29°C for 24 hrs. For time course analysis, media was changed as described in figure legend. To over-express Opi1 above mentioned strategy was employed using BG1805-Opi1-zz.
Replication Protein Analysis
Yeast strains were grown as described above for RNA analysis. A total of 2ml of yeast culture was harvested, the pelleted cells were resuspended in 200 μl of 0.1M NaOH and incubated at 23°C for 10 min. The supernatant was aspirated following a short centrifugation, and the pellet was resuspended in 100μl, 1X SDS-polyacrylamide gel electrophoresis (PAGE) buffer containing 5% β-mercaptoethanol and boiled for 5 min. The supernatant was used for SDS/PAGE and Western blot analysis as described (Panavas et al., 2005a). The primary antibodies were anti-6xHis (Invitrogen) for tombusvirus and anti-FLAG (Sigma) for FHV, and the secondary antibodies were alkaline-phosphatase-conjugated anti-mouse immunoglobulin-G (Sigma).
Membrane fractionation
To check membrane association of p33 in BY4741 and ino2Δ ino4Δ yeast, cells were broken using Fast Prep24 MP Bio. After removing debris by centrifugation at 1,000g for 5 min, membranes were collected at 40,000g for 1 hr. Supernatant and membrane fractions were analyzed for their p33 content by western blotting. In another set of fractionation experiments, we performed alkaline treatment to remove the proteins that bound peripherally to the membranes (Whitley et al., 1996). Briefly, after removal of cell debris, supernatant (250μl) was incubated on ice with Na2CO3 (250 μl) 200mM pH 11.5, for 30 min and then loaded on the 300 μl cushion (200mM sucrose in100mM Na2CO3 pH 11.5) followed by centrifugation for 30 min at 40,000g. Separated soluble and membrane fractions were analyzed for p33 content by western blotting. Different cellular organellar protein markers (Sec61, Ssa1 and Pgk1) were also visualized by using their respective antibodies.
Yeast spheroplasting and membrane flotation
ino2Δino4Δ yeast transformed with pESC-HisY-p33-DI-72 (for inducible expression of YFP-p33) was pre-grown in 2% glucose H- media and then transferred to 2% galactose media and grown to OD600 of 0.8 to 1.0. Spheroplasting was performed using Zymolyase digestion as described (Daum, Bohni, and Schatz, 1982; Wang, Stork, and Nagy, 2009). Briefly, cells were harvested at 3000g, then washed with water and re-suspended in 0.1M TrisSO4 pH 9.4 containing 10mM DTT. This was followed by incubation at 30°C. Cells were collected by centrifugation and washed with 1.2M sorbitol. Pellet was resuspended in 1.2M sorbitol containing 20mM potassium phosphate (pH 7.4) and Zymolyase (5mg per 1 g of wet yeast cells), then incubated at 30°C with very gentle shaking for 50–90 min. Spheroplasting efficiency was checked under a microscope. Yeast spheroplasts were harvested at 500g and washed twice with 1.2M sorbitol.
Using 15 strokes in Dounce homogenizer, 100 milligram of yeast cells was broken in 600 μl of yeast lysis buffer (200 mM sorbitol, 50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mi [Sigma]), followed by centrifugation for 5 min at 100g to pellet unbroken cells. Supernatant was centrifuged at 16,000g for 15 minutes and the pellet containing membranes were collected for the flotation experiments.
For sucrose flotation gradient analysis, samples were adjusted to 52% (wt/wt) sucrose in the lysis buffer, and 400 μl was loaded to the bottoms of ultraclear polycarbonate ultracentrifuge tubes (Beckman), overlaid with 900 μl of 45% sucrose in lysis buffer, topped with 100 μl of 10% sucrose in lysis buffer, and subsequently centrifuged at 40,000 rpm at 4°C for 16 h by using an TLS55 Ti rotor in a Beckman Optima-Max-XP ultracentrifuge. Ten fractions (140 μl each) were manually collected from the top to the bottom, followed by protein analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western-blotting procedures as described previously using antibodies against Sec61p (an integral ER membrane protein) and anti-Pgk1p (cytosolic protein) (Wang, Stork, and Nagy, 2009).
Confocal laser microscopy
Yeast cells were transformed with YFP- or GFP-tagged p33 (pESC-YFP-33 or pYES-HisGFP-33) (Panavas et al., 2005a). To visualize peroxisome and ER, pex13-CFP and pho86-CFP, respectively, were used as markers (Panavas et al., 2005a). Transformed yeast cells were pre-grown overnight at 29°C and then transferred to media containing 2% galactose and samples were collected for microscopy after 6 hrs (Panavas et al., 2005a). To test the effect of Ino2 overexpression on p33 localization pattern, above yeast transformants were re- transformed with pYC-INO2 or pYC (as a control). Samples were collected for confocal microscopy 6 and 24 hrs post induction.
FHV replication assay
BY4741 and ΔINO2/ΔINO4 strains were transformed with pESC-His-Cup-FHV-RNA1-TRSVRz. After pregrowing at 29°C, media (SC-H− with 2% galactose) was supplemented with 50μM Cu++ and harvested after 48 hrs for RNA analysis. To study the effects of Ino2 overexpression, above yeast strains were transformed with pYC-INO2 or pYC-empty plasmids along with pESC-His-Cup-FHV-RNA1-TRSVRz. Cells were grown in media SC-UH − 2% galactose media at 29°C and then supplemented with 50μM Cu++ and harvested after 48 hrs for RNA analysis.
Acknowledgments
The authors thank Drs. Daniel Barajas, and Judit Pogany for valuable comments. pYES- His92 and pYES-His33 were prepared and provided by. T. Panavas. This work was supported by the National Science Foundation (IOB-0517218), NIH-NIAID (5R21AI072170-02) and the Kentucky Tobacco Research and Development Center to PDN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77(15):8181–6. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D, Jiang Y, Nagy PD. A Unique Role for the Host ESCRT Proteins in Replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5(12):e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Ehrenfeld E. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle. 2007;6(1):36–8. doi: 10.4161/cc.6.1.3683. [DOI] [PubMed] [Google Scholar]
- Belov GA, Habbersett C, Franco D, Ehrenfeld E. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J Virol. 2007;81 (17):9259–67. doi: 10.1128/JVI.00840-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106(18):7577–82. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block-Alper L, Webster P, Zhou X, Supekova L, Wong WH, Schultz PG, Meyer DI. IN02, a positive regulator of lipid biosynthesis, is essential for the formation of inducible membranes in yeast. Mol Biol Cell. 2002;13(1):40–51. doi: 10.1091/mbc.01-07-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in yeast. J Lipid Res. 2009;50(Suppl):S69–73. doi: 10.1194/jlr.R800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castorena KM, Stapleford KA, Miller DJ. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics. 2010;11:183. doi: 10.1186/1471-2164-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19(4):445–52. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2(10):e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257(21):13028–33. [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14(16):1471–510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8(1):77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MR. Seeking membranes: positive-strand RNA virus replication complexes. PLoS Biol. 2008;6(10):e270. doi: 10.1371/journal.pbio.0060270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107(40):17345–50. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8(5):422–32. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141(5):799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol. 2006;80(15):7394–404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk M, Pathak KB, Sharma M, Nagy PD. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362(2):320–30. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5(9):e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek BG, Settles EW, Friesen PD, Ahlquist P. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein a, RNA templates, and polymerase activity. J Virol. 2010;84(24):12492–503. doi: 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455(7210):242–5. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100(26):15764–9. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol. 2003;77(23):12819–28. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106(38):16410–5. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. Cdc34p Ubiquitin-Conjugating Enzyme Is a Component of the Tombusvirus Replicase Complex and Ubiquitinates p33 Replication Protein. J Virol. 2008;82(14):6911–26. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2 (4):229–39. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17 (12):3513–31. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendu V, Chiu M, Barajas D, Li Z, Nagy PD. Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology. 2010;406(2):342–51. doi: 10.1016/j.virol.2010.07.022. [DOI] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6(5):363–74. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PD. Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol. 2008;46:217–42. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv Virus Res. 2010;76:123–77. doi: 10.1016/S0065-3527(10)76004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro B, Russo M, Pantaleo V, Rubino L. Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication. J Gen Virol. 2006;87(Pt 3):705–14. doi: 10.1099/vir.0.81325-0. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Zhang SC. Coordination of lipid metabolism in membrane biogenesis. Annu Rev Cell Dev Biol. 2009;25:539–66. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97(2):147–72. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard A, Banerjee M, Johnson JE. Flock house virus: a model system for understanding non-enveloped virus entry and membrane penetration. Curr Top Microbiol Immunol. 2010;343:1–22. doi: 10.1007/82_2010_35. [DOI] [PubMed] [Google Scholar]
- Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005a;338(1):81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T, Nagy PD. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology. 2003;314(1):315–25. doi: 10.1016/s0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Brasher J, Nagy PD. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A. 2005b;102(20):7326–31. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Nagy PD. Role of an internal and two 3'-terminal RNA elements in assembly of tombusvirus replicase. J Virol. 2005;79(16):10608–18. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Serva S, Nagy PD. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol. 2004;78(15):8254–63. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379(2):294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Pogany J, Panavas T, Serviene E, Nawaz-Ul-Rehman MS, PDN A high-throughput approach for studying virus replication in yeast. Curr Protoc Microbiol. 2010;Chapter 16(Unit16J):1. doi: 10.1002/9780471729259.mc16j01s19. [DOI] [PubMed] [Google Scholar]
- Pogany J, Stork J, Li Z, Nagy PD. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A. 2008;105(50):19956–61. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, Ahlquist P, Ball LA. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J Virol. 2002;76(4):1610–6. doi: 10.1128/JVI.76.4.1610-1616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasvari Z, Nagy PD. Making of Viral Replication Organelles by Remodeling Interior Membranes. Viruses-Basel. 2010;2(11):2436–2442. doi: 10.3390/v2112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 2008;29(2):191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187(4):525–36. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9(3):505–14. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol. 2006;80(3):1231–41. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A. 2005;102(30):10545–50. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Sasvari Z, Nagy PD. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol. 2010;84(5):2270–81. doi: 10.1128/JVI.02003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork J, Kovalev N, Sasvari Z, Nagy PD. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology. 2011;409(2):338–47. doi: 10.1016/j.virol.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Blank M, Strohmann B, Schuller HJ. Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast. 1999;15(10A):843–54. doi: 10.1002/(SICI)1097-0061(199907)15:10A<843::AID-YEA424>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol Microbiol. 2001;41 (1):155–66. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- Wang RY, Stork J, Nagy PD. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol. 2009;83(7):3276–87. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Morris TJ. Enhanced competitiveness of tomato bushy stunt virus defective interfering RNAs by segment duplication or nucleotide insertion. J Virol. 1994;68(9):6092–6. doi: 10.1128/jvi.68.9.6092-6096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Nagy PD. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acid Res Mol Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Whitley P, Grahn E, Kutay U, Rapoport TA, von Heijne G. A 12- residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271(13):7583–6. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, Smits GJ, Loewen CJ. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329(5995):1085–8. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]