Abstract
BACKGROUND
Uveal melanoma is the most common intraocular cancer. There are no effective therapies for metastatic disease. Mutations in GNAQ, the gene encoding an alpha subunit of heterotrimeric G proteins, are found in 40% of uveal melanomas.
METHODS
We sequenced exon 5 of GNAQ and GNA11, a paralogue of GNAQ, in 713 melanocytic neoplasms of different types (186 uveal melanomas, 139 blue nevi, 106 other nevi, and 282 other melanomas). We sequenced exon 4 of GNAQ and GNA11 in 453 of these samples and in all coding exons of GNAQ and GNA11 in 97 uveal melanomas and 45 blue nevi.
RESULTS
We found somatic mutations in exon 5 (affecting Q209) and in exon 4 (affecting R183) in both GNA11 and GNAQ, in a mutually exclusive pattern. Mutations affecting Q209 in GNA11 were present in 7% of blue nevi, 32% of primary uveal melanomas, and 57% of uveal melanoma metastases. In contrast, we observed Q209 mutations in GNAQ in 55% of blue nevi, 45% of uveal melanomas, and 22% of uveal melanoma metastases. Mutations affecting R183 in either GNAQ or GNA11 were less prevalent (2% of blue nevi and 6% of uveal melanomas) than the Q209 mutations. Mutations in GNA11 induced spontaneously metastasizing tumors in a mouse model and activated the mitogen-activated protein kinase pathway.
CONCLUSIONS
Of the uveal melanomas we analyzed, 83% had somatic mutations in GNAQ or GNA11. Constitutive activation of the pathway involving these two genes appears to be a major contributor to the development of uveal melanoma. (Funded by the National Institutes of Health and others.)
Uveal melanoma is a neoplasm that arises from melanocytes of the choroid plexus, ciliary body, and iris of the eye.1 Unlike cutaneous melanoma, uveal melanoma lacks mutations in BRAF, NRAS, or KIT2-5 and has characteristic cytogenetic alterations6 and a strong tendency to metastasize to the liver.1,7 The nevus of Ota, a subtle intradermal proliferation of melanocytes resulting in bluish-gray hyperpigmentation in the sclera and periorbital dermis, is a risk factor for uveal melanoma.8
In mice, germline mutations that increase the activity of the closely related GTPases, Gαq (V179M) and Gα11 (I63V), cause dermal hyperpigmentation.9 The microscopical appearance of the skin of these mutant mice is reminiscent of that of human blue nevi,9,10 a finding that prompted us to sequence GNAQ and GNA11 in blue nevi, as well as in a variety of other cutaneous melanocytic neoplasms.11 We found somatic GNAQ mutations in 83% of blue nevi but no mutations in GNA11.11 Because of the link between the nevus of Ota, a form of blue nevus, and uveal melanoma,8 we subsequently genotyped the GNAQ mutational hotspot, predicted to affect the glutamine residue at amino acid position 209 (Q209), in uveal melanomas and observed that 46% of primary lesions carried mutations.11
GNAQ encodes the alpha subunit of heterotrimeric G proteins, which couple seven-transmembrane domain receptors to intracellular signaling machinery.12 Heterotrimeric G proteins are composed of three subunits (alpha, beta, and gamma), of which there are many family members.12 The alpha subunit serves as a molecular switch for the G protein, which is active when bound to guanosine triphosphate (GTP) and shut off when GTP is hydrolyzed to guanosine diphosphate (GDP).13,14 In the alpha subunit, if there are substitutions of specific glutamine or arginine residues that contact the GTP molecule, then its intrinsic GTPase activity is blocked. Thus, the G protein, through its alpha subunit, is locked in a GTP-bound, constitutively active state.14-16 This critical glutamine is at position 209 (Q209) in Gαq and is mutated to either leucine or proline in melanocytic lesions.11,17-20 It is located at position 227 (Q227) in Gαs and is substituted in pituitary and thyroid tumors.14 The GTP-contacting arginines of Gαs (R201) and Gαi2 (R179) have been found to be mutant in pituitary and thyroid tumors (Gαs) and in adrenocortical and ovarian tumors (Gαi2).21 In this study, we broadened our sequencing analysis and revisited the role of GNA11 in uveal melanoma and related neoplasms.
METHODS
We retrieved paraffin-embedded archival biopsy specimens after obtaining approval from the institutional review board at each study center. DNA was extracted and used to sequence GNAQ and GNA11 and to perform array comparative genomic hybridization on a subgroup of samples. Tumorigenicity experiments were carried out in nonobese diabetic mice with severe combined immunodeficiency (NOD SCID) and depletion of the interleukin-2 receptor γ, with the use of melan-a cells22 (immortalized, nontumorigenic mouse melanocytes) transduced with wild-type or constitutively active GNA11 expression constructs or a β-galactosidase control vector. A detailed description of the methods used for sequencing, cell culture, comparative genomic hybridization, Western blotting, and in vivo tumorigenicity studies is provided in the Supplementary Appendix, available with the full text of this article at NEJM.org.
RESULTS
SEQUENCING
We found mutations affecting Q209 in GNA11 in the same specific subgroups of melanocytic tumors that were previously described for GNAQ11 (Tables 1 and 2). The frequency of mutations in GNA11 at the codon encoding Q209 increased progressively from blue nevi (6.5%) to primary uveal melanomas (31.9%) to uveal melanoma metastases (56.5%), a pattern inverse to the distribution of Q209 mutations in GNAQ, which are most common in blue nevi (54.7%) and least common in uveal melanoma metastases (21.7%) (P<0.001) (Table 1). Mutations affecting codon 209 in GNA11 were CAG→CTG (94.5%), CAG→CCG (2.7%), CAG→CTA (1.4%), and CAG→CTT (1.4%). These mutations predicted substitution by leucine (Q209L) in 97.3% of samples that were analyzed and by proline in 2.7% of these samples (Fig. 1 in the Supplementary Appendix).
Table 1.
Frequency of Mutations in Q209 in Exon 5 of GNA11 and GNAQ in 713 Melanocytic Neoplasms.*
| Category and Subtype | GNA11 | GNAQ | Not GNA11 or GNAQ | Total Number |
|||
|---|---|---|---|---|---|---|---|
| no. | % frequency |
no. | % frequency |
no. | % frequency |
||
| Blue nevi | |||||||
| All subtypes | 9 | 6.5 | 76 | 54.7 | 54 | 38.8 | 139 |
| Amelanotic blue nevus | 0 | 0 | 7 | 70.0 | 3 | 30.0 | 10 |
| Cellular blue nevus | 3 | 8.3 | 26 | 72.2 | 7 | 19.4 | 36 |
| Common blue nevus | 4 | 6.7 | 39 | 65.0 | 17 | 28.3 | 60 |
| Nevus of Ito | 0 | 0 | 0 | 0 | 7 | 100.0 | 7 |
| Nevus of Ota | 1 | 5.0 | 2 | 10.0 | 17 | 85.0 | 20 |
| Malignant blue nevus | 1 | 16.7 | 2 | 33.3 | 3 | 50.0 | 6 |
| Ocular melanocytic tumor | |||||||
| All subtypes | 65 | 33.2 | 79 | 40.3 | 52 | 26.5 | 196 |
| Conjunctival melanoma | 0 | 0 | 0 | 0 | 9 | 100.0 | 9 |
| Uveal melanoma | |||||||
| Primary | 52 | 31.9 | 73 | 44.8 | 38 | 23.3 | 163 |
| Metastatic | 13 | 56.5 | 5 | 21.7 | 5 | 21.7 | 23 |
| Uveal nevus | 0 | 0 | 1 | 100.0 | 0 | 0 | 1 |
| Other nevi | |||||||
| All subtypes | 0 | 0 | 0 | 0 | 105 | 100.0 | 105 |
| Common nevus | 0 | 0 | 0 | 0 | 22 | 100.0 | 22 |
| Congenital nevus | 0 | 0 | 0 | 0 | 17 | 100.0 | 17 |
| Deep penetrating nevus | 0 | 0 | 0 | 0 | 27 | 100.0 | 27 |
| Spitz nevus | 0 | 0 | 0 | 0 | 19 | 100.0 | 19 |
| Atypical Spitz tumor | 0 | 0 | 0 | 0 | 20 | 100.0 | 20 |
| Extraocular melanoma | |||||||
| All subtypes | 0 | 0 | 1 | 0.4 | 272 | 99.6 | 273 |
| Acral | 0 | 0 | 0 | 0 | 47 | 100.0 | 47 |
| CSD | 0 | 0 | 1 | 1.4 | 73 | 98.6 | 74 |
| Mucosal | 0 | 0 | 0 | 0 | 62 | 100.0 | 62 |
| Non-CSD | 0 | 0 | 0 | 0 | 90 | 100.0 | 90 |
CSD denotes melanoma located on chronically sun-damaged skin, and non-CSD melanoma located on skin without microscopical signs of chronic sun-induced damage.
Table 2.
Frequency of Mutations in R183 in Exon 4 of GNA11 and GNAQ in 453 Melanocytic Neoplasms.*
| Category and Subtype | GNA11 | GNAQ | Not GNA11 or GNAQ | Total Number |
|||
|---|---|---|---|---|---|---|---|
| no. | % frequency |
no. | % frequency |
no. | % frequency |
||
| Blue nevi | |||||||
| All subtypes | 1 | 1.0 | 1 | 1.0 | 94 | 97.9 | 96 |
| Amelanotic blue nevus | 0 | 0 | 0 | 0 | 9 | 100.0 | 9 |
| Cellular blue nevus | 0 | 0 | 0 | 0 | 25 | 100.0 | 25 |
| Common blue nevus | 1 | 2.4 | 0 | 0 | 40 | 97.6 | 41 |
| Nevus of Ito | 0 | 0 | 0 | 0 | 7 | 100.0 | 7 |
| Nevus of Ota | 0 | 0 | 1 | 9.1 | 10 | 90.9 | 11 |
| Malignant blue nevus | 0 | 0 | 0 | 0 | 3 | 100.0 | 3 |
| Ocular melanocytic tumor | |||||||
| All subtypes | 4 | 2.4 | 5 | 3.0 | 160 | 94.7 | 169 |
| Conjunctival melanoma | 0 | 0 | 0 | 0 | 6 | 100.0 | 6 |
| Uveal melanoma | |||||||
| Primary† | 3 | 2.1 | 4 | 2.8 | 138 | 95.2 | 145 |
| Metastatic | 1 | 5.9 | 1 | 5.9 | 15 | 88.2 | 17 |
| Uveal nevus | 0 | 0 | 0 | 0 | 1 | 100.0 | 1 |
| Other nevi | |||||||
| All subtypes | 0 | 0 | 0 | 0 | 30 | 100.0 | 30 |
| Deep penetrating nevus | 0 | 0 | 0 | 0 | 14 | 100.0 | 14 |
| Spitz nevus | 0 | 0 | 0 | 0 | 8 | 100.0 | 8 |
| Atypical Spitz tumor | 0 | 0 | 0 | 0 | 8 | 100.0 | 8 |
| Extraocular melanoma | |||||||
| All subtypes | 0 | 0 | 0 | 0 | 158 | 100.0 | 158 |
| Acral | 0 | 0 | 0 | 0 | 18 | 100.0 | 18 |
| CSD | 0 | 0 | 0 | 0 | 49 | 100.0 | 49 |
| Mucosal | 0 | 0 | 0 | 0 | 38 | 100.0 | 38 |
| Non-CSD | 0 | 0 | 0 | 0 | 53 | 100.0 | 53 |
CSD denotes melanoma located on chronically sun-damaged skin, and non-CSD melanoma located on skin without microscopical signs of chronic sun-induced damage.
Data are not included for one sample of primary uveal melanoma, which had concomitant T175R and V182I mutations in GNAQ (with unknown functional significance).
We also discovered mutations in GNAQ and GNA11 in exon 4 at arginine 183, which is analogous to R201 in GNAS and R179 in GNAI2 (Table 2).21 Mutations affecting R183 in either GNAQ or GNA11 were present in 2.1% of blue nevi and 4.8% of primary uveal melanomas. Most mutations affecting R183 in GNA11 resulted in a substitution to cysteine, caused by either a single (CGC→TGC) or a double (GTC CGC→GTT TGC) cytosine-to-thymine transition. This double mutation results in a silent mutation in codon 182 (Fig. 2 in the Supplementary Appendix). A single blue nevus had a guanine-to-adenine transition (CGC→CAC) at codon 183, predicting a substitution with histidine. This tumor also carried a concomitant GNA11 Q209L mutation and was the only tumor we analyzed that carried mutations at both codons 183 and 209. In all other samples, mutations at codons 183 and 209 were mutually exclusive, with no concomitant mutations in GNAQ and GNA11 (P<0.001). In GNAQ all mutations in codon 183 were CGA-to-CAA transitions, predicting substitution with glutamine (Fig. 3 in the Supplementary Appendix). A single sample of uveal melanoma had mutations in codons 175 (ACG→AGG) and 182 (GTT→ATT), both of which are of unknown functional significance.
All 11 tumors with mutations at R183 showed a cytosine-to-thymine transition on either the forward or reverse strand. Cytosine-to-thymine transitions on the reverse strand appear as guanine-to-adenine transitions on the forward strand (Fig. 2 and 3 in the Supplementary Appendix). Furthermore, three tumors carried a CC→TT transition. These alterations are characteristic mutational patterns induced by ultraviolet light23,24 and are found with markedly increased frequency in cutaneous melanomas arising on sun-exposed skin.25 By contrast, at codon Q209, in both GNAQ and GNA11, the predominant mutation was an adenine-to-thymine transversion (Fig. 1 in the Supplementary Appendix). The single C→T transitions in the forward strand at Q209 would result in stop codons rather than in activating mutations (CAA→TAA in GNAQ and CAG→TAG in GNA11). Similarly, a C→T transition on the reverse strand in GNA11 at 209 would generate a silent mutation (CAG→CAA). Thus, Q209 does not have a base-pair composition that would reveal a known ultraviolet signature. However, we found two instances of tandem base mutations at Q209 that included C→T transitions (CAA→TTA in GNAQ and CAG→CTA in GNA11).
A total of 63.2% of blue nevi were affected by mutations in either GNAQ or GNA11 (Tables 1 and 2). The two segmental melanocytoses, the nevus of Ito and the nevus of Ota, are sparsely cellular,26 so it is possible that we missed mutations in some samples. If the nevus of Ito and the nevus of Ota are excluded, 74.5% of blue nevi had a mutation in either GNAQ or GNA11. Altogether, 83.0% of all primary uveal melanomas that we examined had oncogenic mutations in GNAQ or GNA11.
CORRELATION WITH PROGNOSTIC INDICATORS AND SURVIVAL
For the 118 samples of uveal melanoma in which it was possible to determine the location of the primary tumor, lesions arising in the ciliochoroidal region had an increased frequency of GNA11 mutations (P=0.048 by Fisher’s exact test) (Fig. 1). Mutations in GNAQ and GNA11 were found more commonly in primary tumors with epithelioid cells or a mixture of epithelioid and spindle cells than in samples comprising only spindle cells, but the difference was not significant (Fig. 4 in the Supplementary Appendix). Using comparative genomic hybridization, we determined whether prognostically relevant chromosomal aberrations7 were present in 35 primary uveal melanomas and observed no association between the presence of such aberrations and the mutational status of GNAQ and GNA11 (Fig. 5 in the Supplementary Appendix). However, this analysis was limited, since the 35 samples did not include any samples without a mutation for comparison.
Figure 1. Anatomical Location and Mutational Status of 118 Samples of Primary Uveal Melanoma.
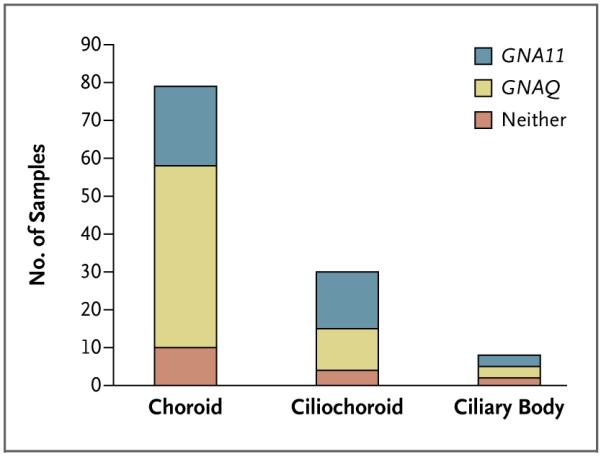
Lesions arising in the ciliochoroidal region had an increased frequency of GNA11 mutations (P=0.048 by Fisher’s exact test).
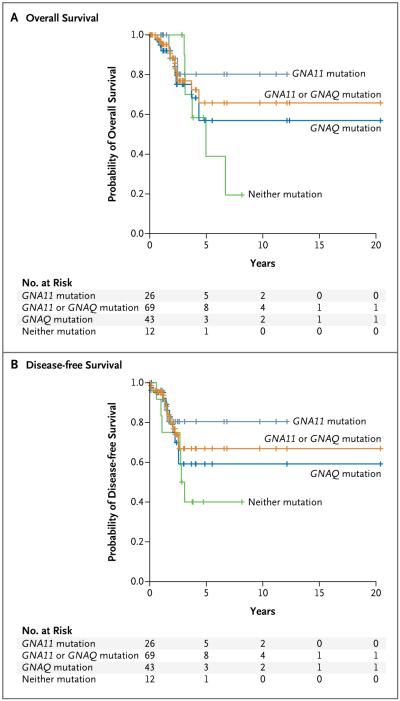
Examination of overall survival and disease-free survival for the 81 patients for whom we had requisite data did not reveal a significant difference between those with tumors bearing a GNAQ mutation and those with tumors bearing a GNA11 mutation (Fig. 2). We observed a trend toward increased survival among patients with tumors carrying a GNA11 mutation, as compared with those carrying a GNAQ mutation or those not carrying either a GNA11 or a GNAQ mutation.
Figure 2. Effect of Mutations in GNAQ and GNA11 on Overall and Disease-free Survival.
There was no significant difference in outcome on the basis of mutational status among 81 patients with primary uveal melanomas for whom follow-up information was available. The characteristics of these patients are provided in Table 1 in the Supplementary Appendix.
FUNCTIONAL VALIDATION OF GNA11
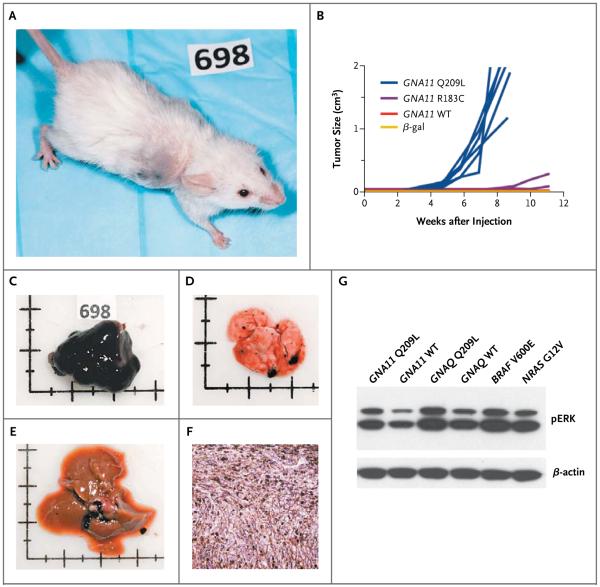
To validate two GNA11 variants, Q209L and R183C, as oncogenes, we transduced mouse melan-a cells22 with one or the other of these mutant genes. We then injected the transduced cells into immuno-compromised mice and monitored the mice for the formation of tumors. Mice that were injected with melanocytes that had been transduced with nonmutated GNA11, or lacZ, encoding β-galactosidase, served as negative controls (Fig. 3A through 3F). Rapidly growing tumors developed at each of the six injection sites for the Q209L variant, as compared with tumors developing with increased latency in only three of eight injection sites for the R183C variant. This finding was consistent with data showing a reduced potency of the R183C variant, as compared with the Q209L variant.27 We observed no tumors at the injection sites in the control groups, including 6 mice with β-galactosidase transduction and 10 with wild-type GNA11. In contrast to the results in mice injected with melanocytes transduced with the GNAQ Q209L variant,11 metastases developed in all 3 mice injected with melan-a cells transduced with the GNA11 Q209L variant, including lung metastases in all 3 and liver metastases in 1.
Figure 3. Induction of Tumors by the GNA11 Q209L and GNA11 R183C Variants in a Mouse Model.
Immortalized mouse melanocytes (melan-a cells) were transduced with GNA11 variant Q209L or R183C, wild-type GNA11, or a β-galactosidase control vector and injected bilaterally into the flank of nonobese diabetic mice with severe combined immunodeficiency and depletion of the interleukin-2 receptor γ chain. By 11 weeks, tumors developed at all 6 injection sites in 3 mice bilaterally injected with melanocytes transduced with the Q209L variant (Panel A). Tumors developed at 3 out of 8 injection sites in 4 mice bilaterally injected with melanocytes transduced with the R183C variant (Panel B). There were no tumors in 5 mice bilaterally injected with melanocytes transduced with wild-type (WT) GNA11 or 3 mice bilaterally injected with melanocytes transduced with the β-galactosidase control vector (β-gal) (Panel B). The graph in Panel B shows the combined results of two independent experiments. Tumors were heavily melanized in all the mice (Panel C). Among the 3 mice with the Q209L variant, multiple lung metastases developed in all 3 mice (Panel D), and liver metastases developed in 1 (Panel E). Tumors were composed of pigmented spindle and epithelioid melanocytes (Panel F, hematoxylin and eosin). On Western blot analysis, melan-a cells that were transduced with GNA11 Q209L, but not their wild-type counterparts, showed activation of the mitogen-activated protein kinase pathway that was similar to the results in GNAQ Q209L and mutant BRAF or NRAS samples used as positive controls (Panel G). The term pERK denotes phosphorylated extracellular signal-regulated kinase.
Western blot analyses of melanocytes transduced with the GNA11 Q209L variant revealed mitogen-activated protein (MAP) kinase activation, as indicated by increased levels of phosphorylated extracellular signal-regulated kinase (ERK) (Fig. 3G). In our previous studies,11 GNAQ Q209L mutant cell lines were highly sensitive to inhibitors of MEK (a component of the MAP kinase pathway). Of the 13 uveal melanoma cell lines available in our laboratory, none carried a GNA11 mutation, so we were unable to test whether the GNA11 Q209L mutant cells are similarly sensitive to MEK inhibitors. However, the close functional relationship between Gαq and Gα11, 9,28 together with the data that we describe here, supports the prediction that the activation of MAP kinase is effected by GNA11 Q209 mutations in uveal melanoma cells and can be countered with MEK inhibitors.11 Of these 13 cell lines, 5 carried the GNAQ Q209 mutation, suggesting that GNA11 mutations compromise growth in culture to a greater extent than do GNAQ Q209 mutations.
DISCUSSION
Although GNA11 somatic mutations are rare in blue nevi,11,18 our data show that they are well represented in uveal melanoma. In our samples, 83.0% of uveal melanomas had a constitutively active mutation in either GNAQ or GNA11, suggesting that activation of the Gαq–Gα11 pathway is the predominant route to the development of uveal melanoma. GNAQ and GNA11 have overlapping functions in melanocytes,9 and both genes upregulate the MAP kinase pathway when constitutively active. Although Gαq and Gα11 have amino acid sequences that are 90% homologous, there appear to be differences in their role in melanocytic neoplasia.
Several lines of evidence suggest that GNA11 mutations may have a more potent effect on melanocytes than do mutations in GNAQ. First, GNA11 mutations were rare in blue nevi, which are benign neoplasms.26 Conversely, there were significantly more GNA11 Q209 mutations than GNAQ Q209 mutations in uveal melanoma metastases. Furthermore, GNA11 mutations were more common in locally advanced primary tumors and in primary tumors originating from the ciliochoroidal region, a prognostically adverse feature.29 Finally, the mouse Gna11 Dsk7 mutation is more tumorigenic than the Gnaq Dsk1 mutation, since it is better able to stimulate melanocyte growth that is impaired by heterozygous mutations in Kit, Pax3, and Ednrb (encoding endothelin receptor type B).9 However, since the mutations found in mice occur at different residues in Gna11 and Gnaq (I63V and V179 M, respectively),9 we cannot dismiss the possibility that the difference is a functional consequence of the mutations themselves, rather than a difference in function between Gna11 and Gnaq.
Although the survival of patients did not differ significantly among those with GNAQ mutations and those with GNA11 mutations in our study, the number of patients who were available for our analysis may have been too small to detect such a difference. In addition, we used enucleated specimens, which could have biased the analysis; tissue for mutation analysis is not routinely available from nonenucleated samples. The samples that we analyzed were typically from large tumors, which may have obscured any association between mutations in GNAQ or GNA11 and prognosis.
The level of activation of Gαq R183 mutants is considerably lower than that of Gαq Q209 mutants in vitro, which raises the possibility that R183-mutated oncoproteins may be less potent.27 Our finding of an increased latency and reduced penetrance in the tumorigenicity assay is consistent with this notion. Of possible clinical relevance is the finding that Gαq R183 but not Gαq Q209 can be inhibited with YM-254890, a naturally occurring toxin from chromobacteria.30
In a recent study of GNAQ and GNA11 in 922 human neoplasms with various histopathological features, the only mutations that were found were in GNAQ in blue nevi.18 Samples of uveal melanoma were not included in the study. Although these results are not exhaustive, together with our data, they suggest that GNAQ and GNA11 mutations are enriched in the melanocytic lineage. Another study showed that GNAQ mutations were present in 37% of melanocytic neoplasms of the central nervous system,17 making it likely that GNA11 mutations will also be found in this category of melanocytic tumors.
The peculiar association among mutations in melanocytic neoplasms of the dermis, uvea, and central nervous system may indicate different cells of origin. A developmental pathway has been described,31 in which a subgroup of melanocytes derives from a precursor shared with Schwann cells. It is possible that Gαq–Gα11 signaling has a critical role in the production of melanocytes through this developmental mechanism. If so, the timing of the occurrence of mutations may determine the localization and extent of the neoplasm. Lesions that are confined to the central nervous system would arise from Gαq–Gα11 mutations in precursors before the onset of migration, and segmental lesions, such as the nevus of Ota, would result from mutations in a precursor early during migration, in contrast to mutations arising later along the migratory pathway, which would result in solitary lesions involving the skin or choroid.
Epidemiologic studies have shown conflicting roles for recreational and occupational exposure to ultraviolet radiation in patients with uveal melanoma.32,33 Fair complexion and light irides are generally considered risk factors for uveal melanoma.34,35 Some studies have shown that uveal melanomas most frequently arise in the macula region, which has the highest level of exposure to ultraviolet radiation.36 Our finding of cytosine-to-thymine transitions in uveal melanomas could suggest a role for ultraviolet radiation.
In summary, our findings suggest that a large majority of uveal melanomas and blue nevi carry mutations in either GNAQ or GNA11.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (R01 CA131524) and the Melanoma Research Alliance (both to Dr. Bastian), the Canadian Institutes of Health Research (MOP-79511) and the Canadian Cancer Society (019055) (both to Dr. Van Raamsdonk), the European Commission (GENINCA, contract no. HEALTH-F2-2008-202230), the Jubiläumsfonds of the Oesterreichische Nationalbank (12480 and 13837), and the Molecular Medicine program of Medical University Graz (to Dr. Obenauf), and Deutsche Forschungsgemeinschaft (research stipend GR 3671/1-1, to Dr. Griewank).
We thank Dr. Dorothy Bennett of St. George’s University, London, for her donation of melan-a cells.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18:75–84. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Saldanha G, Purnell D, Fletcher A, Potter L, Gillies A, Pringle JH. High BRAF mutation frequency does not characterize all melanocytic tumor types. Int J Cancer. 2004;111:705–10. doi: 10.1002/ijc.20325. [DOI] [PubMed] [Google Scholar]
- 3.Zuidervaart W, van Nieuwpoort F, Stark M, et al. Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer. 2005;92:2032–8. doi: 10.1038/sj.bjc.6602598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 6.Horsman DE, White VA. Cytogenetic analysis of uveal melanoma: consistent occurrence of monosomy 3 and trisomy 8q. Cancer. 1993;71:811–9. doi: 10.1002/1097-0142(19930201)71:3<811::aid-cncr2820710325>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 7.White VA, Chambers JD, Courtright PD, Chang WY, Horsman DE. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer. 1998;83:354–9. [PubMed] [Google Scholar]
- 8.Singh AD, De Potter P, Fijal BA, Shields CL, Shields JA, Elston RC. Lifetime prevalence of uveal melanoma in white patients with oculo(dermal) melanocytosis. Ophthalmology. 1998;105:195–8. doi: 10.1016/s0161-6420(98)92205-9. [DOI] [PubMed] [Google Scholar]
- 9.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS. Effects of G-protein mutations on skin color. Nat Genet. 2004;36:961–8. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch KR, McGowan KA, van Raamsdonk CD, et al. Genetics of dark skin in mice. Genes Dev. 2003;17:214–28. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–9. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 13.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science. 1993;262:1895–901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 14.Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989;340:692–6. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- 15.Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature. 1994;372:276–9. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 16.Kalinec G, Nazarali AJ, Hermouet S, Xu N, Gutkind JS. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol Cell Biol. 1992;12:4687–93. doi: 10.1128/mcb.12.10.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Küsters-Vandevelde HV, Klaasen A, Küsters B, et al. Activating mutations of the GNAQ gene: a frequent event in primary melanocytic neoplasms of the central nervous system. Acta Neuropathol. 2009 November 22; doi: 10.1007/s00401-009-0611-3. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamba S, Felicioni L, Buttitta F, et al. Mutational profile of GNAQQ209 in human tumors. PLoS One. 2009;4(8):e6833. doi: 10.1371/journal.pone.0006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onken MD, Worley LA, Long MD, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:5230–4. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer J, Kilic E, Vaarwater J, Bastian BC, Garbe C, de Klein A. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br J Cancer. 2009;101:813–5. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyons J, Landis CA, Harsh G, et al. Two G protein oncogenes in human endocrine tumors. Science. 1990;249:655–9. doi: 10.1126/science.2116665. [DOI] [PubMed] [Google Scholar]
- 22.Bennett DC, Cooper PJ, Hart IR. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer. 1987;39:414–8. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 23.Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat. 2007;28:578–88. doi: 10.1002/humu.20481. [DOI] [PubMed] [Google Scholar]
- 24.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 25.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zembowicz A, Mihm MC. Dermal dendritic melanocytic proliferations: an update. Histopathology. 2004;45:433–51. doi: 10.1111/j.1365-2559.2004.01975.x. [DOI] [PubMed] [Google Scholar]
- 27.Orth JH, Preuss I, Fester I, Schlosser A, Wilson BA, Aktories K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc Natl Acad Sci U S A. 2009;106:7179–84. doi: 10.1073/pnas.0900160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Offermanns S, Zhao LP, Gohla A, Sarosi I, Simon MI, Wilkie TM. Embryonic cardiomyocyte hypoplasia and craniofacial defects in G alpha q/G alpha 11-mutant mice. EMBO J. 1998;17:4304–12. doi: 10.1093/emboj/17.15.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abramson DH, Schefler AC, Dunkel IR, McCormick B, Dolphin KW. Adult ophthalmic oncology: ocular diseases. In: Kufe DW, Pollock RE, Weichselbaum RR, et al., editors. Cancer medicine-6. Vol. 2. BC Decker; Hamilton, ON, Canada: 2003. pp. 1242–4. http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=cmed6&part=A20124. [Google Scholar]
- 30.Takasaki J, Saito T, Taniguchi M, et al. A novel Galphaq/11-selective inhibitor. J Biol Chem. 2004;279:47438–45. doi: 10.1074/jbc.M408846200. [DOI] [PubMed] [Google Scholar]
- 31.Adameyko I, Lallemend F, Aquino JB, et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–79. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 32.Vajdic CM, Kricker A, Giblin M, et al. Sun exposure predicts risk of ocular melanoma in Australia. Int J Cancer. 2002;101:175–82. doi: 10.1002/ijc.10579. [DOI] [PubMed] [Google Scholar]
- 33.Mainster MA, Turner PL. Ultraviolet-B phototoxicity and hypothetical photomelanomagenesis: intraocular and crystalline lens photoprotection. Am J Ophthalmol. 2010;149:543–9. doi: 10.1016/j.ajo.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Mudhar HS, Parsons MA, Sisley K, Rundle P, Singh A, Rennie IG. A critical appraisal of the prognostic and predictive factors for uveal malignant melanoma. Histopathology. 2004;45:1–12. doi: 10.1111/j.1365-2559.2004.01874.x. [DOI] [PubMed] [Google Scholar]
- 35.Singh AD, Rennie IG, Seregard S, Giblin M, McKenzie J. Sunlight exposure and pathogenesis of uveal melanoma. Surv Ophthalmol. 2004;49:419–28. doi: 10.1016/j.survophthal.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Judge H, Gragoudas ES, Seddon JM, Egan KM. Patterns of tumor initiation in choroidal melanoma. Cancer Res. 2000;60:3757–60. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.