Abstract
With the completion of the Genome Sequencing Project, it is now possible to rapidly and accurately determine the frequency and position of a particular repeat sequence in the Caenorhabditis elegans genome. Several repeat sequences with a variety of characteristics have been examined and with few exceptions they show a near-random distribution throughout the genome. We characterized several genes near the left end of Chromosome III in the C. elegans genome, and found a 24-bp minisatellite repeat sequence present in the introns of two unrelated genes. This prompted a search of the databank for other occurrences of this sequence. Multiple copy arrays of this repeat are all located on the same autosome and fall in two clusters: one near the left end, and one in the central region separated by ∼10 Mb. There are >200 copies of this repeat on the chromosome. This euchromatic repeat sequence seems unrelated to gene expression, is absent from homologous sites in a related species, is unstable in Escherichia coli, and is polymorphic between different wild isolates of C. elegans. Most CeRep25B units in the array match the consensus sequence very well, suggesting that either this repeat originated quite recently or its sequence is functionally constrained. Although chromosome-specific repeat sequences have been reported previously in many organisms, such sequences are usually structural and heterochromatic (e.g., centromeric α-satellite) or on the mammalian sex chromosomes. This report describes the first confirmed instance from a whole genome sequencing project of an autosomal euchromatic chromosome-specific minisatellite repeat.
Repetitive elements in eukaryotic genomes have been known for decades (Szybalski 1968) and have been a matter of interest for most of that time (Singer 1982), but apart from classes such as transposable elements or α-satellites at centromeres, no function has been ascribed to them yet. Our knowledge has come largely from three sources: whole genome hybridization [e.g., C0t curves (Britten et al. 1974; Wilson and Thomas 1974) and probing Southern Blots with total genomic DNA], differential buoyant density [e.g., satellite DNAs (Szybalski 1968)], and fortuitous identification while characterizing genomic DNA clones isolated for other purposes (Healy et al. 1988). Repetitive elements have been found in all metazoans examined, yet the sequences are not conserved. Up to 30% of the human genome is repetitive, and repetitive DNA is distributed throughout at least 80% of the genome (Schmid and Deininger 1975). There are several main types of repetitive sequences (for review, see Charlesworth et al. 1994). Satellite sequences are relatively short repeats (5–200 bp) typically arranged in megabase-sized clusters [e.g., α-satellite (Wevrick and Willard 1989; Oakey and Tyler-Smith 1990)] in constitutive heterochromatin. Transposable elements and human Alu sequences are examples of LINEs and SINEs. Also found euchromatically are tens of thousands of short microsatellite loci (∼2–4 bp), as well as minisatellites (∼10–20 bp per repeat), that can be variable in array size. Very little is known about the function of the shorter repetitive sequences, although in Drosophila it is thought that some of these repeats may help regulate global chromatin structure and gene expression (Csink and Henikoff 1998).
Caenorhabditis elegans is not the first eukaryote to have its genome sequence available, but the yeast Saccharomyces cerevisiae is a poor paradigm for the study of evolution and organization of repeated sequences [with certain exceptions (Anderson and Nilsson-Tillgren 1997; Grunstein 1998; Kim et al. 1998)], due primarily to its active homologous recombination system and to its atypical organization of centromere and replication origin sequences. It is now possible to determine the frequency and position of copies of a particular repeat sequence in the C. elegans genome rapidly and accurately because of the near completion of the Genome Sequencing Project. Several repeat sequences with a variety of characteristics have been examined in this species (Emmons et al. 1980; Felsenstein and Emmons 1988; La Volpe et al. 1988; Naclerio et al. 1992), and with few exceptions they show a near-random distribution throughout the genome. Although some repeats are concentrated in particular parts of the genome, such as near telomeres [e.g., RcS5 and CeRep3 (Cangiano and La Volpe 1993)] or outside the “gene clusters” at the center of the genetic map of each chromosome [e.g., CeRep3 (Felsenstein and Emmons 1988; Barnes et al. 1995)], this represents an enrichment in a region, not an absolute restriction.
There have been reports of chromosome-specific repeats in some systems (Das et al. 1987; Stallings et al. 1992; Kogi et al. 1997), but their characterization has been relatively minimal. Low numbers of repeats, or repeats in an array size too small to be detected by in situ hybridization, may still be found elsewhere in the genome. Chromosome-specific sequences include centromeric α-satellites that are either structural and heterochromatic (Haaf and Willard 1992) or on the mammalian sex chromosomes where Y-specific sequences can be conserved between species (Guttenbach et al. 1992). C. elegans has a genome organization that is typical of larger eukaryotes in many ways, with respect to the distribution of repeat sequences. However, the C. elegans genome is unique in that it lacks classical heterochromatin. The diffuse centromeres of C. elegans means that the centromeric heterochromatin (specifically satellite sequences) found in other species is lacking (Sulston and Brenner 1974; Emmons et al. 1980). It is not clear what sequences act in cis to provide the roles that centromeres play in other species.
We have characterized several genes in a small area of the C. elegans genome, at the left end of Chromosome III (Pilgrim 1993) and have commented previously on a novel repeat sequence present in the intron of the sex-determining gene fem-2 (Pilgrim et al. 1995). Recently, when the same repeat was found recently in two different introns of a nearby unrelated gene, unc-45, which is required for normal muscle function (L. Venolia, W. Ao, S. Kim, C. Kim, and D. Pilgrim, in prep.), it prompted a search of the C. elegans genomic DNA sequence for other occurrences of this sequence. The repeat in question is a 24-bp minisatellite that is found at several different locations in the C. elegans genome. With the exception of a single match, multicopy arrays of this repeat are specific to Chromosome III, where they fall into two clusters: one near the left end, and one in the central region. Hence, this is the first instance of an autosomal euchromatic chromosome-specific minisatellite array.
RESULTS
The repeats present in the introns of C. elegans fem-2 have been noted previously (Fig. 1; Pilgrim et al. 1995) and their the palindromic nature was observed. It was only when the same repeats were also found in two introns of unc-45—a nearby but unrelated gene whose sequence was also characterized in the laboratory (L. Venolia, W. Ao, S. Kim, C. Kim, and D. Pilgrim, in prep.)—that it was decided to determine their overall genomic distribution. Because both genes mapped to the same region of the genome (Pilgrim 1993), it was not clear whether these repeats were very rare and occurred coincidentally or were widespread but unrecognized elsewhere in the genome. Repeat sequences from these introns (array sizes of 9, 21, and 27 repeats of 24 bp) were used to produce a consensus sequence (Fig. 1A). As with the individual repeats, the consensus sequence is a perfect palindrome, with no loop. This repeat is a derivative of CeRep25, which has since been described by the C. elegans Genome Sequencing Project (Wilson et al. 1994). CeRep25 is a 31-bp sequence, whereas the sequence described here fits best to a 24-bp consensus, with two degeneracies (see below). Hence, the 24-bp sequence will be referred to as CeRep25B.
Figure 1.
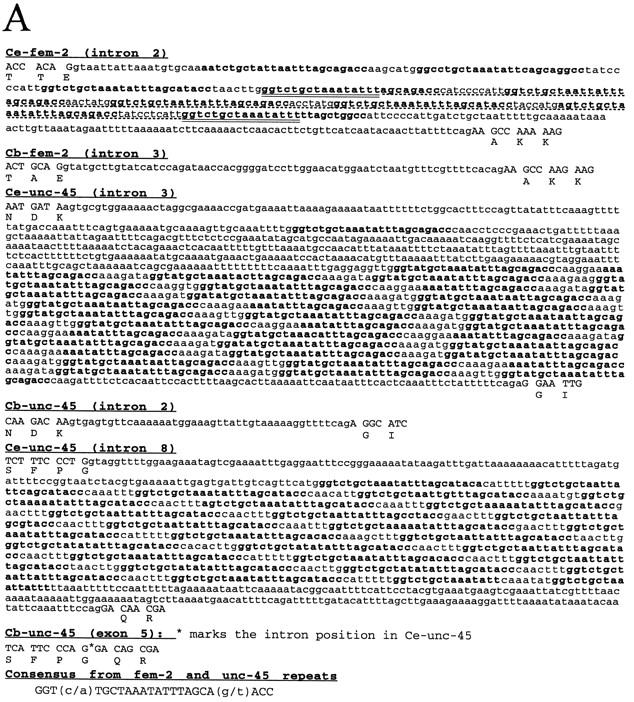
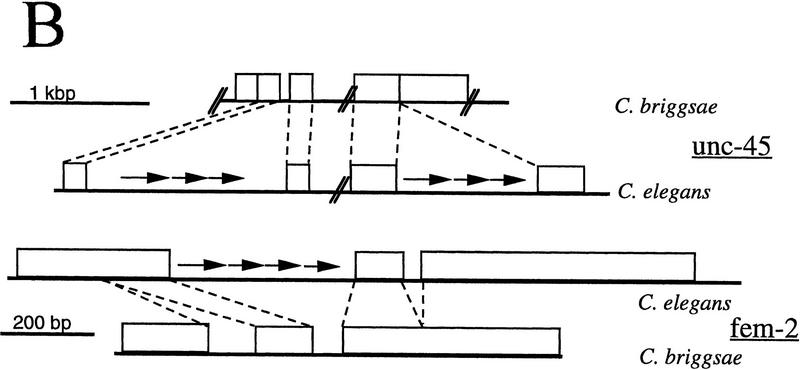
Arrangement of the CeRep25B repeat sequences within the fem-2 and unc-45 genes. (A) Sequences of CeRep25B-containing intron sequences for C. elegans (Ce) fem-2 and unc-45, and the corresponding intron sequences from the related nematode Caenorhabditis briggsae (Cb). Intron sequences are in lowercase, exon sequences in uppercase letters. Translation is shown in single-letter code under the flanking exon sequences. CeRep25B sequences (both full repeats and half elements) are shown in bold. In the C. elegans fem-2 intron sequence, the portion deleted in a recovered E. coli clone is underlined dotted, and the endpoints of the deletions lie within the doubly underlined portion. In the C. briggsae unc-45 sequence, no intron is found in the corresponding position to Ce–unc-45 intron 8, which is marked on the DNA sequence with an asterisk (*). (B) Schematic outline of the C. elegans and C. briggsae homologs. Arrows indicate the position (but not the number) of the CeRep25B sequences. CeRep25B sequences account for most of the difference in size of the introns.
Since they are intronic, these repeats are transcribed but are spliced out of the final mRNA. It is extremely unlikely that these sequences can have any role in gene expression from these loci. First, the genes in which these repeats are found are expressed in different tissues, and the genes have different mutant phenotypes (Pilgrim et al. 1995; L. Venolia, W. Ao, S. Kim, C. Kim, and D. Pilgrim, in prep.). Second, cDNA sequences that lack these repeats are sufficient to rescue the mutant phenotypes when driven by their normal promoters (Pilgrim et al. 1995; L. Venolia, W. Ao, S. Kim, C. Kim, and D. Pilgrim, in prep.). Finally, the Caenorhabditis briggsae homologs of both of these genes (which lack these repeats) have been isolated and can rescue most if not all the phenotypes of the C. elegans mutants (Fig. 1; Pilgrim et al. 1995; Hansen and Pilgrim 1998; L. Venolia, W. Ao, S. Kim, C. Kim, and D. Pilgrim, in prep.). This repeat element is almost completely responsible for the difference in intron sizes between the two homologs of each gene (Fig. 1B). These repeat sequences are unstable in Escherichia coli, and a plasmid with a precise deletion of five (of nine) of the repeat units was fortuitously isolated. The bases at which the deletion occurred cannot be determined precisely because the repeat but fall within the underlined sequences shown in Figure 1A.
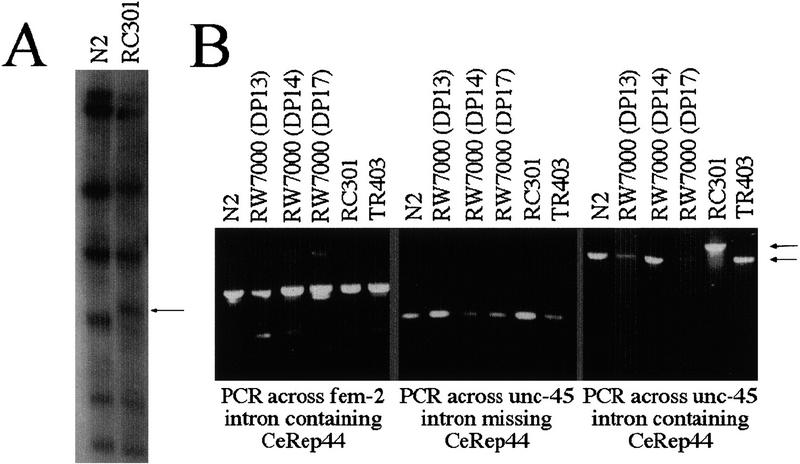
In previous work (Pilgrim 1993) a naturally occurring RFLP was found in a wild isolate (RC301) of C. elegans when a Southern blot was probed with the cosmid W10B10. This is the cosmid that was subsequently shown to contain the unc-45 gene (L. Venolia, W. Ao, S. Kim, C. Kim, and D. Pilgrim, in prep.). With the sequence of the cosmid now available, the pattern of restriction enzyme cut sites can be predicted to see if the polymorphism (eP97) is due to a change in the CeRep25B microsatellite array. The eP97 polymorphism was detected following digestion with ClaI, which produced a 4.5-kb band in the canonical wild-type stain (N2), and a 4.95-kb band in RC301. Analysis of the unc-45 genomic sequence predicts fragment sizes consistent with those seen on the Southern blot. In particular, the CeRep25B elements in unc-45 intron 3 are predicted to lie within a 4531-bp ClaI fragment. Because the sizes of the flanking ClaI fragments are not changed (at least at the resolution of a Southern blot; Fig. 2A), the hybridization results are consistent with an increase in size of this fragment of 400 bp in the RC301 strain. PCR was used to confirm that this polymorphism is found in the same intron as the repeat. Two fragments were amplified: one from exon 3 to exon 5 (1.7 kb), which will contain the CeRep25B element, and an overlapping one from exon 4 to 5 (235 bp) which will not. These primers were used to amplify from genomic DNA of N2 and RC301 (Fig. 2). No difference was seen in the fragment corresponding to the intron that lacks the repeat. However, RC301 showed an increase in the larger PCR fragment, consistent with the 400-bp size increase seen on the Southern blot (Fig. 2B). This size increase must lie in intron 3 or in the flanking exonic sequences. Because RC301 does not have an unc-45 phenotype, the simplest explanation is that the increase is in the intron, where CeRep25B is found. The change in size would require at least 13 CeRep25B units to be added. Thus, neither the presence of the array in an intron nor a 50% increase in its size has a detectable effect on unc-45 gene expression.
Figure 2.
CeRep25B is responsible for the eP97 RFLP. (A) Southern blot of genomic DNA from N2 and RC301 strains probed with cosmid W10B10. The eP97 polymorphism is indicated with an arrow. (B) PCR amplification of genomic DNA from a variety of C. elegans isolates. Genomic DNA, which was used as the template, is indicated. (See Methods for strain and PCR details.) The three panels are aligned such that relative fragment size is not maintained. Arrows indicate positions of wild-type and RC301 PCR fragments for unc-45 intron 3.
Polymorphisms were also detected between different isolates of the Bergerac strain RW7000 (Pilgrim 1993). In particular, the eP64 polymorphism near fem-2 was found to differ in RW7000 isolates from different laboratories (Williams et al. 1992; Pilgrim 1993). With the same PCR primers, a collection of RW7000 isolates was examined, but no differences in CeRep25B length in unc-45 were detected (Fig. 2). PCR was also used to examine the intron in the fem-2 gene that contains CeRep25B; however, no differences in the size of the PCR fragment were observed in any of the wild isolates, including RC301 (Fig. 2B).
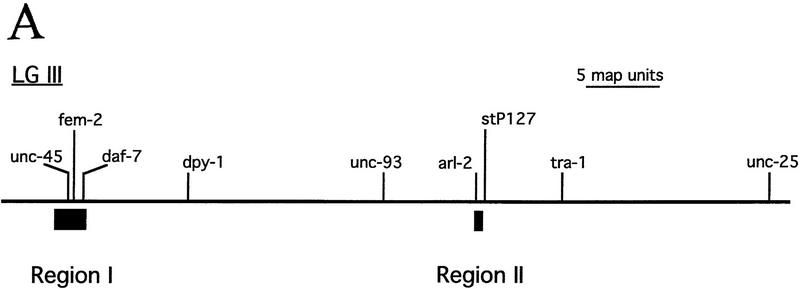
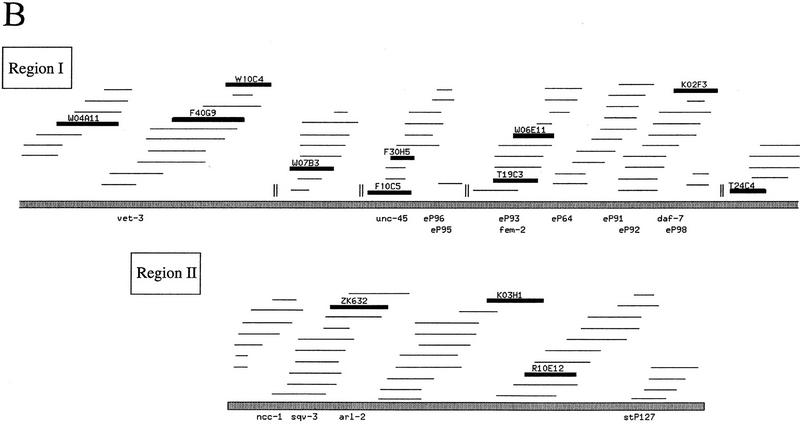
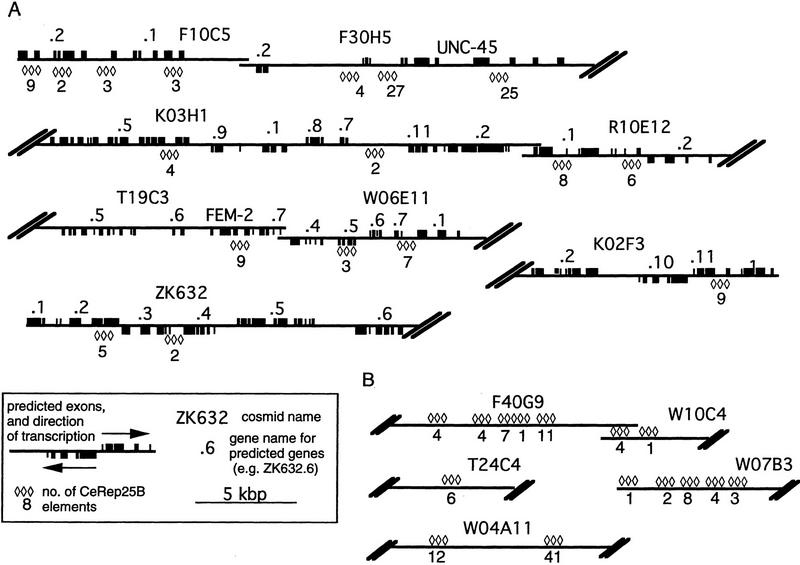
The consensus sequence from Figure 1A was used to search GenBank, as well as for a direct search of the C. elegans genomic DNA sequence, for other occurrences of the repeat. Sequences in which fewer than 22/24 bases matched the consensus were ignored. No match outside C. elegans was found. Apart from a single copy of CeRep25B on cosmid C07E3 (two mismatches to 24-bp consensus) on Chromosome II, 13 different C. elegans cosmids in two large clusters on Chromosome III were detected. One cluster lies at the left end of Chromosome III (including fem-2 and unc-45), and the other in a region in the center of the genetic map (Fig. 3A). The two regions are estimated to span a total of 750 kb, 5% of the total length of the chromosome (Fig. 3B). In these two clusters, there are 32 tandem arrays of the repeat (Fig. 4). Within each of the 32 arrays, a manual search of the flanking sequences for degenerate copies of the full palindromic (24 bp) or half repeat (12 bp) identified 231 recognizable full 24-bp repeats, a further four with insertions or deletions of 1 to 3 bp, and thirty-one 12-bp half elements. Although unc-45 is unusually rich in CeRep25B elements (over one-quarter of all CeRep25B elements are in unc-45 or its regulatory sequences), the remaining elements are relatively evenly spread over the cosmids in these two regions (Fig. 4) and are usually found in several different genes on the same cosmid (Fig. 4A).
Figure 3.
Location of CeRep25B sequences on the physical map. (A) Genetic map of Chromosome III, showing the positions of the cosmids containing CeRep25B sequences and selected genetic markers. (B) Physical map of the two regions of Chromosome III. The short horizontal lines are individual cosmids (only CeRep25B containing cosmids are named). The two short vertical lines represent regions of the physical map that are only contained in YAC clones. The shaded bar represents the genetic map, with known genetic loci given below.
Figure 4.
Positions of CeRep25B sequences on the cosmids. (A) Cosmids for which gene positions are known or have been predicted (Wilson et al. 1994). The positions and numbers of CeRep25B repeats are shown (⋄), but are not to scale. The solid boxes on the line (above or below) are predicted exons. Boxes above the line represent genes transcribed to the right, and boxes below the line represent genes transcribed to the left. Gene identities, when known, are shown. Otherwise, genes are referred to as follows: K02F3.2 represents the second predicted gene from cosmid K02F3. Only the relevant portions of the cosmids are included, and the extent of cosmid overlap is not necessarily to scale. (B) As above, for cosmids in which gene predictions were not available.
Although initially biased by the sequences in the fem-2 and unc-45 introns that were used to search the database in the first instance, the entire set of 235 CeRep25B elements from the 32 different arrays can be used to refine the overall consensus sequence. Because the repeat is palindromic, this is most accurately represented as a consensus for the 12-bp half element (Table 1). As expected, in 10 of the 12 positions, a particular base is found in 95%–99% of all elements. Position 11/14 shows a strong preference for A, whereas position 4/21 is C two-thirds of the time. With the variability of position 4/21, we would expect to see three major CeRep25B variants (Table 2); A4G21 (AG) and its complement C4T21 (CT), A4T21 (AT), and C4G21 (CG). On the basis of abundance, we should see ∼40% of the CeRep25B elements as the AG or CT variant, half as CG, and 10% as AT. Instead, AG and CT variants predominate (Table 2), and there is only one occurrence of an AT variant, where >20 are expected if those two bases vary independently. Given the palindromic nature of CeRep25B, one might expect that perfect palindromes would predominate (e.g., C at position 4 paired in the same repeat with G at position 21). This is not the case. Of the CeRep25B repeats, less than half have complementary bases at these positions (Table 2), and of those, all but one are CG.
Table 1.
Determination of a Consensus Sequence for CeRep25B
| Position in array | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/24 | 2/23 | 3/22 | 4/21 | 5/20 | 6/19 | 7/18 | 8/17 | 9/16 | 10/15 | 11/14 | 12/13 | |
| G | 95 | 96 | 0 | 1 | 0.5 | 98 | 0.5 | 0.5 | 1.5 | 0.5 | 0.5 | 0.5 |
| A | 2.5 | 2.5 | 1 | 30 | 0.5 | 1 | 1 | 1 | 97.5 | 98 | 83 | 0.5 |
| T | 2 | 0.5 | 97.5 | 1 | 99 | 0.5 | 1.5 | 98.5 | 0.5 | 1 | 16 | 96 |
| C | 0.5 | 1 | 1.5 | 68 | 0 | 0.5 | 97 | 0.5 | 1 | 0.5 | 0.5 | 3 |
| G | G | T | C/a | T | G | C | T | A | A | A/t | T | |
Numbers refer to percentage of bases at each position. CeRep25B is represented here as half a palindrome, such that bases 1 and 24, which are normally complementary, are given the same weight. This consensus does not include 12-bp half elements.
Table 2.
CeRep25B Sequence Variants
| Abundance (%) | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. CeRep25B 24-bp elements | ||||||||||||||||||||||||||
| AGa | G | G | T | A | T | G | C | T | A | A | A | T | A | T | T | T | A | G | C | A | G | A | C | C | ||
| CTa | + | G | G | T | C | T | G | C | T | A | A | A | T | A | T | T | T | A | G | C | A | T | A | C | C | 61a |
| AT | G | G | T | A | T | G | C | T | A | A | A | T | A | T | T | T | A | G | C | A | T | A | C | C | <1 | |
| CG | G | G | T | C | T | G | C | T | A | A | A | T | A | T | T | T | A | G | C | A | G | A | C | C | 39 | |
| B. 12-bp half elements | ||||||||||||||||||||||||||
| C1b | C | G | T | C | T | G | C | T | A | A | A | T | n | n | n | |||||||||||
| G2b | + | n | n | n | A | T | T | T | A | G | C | A | G | A | C | C | 88 | |||||||||
| A1c | G | G | T | A | T | G | C | T | A | A | A | T | n | n | n | |||||||||||
| T2c | + | n | n | n | A | T | T | T | A | G | C | A | T | A | C | C | 12 | |||||||||
These are complements of one another and are counted as AG for this purpose.
Complements of one another, counted as CI.
Complements of one another, counted as A1.
There is surprisingly little degeneracy apparent in the CeRep25B repeats that are in these 32 arrays. Of the 24 bp repeats, 38% are identical to two of the three major variants; 45% of the repeats have only one mismatch from this consensus (with a T at position 11, representing 43% of these changes). Thus, >80% of the CeRep25B elements are within 1 bp of matching the consensus and almost 95% within 2-bp changes.
The number of CeRep25B repeat units per array varied from 1 to 41, but more than half the arrays (19/32) contain four or fewer repeats. There is no apparent enrichment for odd or even numbers of repeats within an array. Each array is different not only in the number of the repeat units, but also in the sequence variants present in the array and in the pattern in which they appear, which may suggest models for how these arrays arise and are maintained. A particular sequence variant was always more likely to be found next to a repeat of the same variant, although most of the arrays (especially those over four repeats in length) were composed of more than one variant. In several instances, it was clear that several adjacent repeats were behaving as a single unit. For example, in unc-45, one finds a pattern of three tandem copies of a unit consisting of a 3′-half element, G2 (Table 2), followed by four 24-bp AG repeats and three copies of a unit consisting of an AG repeat followed by a G2 half repeat. This suggests that an earlier ancestor of this array had a G2 half repeat beside an AG repeat, but these two elements can behave independently during amplification or deletion.
The CeRep25B repeats and solo elements within an array are rarely immediately adjacent but, instead, are separated by spacer sequences of from 1 to 30 bp. Because the original CeRep25 element is 31 bp, it prompted an unbiased reexamination of the ability to fit a longer consensus. Again, a pattern emerges, as the vast majority of the spacer sequences examined are 7 bp in length (Table 3). If the 7-bp sequences are aligned and assumed to be themselves palindromic (again, there is no absolute orientation), a loose consensus [(C/A)AA(C/G)TT(G/T)] is apparent (Table 4). Therefore, a more precise consensus for the CeRep25 element can be developed [i.e., TT(G/T)CeRep25B(C/A)AA, in which the first and last 3 bp are more divergent]. Because there is only one instance of two CeRep25B sequences being separated by <6 bp, this element may amplify in units of 31 bp.
Table 3.
Size Distribution of Spacer Sequences between CeRep25B Elements
| No. of bases between CeRep25B repeats | Percent of total spacers |
|---|---|
| >7 | 2.5 |
| 7 | 85.0 |
| 8 | 1.7 |
| 9 | 3.0 |
| 10–15 | 8.5 |
| >15 | 1.3 |
Table 4.
Consensus Sequence for the 7-bp Spacer Sequences between CeRep25B Elements
| Base | Position in spacer | |||
|---|---|---|---|---|
| 1/7 | 2/6 | 3/5 | 4 | |
| A | 34.6 | 86.2 | 74.9 | 12.8 |
| C | 57.8 | 1.8 | 7.8 | 36.7 |
| T | 5.8 | 11.6 | 15.1 | 12.8 |
| G | 0.8 | 0.2 | 1.0 | 36.7 |
| Consensus (C/g)AA(c/g)TT(G/c) | ||||
Numbers refer to percentage of bases at each position. The 7-bp spacer is represented here as half a palindrome, such that bases 1 and 7, which are normally complementary, are given the same weight. This consensus does not include spacers other than 7 bp.
DISCUSSION
Many other repeat families have been described in C. elegans, and at least some of those are found in the same region of the genome as CeRep25B (Naclerio et al. 1992; Cangiano and La Volpe 1993). For example, a CeRep3 element of ∼1 kb is found on cosmid ZK890, just to the left of F30H5 in Figure 3. However, there are only one or two of these elements in the region, and they are also dispersed over the other chromosomes. Clearly, the chromosome-specific CeRep25B repeats can coexist in the same region with more typical longer repeated elements that are more widespread in the genome.
There are a number of models demonstrating how minisatellite repeats may have arisen. For smaller simple repeats, it seems clear that polymorphisms between populations can arise because of polymerase slippage during DNA replication (Streisinger et al. 1966; Schlötterer and Tautz 1992). For minisatellites, a model involving unequal exchange between sequences on sister chromatids is the most compelling, as arrays that seem to show the greatest variability in repeat copy number are those in which the sequence variability between the repeats is lowest (Stephan and Cho 1994); however, there is as yet no direct evidence for this. The arrangement of CeRep25B sequence variants and half elements is not consistent with a model in which they can only be added or deleted one at a time. In scanning other wild isolates of C. elegans, a single CeRep25B polymorphism was detected, which most likely resulted from an increase of 50% in the size of the array. Smaller increases in other strains should have been detected, if some sort of incremental addition process was working. Clearly, some sort of model equivalent to the unequal exchange must be involved, as the repeat variants are not found completely randomly. Also, in this instance and others, alterations in copy number, as well as the pattern of the repeats within an array show that more than one repeat unit at a time is involved (Charlesworth et al. 1994).
The pattern of CeRep25B variants suggests some constraints on its ability to evolve. Except rarely, 12-bp half elements are not found adjacent to one another in these arrays and are most often found on the edges of the arrays. Thus, these elements can amplify as part of a set but apparently not on their own. Whether this is due to their size or their lack of ability to form a hairpin is not clear. The 24-bp palindrome does seem to retain the ability to amplify autonomously, as several of the arrays consist of only one variant. However, three or four adjacent repeats have been treated as a single unit in several of the arrays, as described. Single-base-pair deviations from the consensus are common, but more drastic alterations in the standard 24-bp repeat are rare, except on the boundaries of the arrays. This is consistent with alternating cycles of mutation and amplification or deletion, as first suggested by Southern (1970).
It is intriguing that the most common CeRep25B variants are not complete palindromes. Although there is evidence to suggest that cruciforms are either not found, or are rarely formed in vivo (Courey and Wang 1983; Gellert et al. 1983; Leach 1994), the palindromic nature of CeRep25B must have some role in its ability to propagate. Another paradigm for palindromic sequences is the ability to bind protein dimers, but as these repeats are chromosome specific and seem to play no major role in gene expression (even after a large increase in the size of the array, as in RC301), this mechanism for conservation of their sequences seems unlikely.
There are insufficient data at this time to suggest any functional role for CeRep25B, a problem that is not unique to this minisatellite (Hancock 1996). Clearly many of these repeats are transcribed; on cosmids that have gene identified or predicted, 75% of the arrays (and a much higher percentage of single repeat units) are located in the introns. It is also not clear whether some of the others are within the primary transcript, as the transcription initiation site for most C. elegans genes is difficult to discover, because of trans-splicing (Bektesh and Hirsh 1988).
Most CeRep25B units are found in the telomeric contig, in a region that is known to have higher gene density than elsewhere on that arm of the chromosome (Barnes et al. 1995). This telomeric clustering is a property shared with other C. elegans repeat sequences and is seen in other systems (Royle et al. 1988). On the basis of observations of the genetic behaviour of translocations, the left end of Chromosome III has been proposed to have a role in homolog recognition and pairing (Rosenbluth and Baillie 1981; Wicky and Rose 1996). Because chromosome-specific satellite sequences have been associated with centromere function (Murphy and Karpen 1998), it is intriguing that CeRep25B is a chromosome-specific sequence enriched in the part of the chromosome known to have a meiotic pairing role.
METHODS
Strains
Most strains were obtained from the stock collection of the MRC Laboratory of Molecular Biology (Cambridge, UK) or from the Caenorhabditis Genetics Center (University of Minnesota). N2 is the canonical wild-type strain and is the standard for most genetic manipulations. The Bergerac strain (BO) was initially found to differ from N2 in the number and distribution of Tc1 transposable elements (for review, see Hodgkin and Doniach 1997). The most common BO strain in use is RW7000; however, the RW7000 strain was found to vary in its complement of polymorphisms, depending on its source (Pilgrim 1993; Hodgkin and Doniach 1997). RW7000 variants DP13, DP14, and DP17 were obtained at various times. DP13 is the version of RW7000 in use at the MRC laboratories in 1990. DP14 was from a Cambridge UK stock of RW7000 frozen in liquid nitrogen in 1984. DP17 was a gift from Greg Beitel (Massachusetts Institute of Technology, Cambridge, MA) in 1992. A genomic characterization of wild and laboratory C. elegans isolates, including some of the strains used in this work, has been described (Hodgkin and Doniach 1997). Strains were maintained as described (Wood 1988).
PCR
DNA was prepared as described in (Pilgrim 1993). For PCR analysis, standard reaction conditions were used (Pilgrim et al. 1995), and PCR reactions were carried out in a Robocyler (Stratagene Inc.) as follows: one cycle (94°C for 3 min, 62°C for 1 min, 72°C for 2 min, 15 sec), followed by 32 cycles (94°C for 1 min, 62°C for 1 min, 72°C for 2 min, 15 sec). For the amplification of the unc-45 intron, which contains the CeRep25B repeat, Taq polymerase was supplemented with  the activity of Pfu polymerase. To amplify the fem-2 repeat-containing intron, the primers 5′-CAAAGATCTTGTCCCACCGAAGCCGGTAGTGG-3′ and 5′-TGGAGAATCTTGTCGATCGCCG-3′ were used. For the amplification of exons 3–5 of unc-45, the primers 5′-TGGAAATGTTGGGCCAGC-3′ and 5′-GACTAGTGTCCTTCGCCTCACC-3′ were used. For the amplification of exons 4–5 of unc-45 (no CeRep25B present), primers 5′-GAAGTTCTTCAGCGTCTCG-3′ and 5′-CTCCTGTTGCTCCGGATTC-3′ were used.
the activity of Pfu polymerase. To amplify the fem-2 repeat-containing intron, the primers 5′-CAAAGATCTTGTCCCACCGAAGCCGGTAGTGG-3′ and 5′-TGGAGAATCTTGTCGATCGCCG-3′ were used. For the amplification of exons 3–5 of unc-45, the primers 5′-TGGAAATGTTGGGCCAGC-3′ and 5′-GACTAGTGTCCTTCGCCTCACC-3′ were used. For the amplification of exons 4–5 of unc-45 (no CeRep25B present), primers 5′-GAAGTTCTTCAGCGTCTCG-3′ and 5′-CTCCTGTTGCTCCGGATTC-3′ were used.
Similarity Searches
The initial search for CeRep25B elements was performed using BLAST (Altschul et al. 1990). BLASTN version 1.4.8 was used to search GenBank release 108.0, and BLASTN version 2.0a13MP was used to search the C. elegans genomic sequence database (at http://www.Sanger.ac.uk/Projects/C_elegans/blast_server.shtml) most recently on October 8, 1998. Once cosmids with 22/24 matches to the consensus were identified, the cosmid sequence in the region was searched manually for degenerate elements, or matches to the 12-bp half element.
Acknowledgments
I thank Steve Jones and Marco Marra for their help with the sequence searches, Dave Hansen for the sequence of the deletion plasmid, Greg Beitel for his version of the RW7000 strain, and Ross Hodgetts, John Bell, John Locke, Heather McDermid, Paul Stothard, Dave Hansen, and anonymous reviewers for critical comments on the manuscript. This work was funded by research grants from the Natural Sciences and Engineering Research Council and Medical Research Council of Canada.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson TH, Nilsson-Tillgren T. A fungal minisatellite. Nature. 1997;386:771. doi: 10.1038/386771a0. [DOI] [PubMed] [Google Scholar]
- Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektesh S, Hirsh D. C. elegans mRNAs acquire a spliced leader through a trans-splicing mechanism. Nucleic Acids Res. 1988;16:5692. doi: 10.1093/nar/16.12.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Graham DE, Neufeld BR. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–405. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Cangiano G, La Volpe A. Repetitive DNA sequences located in the terminal portion of the Caenorhabditis elegans chromosomes. Nucleic Acids Res. 1993;21:1133–1139. doi: 10.1093/nar/21.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Courey AJ, Wang JC. Cruciform formation in negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983;33:817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Das HK, Jackson CL, Miller DA, Leff T, Breslow JL. The human apolipoprotein C-II gene sequence contains a novel chromosome 19-specific minisatellite in its third intron. J Biol Chem. 1987;262:4787–4793. [PubMed] [Google Scholar]
- Emmons SW, Rosenzweig B, Hirsh D. Arrangement of repeated sequences in the DNA of the nematode Caenorhabditis elegans. J Mol Biol. 1980;144:481–500. doi: 10.1016/0022-2836(80)90333-2. [DOI] [PubMed] [Google Scholar]
- Felsenstein KM, Emmons SW. Nematode repetitive DNA with ARS and segregation function in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:875–883. doi: 10.1128/mcb.8.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, O’Dea MH, Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci. 1983;80:5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Guttenbach M, Müller U, Schmid M. A human moderately repeated Y-specific sequence is evolutionarily conserved in the Y chromosome of the great apes. Genomics. 1992;13:363–367. doi: 10.1016/0888-7543(92)90254-p. [DOI] [PubMed] [Google Scholar]
- Haaf T, Willard HF. Organization, polymorphism, and molecular cytogenetics of chromosome-specific α-satellite DNA from the centromere of chromosome 2. Genomics. 1992;13:122–128. doi: 10.1016/0888-7543(92)90211-a. [DOI] [PubMed] [Google Scholar]
- Hancock JM. Simple sequences and the expanding genome. BioEssays. 1996;18:421–425. doi: 10.1002/bies.950180512. [DOI] [PubMed] [Google Scholar]
- Hansen D, Pilgrim D. Molecular evolution of a sex determination protein: FEM-2 (PP2C) in Caenorhabditis. Genetics. 1998;149:1353–1362. doi: 10.1093/genetics/149.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy MJ, Russell RJ, Miklos GLG. Molecular studies on interspersed repetitive and unique sequences in the region of the complementation group uncoordinated on the X chromosome of Drosophila melanogaster. Mol & Gen Genet. 1988;213:63–71. doi: 10.1007/BF00333399. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Kogi M, Fukushige S, Lefevre C, Hadano S, Ikeda J-E. A novel tandem repeat sequence located on human chromosome 4p: Isolation and characterization. Genomics. 1997;42:278–283. doi: 10.1006/geno.1997.4746. [DOI] [PubMed] [Google Scholar]
- La Volpe A, Ciaramella M, Bazzicalupo P. Structure, evolution and properties of a novel repetitive DNA family in Caenorhabditis elegans. Nucleic Acids Res. 1988;16:8213–8231. doi: 10.1093/nar/16.17.8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DRF. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Centromeres take flight: Alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- Naclerio G, Cangiano G, Coulson A, Levitt A, Ruvolo V, La Volpe A. Molecular and genomic organization of clusters of repetitive DNA sequences in Caenorhabditis elegans. J Mol Biol. 1992;226:159–168. doi: 10.1016/0022-2836(92)90131-3. [DOI] [PubMed] [Google Scholar]
- Oakey R, Tyler-Smith C. Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics. 1990;7:325–330. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- Pilgrim DB. The genetic and RFLP characterization of the left end of linkage group III in Caenorhabditis elegans. Genome. 1993;36:712–724. doi: 10.1139/g93-096. [DOI] [PubMed] [Google Scholar]
- Pilgrim DB, McGregor A, Jäckle P, Johnson T, Hansen D. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol Biol Cell. 1995;6:1159–1171. doi: 10.1091/mbc.6.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth RE, Baillie DL. The genetic analysis of a reciprocal translocation, eT1(III;V) in Caenorhabditis elegans. Genetics. 1981;99:415–428. doi: 10.1093/genetics/99.3-4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle NJ, Clarkson RE, Wong Z, Jeffreys AJ. Clustering of hypervariable minisatellites in the proterminal regions of human autosomes. Genomics. 1988;3:352–360. doi: 10.1016/0888-7543(88)90127-9. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992;20:211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CW, Deininger PL. Sequence organization of the human genome. Cell. 1975;6:345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Singer MF. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Southern EM. Base sequence and evolution of guinea-pig α-satellite DNA. Nature. 1970;227:794–798. doi: 10.1038/227794a0. [DOI] [PubMed] [Google Scholar]
- Stallings RL, Doggett NA, Okumura K, Ward DC. Chromosome 16-specific repetitive DNA sequences that map to chromosomal regions known to undergo breakage/rearrangement in leukemia cells. Genomics. 1992;13:332–338. doi: 10.1016/0888-7543(92)90249-r. [DOI] [PubMed] [Google Scholar]
- Stephan W, Cho S. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics. 1994;136:333–341. doi: 10.1093/genetics/136.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974;77:95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski W. Use of cesium sulfate for equilibrium density gradient centrifugation. Methods Enzymol. 1968;12B:330–360. [Google Scholar]
- Wevrick R, Willard HF. Long-range organization of tandem arrays of α-satellite DNA at the centromeres of human chromosomes: High frequency array-length polymorphism and meitoic stability. Proc Natl Acad Sci. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky C, Rose AM. The role of chromosome ends during meiosis in Caenorhabditis elegans. BioEssays. 1996;18:447–452. doi: 10.1002/bies.950180606. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DA, Thomas CA. Palindromes in chromosomes. J Mol Biol. 1974;84:115–144. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Wood WB. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]