Abstract
The potent ability of small interfering RNA (siRNA) to inhibit the expression of complementary RNA transcripts is being exploited as a new class of therapeutics for diseases including HIV. However, efficient delivery of siRNAs remains a key obstacle to successful application. A targeted intracellular delivery approach for siRNAs to specific cell types is highly desirable. HIV-1 infection is initiated by the interactions between viral glycoprotein gp120 and cell surface receptor CD4, leading to fusion of the viral membrane with the target cell membrane. Once HIV infects a cell it produces gp120 which is displayed at the cell surface. We previously described a novel dual inhibitory anti-gp120 aptamer-siRNA chimera in which both the aptamer and the siRNA portions have potent anti-HIV activities. We also demonstrated that gp120 can be used for aptamer mediated delivery of anti-HIV siRNAs.
Here we report the design, construction and evaluation of chimerical RNA nanoparticles containing a HIV gp120-binding aptamer escorted by the pRNA of bacteriophage phi29 DNA packaging motor. We demonstrate that pRNA-aptamer chimeras specifically bind to and are internalized into cells expressing HIV gp120. Moreover, the pRNA-aptamer chimeras alone also provide HIV inhibitory function by blocking viral infectivity. The Ab′ pRNA-siRNA chimera with 2′-F modified pyrimidines in the sense strand not only improved the RNA stability in serum, but also was functionally processed by Dicer, resulting in specific target gene silencing. Therefore, this dual functional pRNA-aptamer not only represents a potential HIV-1 inhibitor, but also provides a cell-type specific siRNA delivery vehicle, showing promise for systemic anti-HIV therapy.
Keywords: RNAi, Anti-gp120 aptamer, nanobiotechnology, bionanotechnology, nanotechnology, AIDS Treatment, viral DNA packaging, nanomotors
1. Introduction
The global epidemic of infection by HIV-1 has created a continuing need for new classes of antiretroviral agents [1]. The potent ability of siRNAs to inhibit the expression of complementary RNA transcripts is being exploited as a new class of therapeutics for a variety of diseases [2, 3] including HIV [4–6]. Although novel therapeutic strategies to combat HIV/AIDS by siRNAs show considerable promise as shown in many previous studies [7–10], efficient delivery of siRNAs still remains a key obstacle to its successful therapeutic application and clinical development [11, 12]. In particular, a targeted intracellular delivery approach for siRNAs to specific cell populations or tissues is highly desirable for the safety and efficacy of RNAi-based therapeutics. Moreover, the advent of nanotechnology has greatly accelerated the development of drug delivery, and the field of RNA-based nanotechnology has been emerging [13].
HIV-1 infection is initiated by the interactions between the external envelope glycoprotein gp120 of HIV and the human cell surface receptor CD4, subsequently leading to fusion of the viral membrane with the target cell membrane [14–16]. Thus HIV-1 entry into its target cells represents an attractive target for new anti-HIV-1 drug development [17–21]. We previously described a novel dual inhibitory function anti-gp120 aptamer-siRNA chimera in which both the aptamer and the siRNA portions have potent anti-HIV activities [22]. We also demonstrated that gp120 expressed on the surface of HIV infected cells can be used for aptamer mediated delivery of anti-HIV siRNAs [23]. Therefore the anti-gp120 aptamers represent a promising class of antiviral agents which can also function as siRNA delivery vehicles.
Packaging RNA (pRNA), one component of the bacteriophage phi29 DNA-packaging motor [24, 25], has been developed and manipulated to produce chimeric RNAs that form dimers via interlocking right- and left- hand loops. pRNA monomers can fold into a stable and unique secondary structure that serve as the building blocks to form nanostructures via bottom-up assembly [13, 26–29]. Fusion of the pRNA with a variety of therapeutic and chemical compounds did not impede the formation of dimers or interfere with function [30, 31]. Incubation of cells with dimers, one subunit of which carried a gene-silencing molecule and the other a receptor-binding moiety, resulted in successful binding, entry, and silencing of target genes [28, 31, 32]. Recently, it has been reported that the 2′-F modified pRNA retained its property for correct folding in dimer and hexamer formation, appropriate structure phi29 procapsid binding, authentic function in driving the DNA packaging motors, and biological activity in producing infectious virions [33]. The results suggest that it is possible to construct stable pRNA as siRNA or aptamer carrier.
We have designed, synthesized and evaluated the potential of pRNA-gp120 aptamer chimeras as effective HIV-1 inhibitors and cell-type specific delivery vehicles. In order to ensure the dual functions of both cell-specific targeting and efficient siRNA delivery, our delivery systems contained two structural entities: 1)A Ba′ pRNA-gp120 aptamer portion (Figure 1A) to specifically target the HIV infected cells and 2)An Ab′ pRNA-siRNA portion (Figure 1B) to efficiently transport efficiently anti-HIV siRNAs. Our results demonstrate that the pRNA-aptamer chimeras specifically bind to and are internalized into cells expressing HIV gp120. Moreover, the pRNA-aptamer chimeras provide an HIV inhibitory function which is as effective as the anti-gp120 aptamer. On the other hand, the Ab′ pRNA-siRNA chimera with a 2′-F modified sense strand not only improves the RNA stability in serum, but is functionally processed by Dicer, resulting in sequence specific gene silencing. Furthermore, through Ba′ and Ab′ loop-loop interactions, Ba′ pRNA-aptamer chimeras and Ab′ pRNA-siRNA chimeras formed dimers, which allow the specific delivery of Cy3-labeled siRNAs to cells expressing gp120. This dual functional pRNA-aptamer chimera not only represents a potential HIV-1 inhibitor, but also provides a cell-type specific siRNA delivery vehicle for delivery of anti-HIV siRNAs in systemic anti-HIV therapy.
Figure 1.
The pRNA-aptamer mediated targeted delivery of siRNA. (A) Schematic of the Ba′ pRNA-aptamer RNA chimera. The region of the anti-gp120 aptamer (blue) is responsible for binding to HIV-1 gp120 protein and the Ba′ pRNA is shown with black. (B) Schematic of the Ab′ pRNA-siRNA chimera. The sense and antisense strand are highlighted with blue and red, respectively. (C) Schematic of the dimer of Ba′ pRNA-aptamer/Ab′ pRNA-siRNA. The interlocking interaction through left- and right- hand loops is marked with a red circle.
2. Material and methods
2.1 Materials
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich, all restriction enzymes were obtained from New England BioLabs (NEB) and all cell culture products were purchased from GIBOC (Gibco BRL/Life Technologies, a division of Invitrogen.). Recombinant Human Dicer Enzyme Kit (Genlantis); Lipofectamine 2000 (Invitrogen); HEK 293 (ATCC); CHO-WT gp160, CHO-EE cells [34, 35], and the HIV-1 IIIB virions were obtained from the AIDS Research and Reference Reagent Program. Specific reagents and notes are listed in the Supplementary Table 1.
siRNAs. siRNA and antisense strand RNA were purchased from Integrated DNA Technologies (IDT). Site I (tat/rev) 27 mer:
Sense sequence: 5′- GCG GAG ACA GCG ACG AAG AGC UCA UCA -3′
Antisense: 5′- UGA UGA GCU CUU CGU CGC UGU CUC CGC dTdT -3′
2.2 Description of methods
2.2.1 Nomenclatures of pRNA
The 120-nt full-length pRNA contains two functional domains, the loop-loop interlocking interaction domain and the 5′/3′ end helical domain. The former is located at the central region [36–39] comprising bases 23–97, while the latter is located at the 5′/3′ paired ends [40]. The pRNA molecules were named as previously described [24]. Specifically, the pRNA molecules were identified by the R and/or L loop sequences. A particular R loop sequence is assigned an uppercase letter (i.e., A, B …), and a particular L loop sequence is assigned a lowercase letter with a prime (i.e., a′, b′ …). The same set of letters (i.e., A and a′) designates complementary R/L loop sequences, while different letters indicate non-complementary loop sequences. The intermolecular interaction domains mediate the interactions among pRNA subunits through hand-in-hand loop interactions, facilitating the formation of pRNA dimer, trimer, and hexamer. In our case, pRNA A-b′ represents pRNA where right loop A (5′G45G46A47C48) is complementary to left loop a′ (3′C85C84U83G82) of pRNA B-a′. And left loop b′ (3′U85G84C83G82) of A-b′pRNA is complementary to the right loop B (5′A45C46G47C48) of pRNA B-a′.
2.2.2 Generation of aptamer and chimera RNAs by in vitro transcription
Regular aptamer, pRNA-aptamer chimera and pRNA-siRNA chimera RNAs were generated by in vitro T7 transcription as previously described [22, 23, 41]. Briefly, Transcriptions were carried out in a mixture containing 0.1 μM DNA template, 0.75 mM of each NTP, 0.375 unit/μl of T7 RNA polymerase, 40 mM Tris-HCl (pH 7.5), 15 mM MgCl2, 5 mM DTT, 2 mM spermidine, and 0.01% (v/v) Triton X-100. The mixture was incubated at 37 °C for 3 hours. The transcription products were recovered by ethanol precipitation, and purified using denaturing gels. 2′-F modified aptamer, pRNA-aptamer chimera and pRNA-siRNA chimera RNAs were also transcribed in vitro using DuraScribe® T7 Transcription Kit (EPICENTRE® Biotechnologies).
The sense strands of the pRNA-siRNA chimeras are underlined. The italic UU is the linker between the aptamer and siRNA portions. The bold nucleotides indicate aptamer sequence.
A-1 apatamer (81 nt):
5′- GGG AGG ACG AUG CGG AAU UGA GGG ACC ACG CGC UGC UUG UUG UGA UAA GCA GUU UGU CGU GAU GGC AGA CGA CUC GCC CGA -3′
Ba′ pRNA-aptamer chimera (pRNA-A-1 D3) (152 nt):
5′- GGU UGA UUG UCC GUC AAU CAU GGC GGG AGG ACG AUG CGG AAU UGA GGG ACC ACG CGC UGC UUG UUG UGA UAA GCA GUU UGU CGU GAU GGC AGA CGA CUC GCC CGU CAU GUG UAU GUU GGG GAU UAA CGC CUG AUU GAG UUC AGC CCA CAU AC -3′
Ba′ pRNA-aptamer chimera (pRNA-A-1 D4) (198 nt):
5′- GGG AGG ACG AUG CGG AAU UGA GGG ACC ACG CGC UGC UUG UUG UGA UAA GCA GUU UGU CGU GAU GGC AGA CGA CUC GCC CGA GGA AUG GUA CGG UAC UUC CAU UGU CAU GUG UAU GUU GGG GAU UAA CGC CUG AUU GAG UUC AGC CCA CAU ACU UUG UUG AUU GUC CGU CAA UCA UGG CAA AAG UGC ACG CUA CUU UCC -3′
Ab′ pRNA-Tat/rev siRNA chimera sense strand:
5′- GGU AUG UUG GGG AUU AGG ACC UGA UUG AGU UCA GCC CAC AUA CUU UGU UGA UUG CGU GUC AAU UU GCG GAG ACA GCG ACG AAG AGC UCA UCA UU -3′
Tat/rev Antisense strand: 5′- UGA UGA GCU CUU CGU CGC UGU CUC CGC UU-3′
2.2.3 5′-end P32 labeling
The transcribed aptamer and pRNA-aptamer chimeras were treated by CIP to remove the initiating 5′-triphosphate and labeled with T4 polynucleotide kinase and γ-32P-ATP. 10 pmol of CIP treated RNAs were heat at 95°C for 5 min. and then chilled on the ice. Subsequently, add 2 μL of PNK buffer, 1 μL of T4 polynucleotide kinase, 1 μL of gamma-P32-ATP and water to 20 μL. After incubation at 37 °C for 30 min, add 20 μL of water and reaction was purified by G-50 column. Finally, 40 μL of labeled RNA was obtained at final concentration of 250 nM. The labeled aptamer or pRNA-aptamer chimera was refolded in 1×HBS buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 2.7 mM KCl), heated to 95 °C for 3 min. and then slowly cooled to 37°C. Incubation was continued at 37°C for 10 min. Store refolded RNA at −20 °C until assay. The P32-labeled pRNA-siRNA chimera was obtained by annealing end P32- labeled siRNA antisense to the other piece by heating up to 80 °C for 5 min. and then slowly cooled to room temperature. The annealed pRNA-siRNA chimeras were purified by native polyacrylamide gel and stored at −20 °C.
2.2.4 Determination of binding affinity by gel shift assays
Prepare a 25 mL 5% polyacrylamide gel by mixing 2.5 mL of 10×TBE buffer, with 3.125 mL of 40% acrylamide/bis solution, 19.375 mL water, 150 μL of 10% ammonium persulfate (APS) solution, and 30 μL TEMED. The gel should polymerize in about 30 min. Carefully remove the comb and use a 30-mL syringe fitted with a needle to wash the wells with running buffer (1×TBE). Complete the assembly of the gel unit and connect to a power supply. The gel can be pre-run for one hour at 180 V at 4°C.
The HIV-1Bal gp120 protein was serially diluted with binding buffer to the desired concentrations. The reaction final concentrations of gp120 were 0, 1, 5, 10, 20, 40, 80, 160, 320, 640 nM. A constant amount of 5′-P32-end-labeled RNA (10 nM) was incubated with increasing concentrations of gp120 protein in the binding buffer (total 20 μL reaction) on a rotating platform at room temperature for 30 min. After incubation, 20 μL of binding reaction was mixed with 5 μL native loading buffer and loaded into a 5% non-denaturing polyacrylamide gel. Prepare a native loading buffer (4×) containing 10 mM Tris-HCl, pH 7.5; 1 mM EDTA, 0.1% Bromophenol Blue, 0.1% Xylene Cyanol FF, 0.1% Orange G, 40% Glycerol. Store in aliquots at −20°C. Following electrophoresis (180 V at 4°C for 2 hours, until the secondary dye runs in the middle of the gel), the gel was exposed to a Phosphor image screen and the radioactivity quantified using a Typhoon scanner. The dissociation constants were calculated using non-linear curve regression with a Graph Pad Prism.
2.2.5 Preparation of fluorescent RNAs
Fluorescent aptamer and chimeras were generated using the Silencer siRNA Labeling Kit. Add the following reagents in order: 22.5 μL nuclease-free water; 5 mL 10×Labeling Buffer; 15 μL RNA (5 μg); 7.5 μL Labeling Dye. Total 50 μL labeling reaction was incubated at 37°C for 1 hour. After incubation, add 5.0 μL (0.1 vol) 5 M NaCl and 125 μL (2.5 vol) cold 100% EtOH, and mix thoroughly. Incubate at −20°C for 60 min. then centrifuge at top speed at 4°C for 20 min. Remove the supernatant and wash the pellet with 175 μL 70% EtOH. Air dries the pellet in the dark. Suspend labeled RNA in 15 μL of nuclease-free water. Measure the absorbance of the labeled RNA at 260 nm and at the absorbance maximum for the fluorescent dye. Calculate the base:dye ratio and RNA concentration according to the calculator provided by http://www.ambion.com/techlib/append/base_dye.html. Cy3-labeled chimeras sense stand and antisense strands were mixed and refolded in refolding buffer as described above.
2.2.6 Cell culture
HEK 293 cells were cultured in DMEM supplemented with 10% FBS. CHO-WT and CHO-EE cells were grown in GMEM-S medium (Glutamine-deficient minimal essential medium with 400 μM methionine sulfoximine). Peripheral blood mononuclear samples were obtained from healthy donors. PBMCs were isolated from whole blood by centrifugation through a Ficoll-Hypaque solution (Histopaque-1077, Sigma). CD8 cells (T-cytotoxic/suppressor cells) were depleted from the PBMCs by CD8 Dynabeads (Invitrogen, CA) according to the manufacturer’s instructions. CD8+ T cell-depleted PBMCs were washed twice in PBS and resuspended in culture media (RPMI 1640 with 10% FBS, 1×PenStrep and 100 U/mL interleukin-2). Cells were cultured in a humidified 5% CO2 incubator at 37 °C.
2.2.7 Cell-surface binding studies by flow cytometry
The CHO-WT gp160 or CHO-EE control cells were washed with prewarmed washing buffer (DPBS), trypsinized and detached from the plates. After washing cells twice with 500 μL binding buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 2.7 mM KCl and 0.01% BSA), cell pellets were resuspended in binding buffer and incubated at 37°C for 30 min. Cells were then pelleted and resuspended in 50 μL of prewarmed binding buffer containing 400 nM Cy3-labeled experimental RNAs. After incubation at 37°C for 40 min, cells were washed three times with 500 μL of prewarmed binding buffer, and finally resuspended in 350 μL of binding buffer prewarmed to 37°C and immediately analyzed by flow cytometry.
2.2.8 Cell internalization and localization studies by real-time confocal microscopy
The CHO-WT gp160 and CHO-EE cells were grown in 35 mm plate (Glass Bottom Dish, MatTek, Ashland, MA) with seeding at 0.3×106 in GMEM-S medium to allow about 70% confluence in 24 h. On the day of the experiments, cells were washed with 1 mL of prewarmed PBS and were incubated with 1 mL of pre-warmed complete growth medium for 30 min at 37°C. Cy3-labeled RNAs at a 100 nM final concentration were added to the media and incubated for live-cell confocal microscopy in a 5% CO2 microscopy incubator at 37 °C. The images were collected every 20 min using a Zeiss LSM 510 Meta Inverted 2 photon confocal microscope under water immersion at 40×magnification. After 5 hours of incubation and imaging, the cells were stained by treatment with 0.15 mg/mL Hoechst 33342 (nuclear dye for live cells, Molecular Probes, Invitrogen, CA) according to the manufacturer’s instructions. The images were collected as described previously.
2.2.9 In vitro HIV-1 challenge and p24 antigen assay
HIV-1 NL4-3 virus was obtained from the AIDS Research and Reference Reagent Program. After propagation of virus, store in aliquots at −80°C. Human PBMCs were infected with HIV NL4-3 virus (MOI 0.001). After 24 hour of infection, cells were gently washed with PBS three times to remove free virus. Continue to culture the infected cells in a 5% CO2 microscopy incubator at 37°C for 4 days.
Prior to RNA treatments the infected cells were gently washed with PBS three times to remove free virus. 2×104 infected cells and 3×104 uninfected cells were incubated with refolded experimental RNAs at 400 nM final concentration in 96-well plates at 37°C (100 μL per well, triplex assay). The culture supernatants (10 μL per well) were collected at different times (4 d, 6 d, 8 d and 10 d) and stored at −20°C until p24 assay. The p24 antigen analyses were performed using a HIV-1 p24 Antigen ELISA kit.
2.2.10 Serum stability assay
Five micrograms of both regular and 2′ F modified pRNA-siRNA chimera were incubated at 37°C in 100 μL RPMI 1640 Medium containing 10% (v/v) FBS, resulting in a concentration of 50 ng/μL RNA. At times 0 min to 12 hours, 10 μL aliquots of the reaction was withdrawn and 2 μL of 6 × loading buffer added to each sample. 12 μL of each mixture was analyzed by electrophoresis in an 8% native polyacrylamide gel and the RNAs were visualized following ethidium bromide staining using a UV-transilluminator.
2.2.11 The siRNA function detection by psiCHECK dual luciferase assay
Before day one of the assay, the HEK293 cells were grown in 24-well plate with seeding at 0.8×105 in 400 μL DMEM medium to allow about 70% ~ 80%confluence in 24 h. Nucleic acid mixtures containing 100 ng of siCHECK target derivative, 25 nM of experimental RNAs in 50 μL OptiMEM were cotransfected into HEK 293 cells. After 24 h of post-transfection, the reporter gene expression was tested by Dual-luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instructions. All samples were cotransfected in triplicate and the experiment performed a minimum of three times. For each replicate, the Renilla-luc target reading was normalized internally to the Firefly-luc value, and an average value was calculated from the replicates.
2.2.12 In vitro Dicer assays
The antisense strands were end-labeled with T4 polynucleotide kinase and γ-32P-ATP. Subsequently, corresponding chimera sense strands are annealed with equimolar amounts of 5′-end-labeled antisense strands in HBS buffer to form the chimeras. The annealed pRNA-siRNA chimeras (1 pmol) were incubated at 37°C for 40 min in the presence or in the absence of 1 U of human recombinant Dicer enzyme following the manufacturer’s recommendations (Ambion, Austin, TX). Reactions were stopped by phenol/chloroform extraction and the resulting solutions electrophoresed in a denaturing 15% polyacrylamide gel. The gels were subsequently exposed to X-ray film.
2.2.13 Formation of Dimer of pRNA-aptamer chimera/pRNA-siRNA chimera (gel shift assay)
8% native polyacrylamide gels were prepared in TBM buffer (89 mM Tris, 200 mM boric acid, 1 mM MgCl2, pH 7.6). Ba′ pRNA-A-1 chimeras and Ab′ pRNA-siRNA chimeras were refolded in 1×HBS buffer as described above. Equal amounts of the refolded Ba′ pRNA-A-1 chimeras and Ab′ pRNA-siRNA chimeras were mixed and directly analyzed by electrophoresis in an 8% native polyacrylamide gel. After running at 4 °C for 2 h using TBM running buffer, the RNAs were visualized following ethidium bromide staining using a UV-transilluminator.
2.2.14 Statistical Methods for HIV-1 challenge p24 assay
We tested for differences in the mean and areas under the curve with the student t-test. To determine if the Ba′ pRNA-gp120 aptamer chimeras would also block HIV infectivity in cell culture, we tested for parallel slopes with the Chow test for heterogeneous linear regression.
3. Results
3.1 Ba′ pRNA-gp120 aptamer chimeras bind to the HIV-1 gp120 protein and the cells expressing HIV gp160
Two different chimeric Ba′ pRNA-anti gp120 aptamer constructs (pRNA-A-1 D3 and pRNA-A-1 D4) were designed as shown in Figure 2A. Specifically, for the pRNA-A-1 D3 construct, the A-1 aptamer sequence was linked to the base 23 and 97 of Ba′ pRNA since the bases 23–97 are the minimum fragment required for stable dimer formation [41]. The constructed chimeric pRNA-aptamer sequence has the new 5′/3′ end relocated to bases 71/75 of the pRNA [42]. For the pRNA-A-1 D4 construct, the A-1 aptamer was directly appended to the 5′-end of the Ba′ pRNA sequence. Both constructs are shown to fold properly and maintain the capability to interact with its Ab′ pRNA partners.
Figure 2.

The design and binding activity of the pRNA-aptamer chimeras. (A) Schematic of Ba′ pRNA-Aptamer RNAs (pRNA-A-1 D3 and pRNA-A-1 D4) and RNA sequences. The A-1 aptamer sequence was inserted into the 3′/5′ double helical domain (23 nt fragment) and loop domain (97 nt fragment) of Ba′ pRNA to obtain pRNA-A-1 D3 construct. The intact Ba′ pRNA sequence was directly appended to the 3′-end of the A-1 aptamer to obtain pRNA-A-1 D4 construct. The pRNA fragments (5′/3′ double stranded helical domain; right hand loop and left hand loop) are highlighted by a clear box; the intermolecular interacting domain is highlighted by a grey box. (B) Binding affinity detected by a gel shift assay. The 5′-end P32 labeled aptamers or Ba′ pRNA-aptamer chimeras were incubated with increasing amounts of gp120 protein. The binding reaction mixtures were analyzed by a gel mobility shift assay and the Kd determinations are indicated. The Ba′ pRNA-aptamers showed a comparable binding affinity with the target protein as well as aptamer alone. (C) Cell-type specific binding studies of pRNA-aptamer chimeras. Cy3-labeled Ba′ pRNA-aptamers were incubated with CHO-WT gp160 cells and CHO-EE control cells and cell surface binding of Cy3-labeled chimeras was assessed by flow cytometry. (D, E) Internalization and localization analysis. CHO-WT gp160 cells (D) or CHO-EE cells (E) were grown in 35 mm plates and incubated with a 100 nM concentration of Cy3-labeled pRNA-A-1 D4 chimera in culture media for real-time live-cell-confocal microscopy analysis. The images were collected at 20 min. intervals using 40 × magnification. After 5 hours incubation with 100 nM of Cy3-labeled chimera, cells were subsequently stained with Hoechst 33342 (nuclear dye for live cells) and then imaged using real-time confocal microscopy.
To enhance the stability of these chimeric RNAs in cell culture and in vivo, the Ba′ pRNA-aptamer contained nuclease-resistant 2′-Fluoro UTP and 2′-Fluoro CTP and were synthesized from the corresponding double stranded DNA templates by in vitro T7 bacteriophage transcription. The binding affinities of the Ba′ pRNA-aptamer chimeric RNAs for HIV-1 gp120 protein was assessed by using a gel shift assay and flow cytometry.
First, the dissociation constants (Kd) for Ba′ pRNA-aptamer chimeras with the target protein HIV-1Bal gp120 were calculated from a native gel mobility shift assay (Figure 2B). They showed good binding kinetics to the gp120 protein. The apparent Kd values of the A-1 aptamer alone, pRNA-A-1 D3 and pRNA-A-1 D4 were about 48 nM, 49 nM and 79 nM, respectively.
Next, CHO-WT cells stably expressing the HIV envelope glycoprotein gp160 (of which gp120 is a component) were used to test for binding of the pRNA-aptamer chimeras. These cells do not process gp160 into gp120 and gp41 since they lack the gag encoded proteases required for envelope processing. As a control we used the parental CHO-EE cell line which does not express gp160. The anti-gp120 aptamer A-1 and the Ba′ pRNA-aptamer chimeras were labeled with Cy3 to follow their binding and uptake. Flow cytometric analyses (Figure 2C) revealed that the aptamer and chimeras specifically bind to the CHO-gp160 cells. These data indicate that the Ba′ pRNA-aptamer chimeras maintain approximately the same binding affinities as the parental aptamers alone. In order to determine if the chimeras were internalized in the gp160 expressing cells, we also carried out real-time live-cell Z-axis confocal microscopy with the CHO-WT gp160 cells incubated with the Cy3-labeled pRNA-A-1 D4 chimera (Figure 2D). These cells express the precursor for gp120, gp160 in which the gp41 portion has not been processed by the HIV-1 gag protease since the cells only are transfected with the envelope gene. The gp120 glycoprotein is exposed on the cell surface whereas the gp41 segment is within the cell membrane. After 5 hours of incubation, the Cy3-labeled construct was selectively internalized within the CHO-WT gp160 cells, but not the CHO-EE control cells (Figure 2E). To visualize the nucleus, the cells were stained with the nuclear dye Hoechst 33342 after incubation with the Cy3 labeled chimera. Figure 2D showed that the chimera aggregated within the cytoplasm suggesting that the gp120 aptamers maybe enter cells via receptor-mediated endocytosis.
3.2 Ba′ pRNA-gp120 aptamer chimeras inhibit HIV-1 infection of human PBMCs
In a previous study investigators demonstrated that anti-gp120 aptamers provided significant anti-HIV function via binding to the viral envelope and preventing viral interaction with the cellular CD4 receptor [43–45]. We therefore carried out an assay to determine if the Ba′ pRNA-gp120 aptamer chimeras would also block HIV infectivity in cell culture. In this assay, the aptamer and chimeras were incubated with HIV-1 infected primary human PBMCs four days after the cells were challenged with the virus. At different days post treatment with the aptamers, aliquots of the media were assayed for viral p24 antigen levels (Figure 3). The results showed that the Ba′ pRNA-gp120 aptamers (pRNA-A-1 D4) inhibited HIV-1 p24 production and provided comparable inhibitory potency as well as the original A-1 aptamer, reaching statistical significance (P = 0.0302 and P = 0.0055, respectively).
Figure 3.
The inhibition of HIV-1 infection mediated by pRNA-aptamer chimeras. Both anti-gp120 aptamer and pRNA-aptamer chimera neutralized HIV-1 infection in HIV infected human PBMCs (NL4-3 strain) culture. Data represent the average of triplicate measurements of p24. Ba′ pRNA-gp120 aptamer chimera D4 inhibits HIV-1 infection of human PBMCs with comparable inhibitory potency as original A-1 aptamer. Aptamer A-5 that has previously been shown to have poor affinity for gp120 is used as a negative control. P values for the effects of A-1 and pRNA-aptamer chimera are indicated and were calculated as described in Materials and Methods.
3.3 Ab′ pRNA-siRNA chimeras improve RNase resistance and specifically knock-down target gene expression
It has been demonstrated that the double-stranded helical domain at the 5′/3′ end and the intermolecular interaction domain (Loop-loop region) in the pRNA can fold independently of each other, and altering the primary sequences of the 5′/3′ helical domain in the pRNA does not affect its folding structure as long as the two strands are base-paired [24, 27]. Because of this tolerability, the double-stranded helical region of the pRNA can be replaced with a duplex RNA (e.g. siRNA) (Figure 1B), to introduce additional functionalities to pRNA nanoparticles. Since the synthetic Dicer substrate duplexes of 25–30 nt have been shown to enhance RNAi potency and efficacy [46], we chose a 27 mer Dicer substrate RNA as the siRNA portion of the chimeric molecule. The 27 mer siRNA portion of chimeras targets the HIV-1 tat/rev common exon sequence. In the design we inserted a two nucleotide linker (UU) between the Ab′ pRNA and siRNA portion to minimize steric interference of the pRNA portion with Dicer.
As described above, the Ab′ pRNA and sense strand segment of the siRNAs contained nuclease-resistant 2′-Fluoro UTP and 2′-Fluoro CTP and were synthesized from corresponding dsDNA templates by in vitro T7 transcription. To prepare the siRNA containing chimeras, in vitro transcribed chimeric Ab′ pRNA-sense strand polymers were annealed with equimolar concentrations of an unmodified antisense strand RNA (Figure 4A). In the design, 2′-Fluoro modified chimeras were stable in cell-culture media containing 10% FBS for up to 12 hours whereas the unmodified chimeric RNAs were degraded within several minutes (Figure 4B).
Figure 4.
The design and evaluation of Ab′ pRNA-siRNA chimeras. (A) Schematic of the Ab′ pRNA-siRNA chimera. The siRNA part of the chimera consists of 27 bps. As an example here, the siRNA targets HIV-1 tat/Rev. The chimeric pRNA-siRNA sense strand was transcribed in vitro with T7 RNA polymerase and then annealed with the antisense strand to get the whole pRNA-siRNA chimera. A linker (UU) between the aptamer and siRNA is indicated. (B) The stability assay in cell culture medium containing 10% FBS of the 2′-F modified pRNA-siRNA chimeras vs. unmodified pRNA-siRNA chimeras. (C) Gene silencing activity and strand selectivity of pRNA-siRNA chimeras. Dual luciferase assays of psiCHECK sense and anti-sense targets are shown. All RNAs were normalized to the value of the corresponding buffer control. All the data were represent the averages of triplicate assays.
Because of competition by the sense (passenger) strand with the anti-sense (guide) strand for RISC entry, the strand selectivity is an important factor for evaluating siRNAs. Therefore, we evaluated the Ab′ pRNA-tat/rev siRNA chimera RNA using the pSiCheck reporter system, which readily allows screening of the potencies of candidate sh/siRNAs. The gene silencing of both the sense target (corresponding to the mRNA) and the anti-sense target were tested independently and the selectivity ratios calculated as a measure of the relative incorporation of each strand into the RISC. The comparison (Figure 4C) demonstrated that the Ab′ pRNA-tat/rev siRNA chimera mediated ~83% knockdown of the sense target; however, knockdown of the anti-sense target is much less (~63 %), indicative of good strand selection (R = 2.3).
3.4 Ab′ pRNA-siRNA chimeras are processed by Dicer
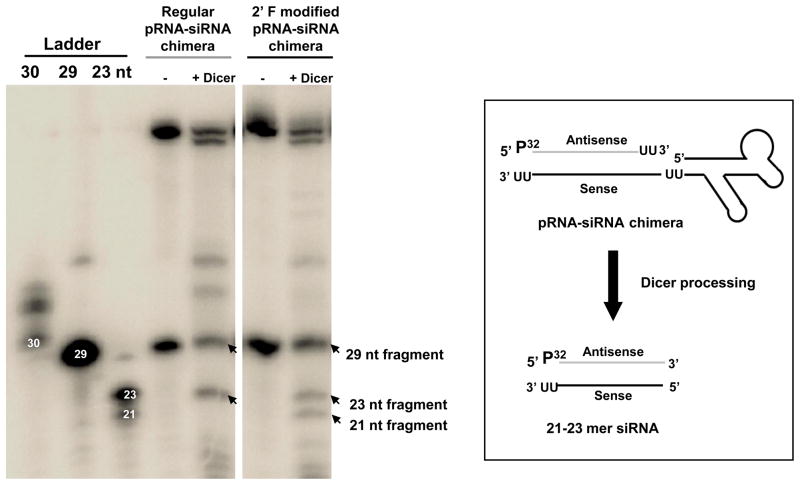
We next detected whether or not the siRNAs could be processed from the chimeric pRNA-siRNA in vitro by recombinant human Dicer. This set of experiments used a 5′-end P32 labeled antisense strand to follow Dicer processing (Figure 5). The size of the P32 labeled cleavage product(s) indicates the direction Dicer enters the siRNA and cleaves [47]. When pRNA-siRNA chimeras were incubated with the recombinant human Dicer we observed that the 29 nt antisense strand (containing 2 nt 3′-overhang) was processed into a 21–23 nt siRNA. This result suggests Dicer processing preferentially enters from the 5′ end of the antisense strand and cleaves 21 to 23 nt downstream, leaving the 3′ end of the antisense strand intact. These data suggest that the Ab′ pRNA-siRNA chimeras can be processed by Dicer and result in an siRNA that guides target gene silencing.
Figure 5.
In vitro Dicer processing. Dicer cleavage of 5′-end P32 antisense labeled RNAs [47]. The RNA sense strands (2′-F modified or regular RNA) were annealed with equal molar equivalents of 5′-end P32-labeled complementary antisense RNA strands. The cleavage products or un-cleaved, denatured strands were visualized following 15% denaturing polyacrylamide-gel electrophoresis.
3.5 Ba′ pRNA-aptamer chimeras and Ab′ pRNA-siRNA chimeras form dimers through Ba′ and Ab′ loop interaction
In order to determine whether or not the Ba′ pRNA-aptamer chimeras and Ab′ pRNA-siRNA chimeras can form dimers, we carried out the gel shift assay to monitor dimerization. In this study, the mixing of one pair of pRNAs with interlocking loops was carried out to test whether the Ba′ pRNA-aptamer and Ab′ pRNA-siRNA chimeras were still able to form dimers through loop-loop interactions. As shown in Figure 6A, dimers could form between two loops (Ab′ pRNA and Ba′ pRNA) at different concentration of Mg2+ (1 mM is shown as example). Compared with the pRNA alone, the additional aptamer or siRNA segments in the chimeras somewhat reduced the efficiency of dimerization (Figure 6A).
Figure 6.
Dimerization study of Ba′ pRNA-aptamer and Ab′ pRNA-siRNA chimeras. (A) Formation of dimmers composed of Ba′ pRNA-aptamer chimeras and Ab′ pRNA-siRNA chimeras through the interaction of right and left hand loops of Ba′ and Ab′ in gel shift analysis at 1 mM Mg2+. (B) In vitro Dicer processing of dimers derived from 5′-end P32 RNAs [47]. Ba′ pRNA-aptamer D4 and Ab′ pRNA-siRNA chimers were mixed to form dimers and incubated with Dicer. “D” means dimer; “M” means monomer; “+” means with Dicer; “−” means without Dicer. The cleavage products (arrows) or un-cleaved, denatured strands were visualized following 15% denaturing polyacrylamide-gel electrophoresis. (C) Flow cytometry assay for cell-type specific binding of dimer. Ba′ pRNA-aptamers or Ab′ pRNA-siRNA chimeras with Cy3 label at either the pRNA-sense or antisense strands were tested for binding to CHO-WT gp160 cells and CHO-EE control cells.
The in vitro Dicer assay [47] was also performed to verify that Dicer can process the siRNA portion after dimer formation (Figure 6B). Similarly, the dimers of Ba′ pRNA-aptamer and Ab′ pRNA-siRNA chimeras wer preferentially cleaved from the 5′-end labeled antisense side resulting in the 32P- labeled 21–23 nt species.
Next, to examine whether or not the Ba′ pRNA-aptamer selectively delivers Ab′ pRNA-siRNA chimeras to the cells expressing gp160, we performed the flow cytometric analysis (Figure 6C) as previous described. As expected, the Cy3-labeled Ba′ pRNA-aptamer specifically bound to the CHO-gp160 cells but did not bind to the control CHO-EE cells. The Ab′ pRNA-siRNA chimeras in which either the chimera pRNA-sense or antisense strands were labeled with Cy3 and subsequently used in formation of the chimeras, didn’t show differences between the two cell lines. However, the combination of the Ba′ pRNA-aptamer D4 with the Cy3-labeled Ab′ pRNA-siRNA chimera resulted in a three-fold binding specificity with the CHO-gp160 cells as compared with the CHO-EE control cells. These results indicate that the pRNA-siRNA chimera is delivered by the pRNA-aptamer to gp160 expressing cells via dimerization.
4. Discussion
The essential feature of the RNAi mechanism is the sequence-specificity, which derives from complementary Watson-Crick base pairing of a target messenger RNA (mRNA) and the guide strand of the siRNA [48]. Within the past decade, RNA interference has shown great potential as a therapeutic modality for various diseases. HIV-1 became one of the first infectious agents targeted by RNAi due to its well-understood life cycle and pattern of gene expression. However, getting the technique to work in vivo will require targeted delivery of siRNAs to virally infected cells or cells which are targets for HIV-1 infection.
We have capitalized upon the exquisite specificity of an anti-gp120 aptamer to deliver anti-HIV-1 siRNAs to HIV-1 infected cells, thereby resulting in a dual inhibitory function of replication and spread of HIV by the combined action of the aptamer and siRNA targeting the tat/rev common exon of HIV-1. This system provides a highly selective approach for inhibiting HIV-1 replication that combines siRNA delivery with the inhibitory properties of an anti-HIV-1 envelope aptamer. On the other hand, pRNA has been reported to have novel applications in nanotechnology and nanomedicine [13]. Through the interaction of two reengineered interlocking loops, pRNAs are able to form nanoscale dimers, trimers, hexamers and patterned superstructures. This unique ability of pRNA makes it a versatile polyvalent vehicle to combine siRNAs and other therapeutic molecules and may be applied as a therapeutic nano-platform which can improve drug solubility and pharmacokinetic (PK)/distribution properties.
In the present study, we therefore took advantages of the gp120 aptamer binding affinity for HIV-1 gp120 and the ability of pRNA to form dimers to explore the potential use of chimeric pRNA-aptamers for delivery of anti-HIV siRNAs into HIV-1 infected cells. In this delivery system, the Ba′ pRNA and Ab′ pRNA were successfully fused with an anti-gp120 aptamer and an HIV siRNA against HIV-1 tat/rev, respectively. The resultant chimeric pRNA-aptamer and pRNA-siRNA did not interfere with the function of either moiety. We designed two different pRNA-aptamer chimeras D3 and D4 (as shown in Figure 2A). Although the linkage strategy is different, both constructs are shown to fold properly and maintain the capability to interact with its Ab′ pRNA partners. Moreover, our results demonstrate that the pRNA-aptamer chimera specifically binds to cells expressing HIV gp120 and provides inhibition of HIV-1 replication comparable to the original anti-gp120 aptamer, suggesting fusion of the pRNA with aptamers does not impede the formation of dimers or interfere with binding affinity of the aptamer.
In our design of the Ab′ pRNA-siRNA chimera 2′-Fluoro backbone modifications of pyrimidines were incorporated into the pRNA, aptamer and siRNA sense strand. Although the siRNA antisense has no chemical modification, the pRNA-siRNA chimeras were stabilized by virtue of the non-modified guide strand base pairing to the modified sense strand. In contrast, unmodified pRNA-siRNA chimeras were quickly degraded in several minutes in sera. Moreover, the Ab′ pRNA-siRNA chimera also is functionally processed by Dicer and specifically induced target gene silencing with reasonably good strand selectivity. Through the Ba′ and Ab′ loop-loop interactions, the Ba′ pRNA-aptamer chimeras and Ab′ pRNA-siRNA chimeras formed dimers, which facilitate the specific delivery of Cy3-labeled chimeric pRNA-siRNA to cells expressing gp120. However, compared with pRNA alone, the additional aptamer or siRNA segments in the chimeras reduced somewhat the efficiency of dimerization. Although our results indicate the potential of pRNA-aptamer chimera as an anti-HIV agent and a targeted delivery vector for siRNA, further optimization and evaluation, for example using crosslinking of the dimers to improve stability will be necessary to optimize the dual function inhibition afforded by the aptamer and siRNA.
In conclusion, this dual functional pRNA-aptamer chimera not only represents a potential HIV-1 inhibitor, but also provides a cell-type specific siRNA delivery vehicle, showing considerable promise for systemic anti-HIV therapy. Future work will aim at optimizing the dimerized delivery system. This system might be precisely tailored as a multifunctional nanomedicine with optimal size and pharmacokinetic properties. Although the HIV-1 gp120 aptamer and anti-HIV-1 siRNA were discussed as representative examples for fusion with the pRNAs, to take advantage of pRNA-assembled nanoparticles, this combinatorial approach also can be generalized with different cell internalizing aptamers and therapeutics for the treatment of other diseases.
Supplementary Material
Acknowledgments
We thank Haitang Li and Jang Zhang for HIV-1 challenge assay. This work was supported by grants from the National Institutes of Health awarded to J.J.R and P.X.G. This work was supported by the NIH Nanomedicine Development Center on Phi29 DNA-packaging Motor for Nanomedicine, through the NIH Roadmap for Medical Research (PN2 EY 018230) as well as NIH grants AI29329 to JJR and EB003730 to PG. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: The CHO-EE and CHO-gp160 cell line; HIV NL4-3 virus; HIV-1BaL gp120.
Abbreviations
- RNAi
RNA interference
- siRNA
Small interfering RNA
- pRNA
Phi29 motor packaging RNA
- HIV
human immunodeficiency virus
- HIV-1 gp120
glycoprotein 120
Footnotes
Disclosures (Competing interests)
J. R and J. Z. have a patent pending on “Cell-type specific aptamer-siRNA delivery system for HIV-1 therapy”. USPTO, No. 12/328994, publication date: June 11, 2009. PG is the inventor of patents on phi29 motor pRNA chimera and a co-founder of Kylin Therapeutics, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jiehua ZHOU, Email: Jzhou@coh.org.
Yi SHU, Email: shuyi@mail.uc.edu.
Peixuan GUO, Email: guopn@ucmail.uc.edu, guop@purdue.edu.
David D. Smith, Email: Dsmith02@coh.org.
John J ROSSI, Email: Jrossi@coh.org.
References
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. Science. 2009:323. 1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossi JJ, June CH, Kohn DB. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherer L, Rossi JJ, Weinberg MS. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- 6.Subramanya S, Kim SS, Manjunath N, Shankar P. Expert Opin Biol Ther. 2010;10:201–213. doi: 10.1517/14712590903448158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Winters M, Holodniy M, Dai H. Angew Chem Int Ed Engl. 2007;46:2023–2027. doi: 10.1002/anie.200604295. [DOI] [PubMed] [Google Scholar]
- 9.Weber N, Ortega P, Clemente MI, Shcharbin D, Bryszewska M, de la Mata FJ, Gomez R, Munoz-Fernandez MA. J Control Release. 2008;132:55–64. doi: 10.1016/j.jconrel.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Eguchi A, Meade BR, Chang YC, Fredrickson CT, Willert K, Puri N, Dowdy SF. Nat Biotechnol. 2009;27:567–571. doi: 10.1038/nbt.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castanotto D, Rossi JJ. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo P. Nat Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 16.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ugolini S, Mondor I, Sattentau QJ. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 18.Wyatt R, Sodroski J. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 19.Chan DC, Kim PS. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 20.Blair WS, Lin PF, Meanwell NA, Wallace OB. Drug Discov Today. 2000;5:183–194. doi: 10.1016/s1359-6446(00)01484-7. [DOI] [PubMed] [Google Scholar]
- 21.Moore JP, Doms RW. Proc Natl Acad Sci U S A. 2003;100:10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo P, Zhang C, Chen C, Garver K, Trottier M. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 25.Guo PX, Erickson S, Anderson D. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 26.Hoeprich S, Guo P. J Biol Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- 27.Shu D, Huang LP, Hoeprich S, Guo P. J Nanosci Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 28.Khaled A, Guo S, Li F, Guo P. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu D, Moll WD, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo P. J Nanosci Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo S, Tschammer N, Mohammed S, Guo P. Hum Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Liu J, Diao Z, Shu D, Guo P, Shen G. Mol Biosyst. 2009;5:1361–1368. doi: 10.1039/b903428e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Guo S, Cinier M, Shlyakhtenko LS, Shu Y, Chen C, Shen G, Guo P. ACS Nano. 2010 doi: 10.1021/nn1024658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss CD, White JM. J Virol. 1993;67:7060–7066. doi: 10.1128/jvi.67.12.7060-7066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 36.Reid RJ, Bodley JW, Anderson D. J Biol Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 37.Reid RJ, Zhang F, Benson S, Anderson D. J Biol Chem. 1994;269:18656–18661. [PubMed] [Google Scholar]
- 38.Chen C, Sheng S, Shao Z, Guo P. J Biol Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 39.Garver K, Guo P. Rna. 1997;3:1068–1079. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C, Lee CS, Guo P. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 41.Chen C, Zhang C, Guo P. Rna. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Tellinghuisen T, Guo P. Rna. 1997;3:315–323. [PMC free article] [PubMed] [Google Scholar]
- 43.Khati M, Schuman M, Ibrahim J, Sattentau Q, Gordon S, James W. J Virol. 2003;77:12692–12698. doi: 10.1128/JVI.77.23.12692-12698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dey AK, Khati M, Tang M, Wyatt R, Lea SM, James W. J Virol. 2005;79:13806–13810. doi: 10.1128/JVI.79.21.13806-13810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen C, Forzan M, Sproat B, Pantophlet R, McGowan I, Burton D, James W. Virology. 2008;381:46–54. doi: 10.1016/j.virol.2008.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 47.Guo S, Huang F, Guo P. Gene Ther. 2006;13:814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.