Abstract
Purpose of review:
The identification of mutations in signal transduction pathways that are central in melanoma pathophysiology has provided new therapeutic targets for drug development. The purpose of this review is to define those oncogenes for which there is preclinical data supporting clinical trials and to summarize results from clinical investigations.
Recent findings:
CKIT mutations were first reported in 2005, but are present in only a small subpopulation of melanoma patients. The validation of inhibitors developed in gastrointestinal stromal tumors has taken several years, but recent evidence suggests that responses can be seen in CKIT mutant melanoma. First reported in 2002, BRAF is mutated in 50% of all melanomas and subsets of other cancers. The melanoma field is leading the clinical trials evaluating the value of targeting BRAF and MEK in BRAF mutant tumors. Results from the first clinical trial with a potent and selective BRAF inhibitor clearly show the therapeutic promise of this approach.
Summary:
Larger clinical trials are needed to fully define the efficacy of BRAF and CKIT directed therapy in melanoma, but early results suggests that this strategy will transform treatment options. Additional potential targets have been identified and clinical trials evaluating novel drugs against them are underway.
Keywords: melanoma, BRAF, CKIT, oncogene
Introduction
The melanoma field has struggled to develop a therapy that is capable of altering the natural history of advanced disease. The same chemotherapies that are effective in numerous types of cancer are largely ineffective in melanoma. (1) Even immunotherapies fail to benefit more than small subset of patients, though melanoma is thought to be uniquely susceptible to such treatments. (2) In the past decade, advances have been made in understanding the molecular basis of cancer and several therapies have been developed that directly antagonize the mediators of cancer pathophysiology. A portfolio of pharmacologic inhibitors targeting several of the recently identified mutated signal transduction molecules is being explored in clinical trials in genetically-defined subgroups of melanoma patients. [Table 1] Early results suggest that this approach will change the paradigm for melanoma therapy.
Table 1.
Drugs in clinical development for mutated oncogenes in melanoma
| Oncogene | Drug | Phase of clinical trial in melanoma |
|---|---|---|
| c-KIT | imatinib | phase II |
| nilotinib | phase II | |
| dasatinib | phase II | |
|
| ||
| NRAS | R115777 | phase II (completed) |
| BRAF | ||
| non-selective | sorafenib | phase III (completed) |
| XL-281 | phase I (completed) | |
| RAF-265 | phase I | |
| selective | PLX4032 | phase I (completed) |
| GSK2118438 | phase I | |
| MEK | AZD6244 | phase II (completed) |
| PD0325901 | phase I (completed) | |
| GSK1120212 | phase I | |
|
| ||
| PI3K | GDC0941 | - |
| XL147 | - | |
| Akt | MK-2206 | - |
| GSK690693 | - | |
| mTOR | temsirolimus | phase II (completed) |
|
| ||
| CDK4 | PD032991 | - |
KIT
It has long been described that patients with melanomas that originate in acral, mucosal, or chronically sun-damaged skin have different clinical and histopathological features than patients with more common cutaneous melanomas arising on the remainder of the integument. More recently, differences in the patterns of chromosomal aberrations and mutation frequency in BRAF have supported that these types are biologically distinct. (3) Recently, activating mutations and/or gene amplification of KIT have been found in 39% of mucosal, 36% of acral, and 28% of melanomas that arise in chronically sun damaged skin (defined by the presence of solar elastosis on pathology review) (4) Mutations most commonly affect the juxtamembraneous region of the receptor with L576P in exon 11 representing the most common alteration. (5) Many of the CKIT mutations found in melanoma are known oncogenic mutations observed in other cancer types and respond to tyrosine kinase inhibitors.
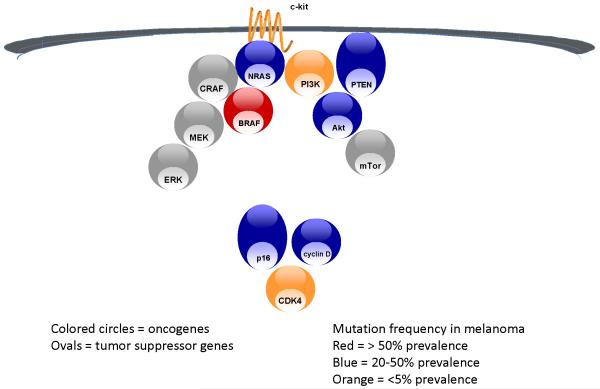
Activating KIT mutations have been demonstrated to initiate a series of signaling events that result in cellular proliferation and propagation of cancer. (6) [Figure 1] Preclinical studies in vitro with cultured melanoma cells that possess a KIT mutation revealed potent effects for the tyrosine kinase inhibitor imatinib on proliferation, induction of apoptosis, as well as decreased MAPK, PI3K/AKT, and STAT signaling. (7)
Figure 1.
Mutation & deletion in MAP kinase, PI3 kinase & p16 pathways in melanoma
Prior to the identification of KIT mutations in melanoma, phase II studies of imatinib in patients with metastatic melanoma reported a lack of clinical benefit. (8, 9) Only since the discovery of KIT mutations and amplification in specific subsets of melanomas has rational targeting of KIT in these patients has been investigated. To date, the clinical activity of imatinib in patients whose melanomas harbor KIT mutations has been described in a series of case reports. (10, 11) While limited in numbers thus far, these clinical experiences confirm KIT as a melanoma therapeutic target, with patients experiencing dramatic and durable responses to treatment. Interestingly, even with limited clinical data, there is the suggestion that durability of clinical responses can be dose-dependent.
As the result of the discovery of KIT genomic aberrations in melanoma subtypes and initial clinical reports targeting KIT, several multicenter phase II trials for the treatment of metastatic acral, mucosal, and chronically sun-damaged melanoma have been initiated. Such efforts include the evaluations of imatinib, sunitinib, nilotinib, and dasatanib in melanoma patients whose tumors harbor KIT genomic aberrations. It will be important in the near future to determine whether there exist differences in clinical efficacy amongst these agents, and furthermore whether the genomics of the tumor, as defined by activating KIT mutations alone versus amplification alone versus tumors with both mutations and amplification, influence the likelihood or durability of responses. One principle question that remains is whether tyrosine kinase inhibition of KIT can effectively target wild-type, amplified tumors in patients.
Already an active area of investigation s is the search for resistance mechanisms to KIT inhibition. Acquiring additional CKIT exon mutations is a common mechanism for treatment resistance in gastrointestinal stromal tumor (GIST) patients receiving a tyrosine kinase inhibitor. Whether such resistance mechanisms will hold true for KIT melanomas or whether alternative resistant mechanisms, such as KIT amplification or activation of additional signaling pathways, will emerge is yet to be determined.
NRAS
Mutations in NRAS were described long before activating mutations in other signal transduction molecules were found. (12, 13) In large series of melanomas analyzed for NRAS mutations, the frequency is approximately 20%. (14, 15) However, RAS remains an elusive target in cancer, with no drugs being available that can directly antagonize its signaling activity. Attempts have been made to inhibit to activation of RAS by blockade of a key post-translational modification required for membrane localization, farnesylation. However, the farnesylation transferase inhibitors that have been evaluated clinically produce dose-limiting toxicities without significant evidence of antitumor activity in tumors known to harbor RAS mutations. In melanoma, only a single such trial has been conducted and it was terminated for lack of efficacy in 11 patients with metastatic disease treated with single-agent therapy. (16) Notably, this small cohort of patients was not selected on the basis of NRAS mutations and their mutation status was not even determined retrospectively. Lacking advances in the development of direct RAS inhibitors, it is hypothesized that targeting one or more of the downstream RAS-effecter pathways might prove to be efficacious in this subset of patients. [Figure 1] Preclinical evidence in KRAS mutant tumors suggests that dual targeting of the MAP kinase and PI3 kinase pathways might abrogate the effect of RAS mutation.(17) However, this remains to be proven clinically.
BRAF
The discovery of activating mutations in BRAF generated hope that mutation-directed therapy might become a reality for the majority of melanoma patients. [Figure 1] Amongst a large series of tumors screened for the presence of BRAF mutations, melanoma was found to have the highest prevalence.(18) This finding was confirmed in subsequent series (19, 20) and found to primarily affect melanomas of younger individuals, originating on non-chronically sun-damaged skin.(21) Since BRAF is a serine-threonine kinase, it was immediately plausible that an inhibitor could developed.
Sorafenib, a broad-spectrum kinase inhibitor, was originally selected for clinical development as a CRAF inhibitor, and was known to have some activity against BRAF.(22) In preclinical studies, sorafenib demonstrated an ability to inhibit the MAP kinase pathway and proliferation in melanoma cells, but that activity was independent of BRAF mutations. Sorafenib proved to be disappointing in clinical trials, both as a single-agent and in combination with chemotherapy.(23, 24) Investigations into the mechanism of action of sorafenib in melanoma patients suggested that an anti-angiogenic effect was achieved, but incomplete inhibition of the MAP kinase pathway. (25)
Several years passed between the identification of BRAF mutations and the development of potent and selective inhibitors of BRAF. However, the first selective BRAF inhibitor to enter clinical development, PLX4032, proved to have single-agent activity in patients with metastatic melanoma whose tumors harbored the most common BRAF mutation (V600EBRAF)(26). Throughout the phase I trial of this agent, patients with metastatic melanoma, and preferentially those with V600EBRAF mutations were enrolled. Amongst the sixteen patients with V600EBRAF mutant metastatic melanoma enrolled at the five highest dose cohorts, nine patients achieved objective responses. This provided clear evidence that even single-agent therapy against a mutated oncogene in melanoma could have therapeutic value.
GNAQ
While it has become clear that the prevalence of activating mutations in oncogenes varies widely based on the site of primary melanoma, it has been known for some time that the genomic alterations in uveal melanoma differ substantially from any other anatomic site of melanoma(27, 28). Uveal melanomas lack mutations in CKIT, NRAS and BRAF, but they uniquely harbor activating mutations in the alpha subunit of a G-protein of the Gq family, GNAQ. In melanoma activated GNAQ results in MAP kinase pathway activation.(29) and melanoma cell lines that harbor GNAQ mutations are sensitive to MEK inhibition. MAP-kinase pathway inhibtion thus appears capable of inhibiting cell proliferation and induce cell death and, thus, may be an effective therapeutic strategy for this subset of tumors. Clinical trials evaluating this hypothesis are just underway.
CDK4
Activating mutations and allelic amplification in CDK4 have been described in melanoma (3, 30). [Figure 1] However, the possibility that CDK4 represents a unique therapeutic target in melanoma has not been explored preclinically or clinically, despite the emergence of inhibitors of cyclin dependent kinases, including some with relative selectivity for CDK4. More commonly than CDK4 genetic alteration, melanoma has been found to have deletion of p16 or amplification of cyclin D.(3) Neither of these alterations are amenable to direct pharmacologic targeting, but support CDK4 inhibition as a strategy.
Erbb4
Genetic alterations in ERBB1 and ERBB2 are not found in melanoma, but there is evidence that signaling among ERBB family members is important in melanomagenesis and may serve as therapeutic targets. (31, 32) Given the prominent role of heterodimerization among ERBB receptors, it has been difficult to implicate the critical node of signaling in melanoma amongst these molecules. ERBB4 was recently found to harbor mutations in melanoma, having previously been described in small subsets of other cancers.(33, 34) Mutations in ERBB4 were found in 19% of cohort of 79 uncultured melanoma tumors.(33) Unlike BRAF, NRAS and GNAQ, there was no mutation hotspot. Seven of the 24 different mutations identified were characterized for functional significance and all but one appeared associated with hyperphosphorylation of errb4 and transformation of transfected fibroblasts. Furthermore, erbb4-targeted siRNA inhibited growth of melanoma cell lines that harbored ERBB4 mutations. While more work is needed to understand the relative importance of ERBB4 mutations in the context of other oncogenic events in the same cells, the availability of several “pan-erb” receptor tyrosine kinase inhibitors in clinical trials for other cancers provides the possibility of rapid translation of this finding.
Loss of tumor suppressors as modifiers of response to mutation-directed therapy
Many of the key genetic alterations that facilitate melanoma, and more generally cancer formation, are loss of function mutations or allelic deletions of tumor suppressor genes. While p53 if the quintessential tumor suppressor in cancer, being lost in approximately 50% of all tumors, it is infrequently mutated or deleted in melanoma.(35) But as the integrator of signals regarding DNA damage and metabolic stress, p53 function is clearly aberrant in melanoma. (36) The predominant genetic cause for this is mutation of p16 or p14, both of which are encoded for by INK4A/ARF. [Figure 1] Mutations in this gene underlie the most highly penetrant form of familial melanoma described. Estimates of the frequency of these events in sporadic melanoma were likely inflated by studies focused on melanoma cell lines, owing to either inherent genetic instability once cells are cultured or the loss of p16 and p14 being selection factors for survival in vitro. (37) In uncultured tumors, it still appears that the majority of tumors have lost expression or function of either of these proteins due to deletion or mutation. (38) Restoring the function of tumor suppressor genes is beyond the reach of current pharmaceutical technology. But, targeting the interaction of mdm2 with p53 appears to be a viable strategy in tumors in which p53 is present, such as melanoma.(39) Mdm2 is a negative regulator of p53, and thus blocking the interface with p53 upregulates p53 activity. Mdm2 antagonists are nearing clinical evaluation, but have not yet been investigated in melanoma.
Resistance & combinations of mutation-targeted therapy
Resistance to oncogene-targeted therapy has already been established as an issue in others cancers. In CML and GIST, disease progression following chronic administration of abl or c-kit targeted therapy is associated with novel mutations in the primary oncogenic target.(40, 41) In these cases, disease control can be regained by switching to agents that inhibit targeting the mutant isoforms.(42) In epidermal growth factor receptor (EGFR) mutant non-small cell lung cancer, tumors that emerge following an initial response to an EGFR tyrosine kinase inhibitor manifest amplification of the CMET receptor allele which appears to restore survival-promoting signal transduction.(43)
The issues to consider regarding resistance to targeted therapy start with the fundamental issue of adequate target inhibition. With sorafenib it became clear that adequate BRAF inhibition was not being achieved. It is possible that even the most highly selective BRAF inhibitors will be limited in their impact on V600EBRAF because of the dose-limiting toxicities resulting from chronic inhibition of wild-type BRAF or other targets. A strategy that might overcome such limitations is administering high doses of an inhibitor in an intermittent fashion to provide above-threshold levels of inhibition and greater induction of cell death. There is preclinical evidence suggesting that this approach has merit in CML and EGFR mutant non-small cell lung cancer, but has not been thoroughly evaluated clinically in any setting.(40, 44)
None of the genetic changes described above are stand-alone events in melanoma. Even amongst this set of defined oncogenes and tumor suppressor genes there is overlap, such as BRAF mutation occurring concomitantly with PTEN loss or CDK4 mutation. As a first consideration, these co-existent genetic changes might mediate resistance to targeted therapies inhibiting any one mutation or upregulated pathway.(45) But they these may also provide a map for combinations of targeted therapy, countering the primary and secondary drivers of melanoma.(46) Creating such hierarchies of therapeutic targets within any one cancer is the goal of current laboratory and clinical translational research.
The complexities of evaluating numerous potential combination strategies in relatively small, genetically-defined patient subpopulations is onerous. For each signaling pathway and, in some cases, for each particular molecular target, there is diversity with regard to point of intervention and properties of the pharmacologic agents. Taking BRAF as an example, there are broadly cross-reactive and highly selective agents in clinical trials currently. Which of these agents to incorporate into combination regimens has not been sorted out preclinically. Intuitively it is reasonable to consider that the most highly selective agents will produce the least toxicity when combined with other targeted therapies, but it is still possible that there might be serendipitous benefit derived from the effects of any given agent’s against its “secondary” targets (other than the one for which is first being selected for use in melanoma). And, in each pathway the optimal point of intervention has not been defined. This is the case for BRAF versus MEK inhibition in the MAP kinase pathway, as well as, PI3 kinase, Akt, or mTor in the PI3 kinase pathway. The number of potential combinations is daunting even considering only these two pathways. Rationalizing approaches for clinical testing will require unprecedented and coordinated preclinical investigations, bringing to bear the best in vitro and animal models.
Conclusion
The era of oncogene-directed therapy in melanoma has begun, most convincingly for BRAF and c-kit targeted agents. These examples alone indicate that melanoma can be susceptible to this strategy to a similar degree as other oncogene-defined cancer subpopulations. But, the challenges that remain in realizing maximum clinical benefit are numerous. Nearly half of all melanomas lack a BRAF or KIT mutation, leaving a large fraction of patients without a therapeutic strategy to investigate. In the context of clinical trials it already the case that the there is a proliferation of agents and trials for those patients whose tumors harbor a BRAF or KIT mutation. Further genetic exploration remains a high priority in melanoma. Optimizing drugs against each target is time-consuming at the chemical and biological level, much less in terms of pharmacodynamic validation in the clinic. And lastly, assembling combination regimens that include solid building blocks that are matched to the appropriate subpopulation is a task that will require iterative clinical trials simultaneously evaluating the predictive biomarker and the therapy.
References
- 1.Huncharek M, Caubet JF, McGarry R. Single-agent DTIC versus combination chemotherapy with or without immunotherapy in metastatic melanoma: a meta-analysis of 3273 patients from 20 randomized trials. Melanoma Res. 2001 Feb;11(1):75–81. doi: 10.1097/00008390-200102000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999 Jul;17(7):2105–16. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 3.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005 Nov 17;353(20):2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 4.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006 Sep 10;24(26):4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- *5.Beadling C, Jacobson-Dunlop E, Hodi FS, Le C, Warrick A, Patterson J, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008 Nov 1;14(21):6821–8. doi: 10.1158/1078-0432.CCR-08-0575. This paper provided the definitive catalog of KIT mutation and amplification frequency in melanoma.
- 6.Webster JD, Kiupel M, Yuzbasiyan-Gurkan V. Evaluation of the kinase domain of c-KIT in canine cutaneous mast cell tumors. BMC Cancer. 2006;6:85. doi: 10.1186/1471-2407-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Jiang X, Zhou J, Yuen NK, Corless CL, Heinrich MC, Fletcher JA, et al. Imatinib targeting of KIT-mutant oncoprotein in melanoma. Clin Cancer Res. 2008 Dec 1;14(23):7726–32. doi: 10.1158/1078-0432.CCR-08-1144. Preclinical validation that CKIT was a target in melanoma.
- 8.Wyman K, Atkins MB, Prieto V, Eton O, McDermott DF, Hubbard F, et al. Multicenter Phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006 May 1;106(9):2005–11. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 9.Ugurel S, Hildenbrand R, Zimpfer A, La Rosee P, Paschka P, Sucker A, et al. Lack of clinical efficacy of imatinib in metastatic melanoma. Br J Cancer. 2005 Apr 25;92(8):1398–405. doi: 10.1038/sj.bjc.6602529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodi FS, Friedlander P, Corless CL, Heinrich MC, Mac Rae S, Kruse A, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008 Apr 20;26(12):2046–51. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 11.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008 Aug;21(4):492–3. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 12.Albino AP, Le Strange R, Oliff AI, Furth ME, Old LJ. Transforming ras genes from human melanoma: a manifestation of tumour heterogeneity? Nature. 1984 Mar 1-7;308(5954):69–72. doi: 10.1038/308069a0. [DOI] [PubMed] [Google Scholar]
- 13.Padua RA, Barrass NC, Currie GA. Activation of N-ras in a human melanoma cell line. Mol Cell Biol. 1985 Mar;5(3):582–5. doi: 10.1128/mcb.5.3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006 Dec;16(6):471–8. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 15.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006 Jan;126(1):154–60. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 16.Gajewski TK, Johnson J, Linette G, Bucher C, Blaskovich M, Sebti S, Haluska F. Phase II study of the farnesyltransferase inhibitor R115777 in advanced melanoma: CALGB 500104. J Clin Oncol. 2006;24(18S) doi: 10.1186/1479-5876-10-246. ND. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaiswal BS, Janakiraman V, Kljavin NM, Eastham-Anderson J, Cupp JE, Liang Y, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS One. 2009;4(5):e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 19.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002 Dec 1;62(23):6997–7000. [PubMed] [Google Scholar]
- 20.Gorden A, Osman I, Gai W, He D, Huang W, Davidson A, et al. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res. 2003 Jul 15;63(14):3955–7. [PubMed] [Google Scholar]
- 21.Maldonado JL, Fridlyand J, Patel H, Jain AN, Busam K, Kageshita T, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003 Dec 17;95(24):1878–90. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 22.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004 Oct 1;64(19):7099–109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 23.Eisen T, Ahmad T, Flaherty KT, Gore M, Kaye S, Marais R, et al. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br J Cancer. 2006 Sep 4;95(5):581–6. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauschild A, Agarwala SS, Trefzer U, Hogg D, Robert C, Hersey P, et al. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J Clin Oncol. 2009 Jun 10;27(17):2823–30. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 25.Flaherty KT, Redlinger M, Schuchter LM, Lathia CD, Weber BL, O’Dwyer PJ. Phase I/II, pharmacokinetic and pharmacodynamic trial of BAY 43-9006 alone in patients with metastatic melanoma. J Clin Oncol. 2005 June 1;23(16_suppl):3037. Meeting Abstracts. 2005. [Google Scholar]
- **26.Flaherty KT, Puzanov I, Sosman J, Kim K, Ribas A, McArthur G, Lee RJ, Grippo JF, Nolop K, Chapman P. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009;27(15S) First report of clinical activity with a selective BRAF inhibitor in BRAF mutant melanoma.
- 27.Cruz F, 3rd, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003 Sep 15;63(18):5761–6. [PubMed] [Google Scholar]
- 28.Blasi MA, Roccella F, Balestrazzi E, Del Porto G, De Felice N, Roccella M, et al. 3p13 region: a possible location of a tumor suppressor gene involved in uveal melanoma. Cancer Genet Cytogenet. 1999 Jan 1;108(1):81–3. doi: 10.1016/s0165-4608(98)00114-9. [DOI] [PubMed] [Google Scholar]
- **29.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009 Jan 29;457(7229):599–602. doi: 10.1038/nature07586. First paper to identify an activated oncogene in uveal melanoma and provides basis for clinical trials with MEK inhibitors in this patient subset.
- 30.Smalley KS, Contractor R, Nguyen TK, Xiao M, Edwards R, Muthusamy V, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008 Jul 15;68(14):5743–52. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Djerf EA, Trinks C, Abdiu A, Thunell LK, Hallbeck AL, Walz TM. ErbB receptor tyrosine kinases contribute to proliferation of malignant melanoma cells: inhibition by gefitinib (ZD1839) Melanoma Res. 2009 Jun;19(3):156–66. doi: 10.1097/CMR.0b013e32832c6339. [DOI] [PubMed] [Google Scholar]
- 32.Ueno Y, Sakurai H, Tsunoda S, Choo MK, Matsuo M, Koizumi K, et al. Heregulin-induced activation of ErbB3 by EGFR tyrosine kinase activity promotes tumor growth and metastasis in melanoma cells. Int J Cancer. 2008 Jul 15;123(2):340–7. doi: 10.1002/ijc.23465. [DOI] [PubMed] [Google Scholar]
- **33.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009 Oct;41(10):1127–32. doi: 10.1038/ng.438. First report of activating mutations in ERBB4 in melanoma and evidence that ERB inhibition may have therapeutic value in this subset of melanoma patients.
- 34.Soung YH, Lee JW, Kim SY, Wang YP, Jo KH, Moon SW, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer. 2006 Mar 15;118(6):1426–9. doi: 10.1002/ijc.21507. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson G, Dahl C, Staaf J, Sandberg T, Bendahl PO, Ringner M, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007 Jul 12;26(32):4738–48. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 36.Yang G, Rajadurai A, Tsao H. Recurrent patterns of dual RB and p53 pathway inactivation in melanoma. J Invest Dermatol. 2005 Dec;125(6):1242–51. doi: 10.1111/j.0022-202X.2005.23931.x. [DOI] [PubMed] [Google Scholar]
- 37.Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004 Aug 5;23(35):5968–77. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 38.Rakosy Z, Vizkeleti L, Ecsedi S, Begany A, Emri G, Adany R, et al. Characterization of 9p21 copy number alterations in human melanoma by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2008 Apr 15;182(2):116–21. doi: 10.1016/j.cancergencyto.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Shangary S, Ding K, Qiu S, Nikolovska-Coleska Z, Bauer JA, Liu M, et al. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol Cancer Ther. 2008 Jun;7(6):1533–42. doi: 10.1158/1535-7163.MCT-08-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008 Dec 9;14(6):485–93. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Heinrich MC, Corless CL, Blanke CD, Demetri GD, Joensuu H, Roberts PJ, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006 Oct 10;24(29):4764–74. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 42.Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006 Oct 14;368(9544):1329–38. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 43.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solit DB, She Y, Lobo J, Kris MG, Scher HI, Rosen N, et al. Pulsatile administration of the epidermal growth factor receptor inhibitor gefitinib is significantly more effective than continuous dosing for sensitizing tumors to paclitaxel. Clin Cancer Res. 2005 Mar 1;11(5):1983–9. doi: 10.1158/1078-0432.CCR-04-1347. [DOI] [PubMed] [Google Scholar]
- 45.Krasilnikov M, Ivanov VN, Dong J, Ronai Z. ERK and PI3K negatively regulate STAT-transcriptional activities in human melanoma cells: implications towards sensitization to apoptosis. Oncogene. 2003 Jun 26;22(26):4092–101. doi: 10.1038/sj.onc.1206598. [DOI] [PubMed] [Google Scholar]
- 46.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006 May;5(5):1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]